Summary
We used an integrative approach to probe the significance of the interaction between the relay loop and converter domain of the myosin molecular motor from Drosophila melanogaster indirect flight muscle. During the myosin mechanochemical cycle, ATP-induced twisting of the relay loop is hypothesized to reposition the converter, resulting in cocking of the contiguous lever arm into the pre-power stroke configuration. The subsequent movement of the lever arm through its power stroke generates muscle contraction by causing myosin heads to pull on actin filaments. We generated a transgenic line expressing myosin with a mutation in the converter domain (R759E) at a site of relay loop interaction. Molecular modeling suggests that the interface between the relay loop and converter domain of R759E myosin would be significantly disrupted during the mechanochemical cycle. The mutation depressed calcium as well as basal and actin-activated MgATPase (Vmax) by ~60% compared to wild-type myosin, but there is no change in apparent actin affinity (Km). While ATP or AMP-PNP binding to wild-type myosin subfragment-1 enhanced tryptophan fluorescence ~15% or ~8%, respectively, enhancement does not occur in the mutant. This suggests that the mutation reduces lever arm movement. The mutation decreases in vitro motility of actin filaments by ~35%. Mutant pupal indirect flight muscles display normal myofibril assembly, myofibril shape, and double-hexagonal arrangement of thick and thin filaments. Two-day-old fibers have occasional “cracking” of the crystal-like array of myofilaments. Fibers from one-week-old adults show more severe cracking and frayed myofibrils with some disruption of the myofilament lattice. Flight ability is reduced in two-day-old flies compared to wild-type controls, with no upward mobility but some horizontal flight. In one-week-old adults, flight capability is lost. Thus altered myosin function permits myofibril assembly, but results in a progressive disruption of the myofilament lattice and flight ability. We conclude that R759 in the myosin converter domain is essential for normal ATPase activity, in vitro motility and locomotion. Our results provide the first mutational evidence that intramolecular signaling between the relay loop and converter domain is critical for myosin function both in vitro and in muscle.
Keywords: myosin, muscle, Drosophila, ATPase, myofibril
The structure of the motor domain of myosin II has been determined at atomic-level resolution for skeletal muscle, smooth muscle and non-muscle isoforms.1–4 Co-crystallization of myosin with nucleotide analogs permitted visualization of various states of the mechanochemical cycle and reconstruction of myosin lever arm movement from pre-power stroke through post-power stroke.5–7 These structural studies, along with biochemical and biophysical approaches, resulted in models for ATP’s role in cocking the lever arm and for the release of ATP hydrolysis products following the lever arm power stroke.8 Mutations in amino acid residues that serve as communication pathways between the nucleotide binding site and the lever arm show that domain interaction is critical for myosin function in vitro and in the cellular slime mold Dictyostelium.9,10
The importance of myosin domain interactions has not been addressed in regard to the assembly, stability or function of muscle, largely due to the difficulty in creating transgenic organisms with altered myosin heavy chain (Mhc) genes. In muscle sarcomeres, myosin-containing thick filaments and actin-containing thin filaments are transiently connected by myosin cross-bridges. The myosin lever arm located at the base of the cross-bridge ratchets thin filaments past thick filaments, resulting in muscle contraction. The muscle mechanochemical cycle is an ATP-dependent process, with the nucleotide state coupled to conformational changes at the actin-binding site and in the lever arm.8
Here we probe the importance of myosin inter-domain communication in muscle by employing an integrative approach using transgenic Drosophila melanogaster. This organism has a single Mhc gene that encodes all MHC isoforms by alternative RNA splicing,11,12 as well as Mhc null alleles that specifically knock out myosin accumulation in indirect flight muscles.13–15 Thus an Mhc transgene can be used to replace endogenous Mhc gene expression in a muscle type that is not required for viability. We employed site-directed mutagenesis to disrupt the interface between the myosin relay loop and the converter, two domains that are hypothesized to serve as a communication pathway between the ATP binding site and the lever arm. We show that this decreases ATPase activity and actin motility. Further, we demonstrate that the normal relay-converter interface is essential for wild-type muscle function.
Communication to the converter domain
The ATP-dependent reorientation of the relay helix and the SH1 helix are proposed to drive converter movement during the recovery step of the mechanochemical cycle.16–17 This results in rotation of the linked lever arm into its cocked position. ATP acts by changing the conformation of residues in the phosphate tube, which contains the P loop, Switch-1 and Switch-2. The rearrangement of Switch-2 residues appears to be critical for kinking and tilting of the relay helix, which changes the orientation of the relay loop. Through its contacts with the relay loop, the converter domain is reoriented and drives the lever arm through a 25° rotation.18 A subsequent Switch-2 and wedge-loop induced see-saw motion of the SH1 helix results in a further 40° tilt of the converter and lever arm.18
The interaction of the relay loop with the converter domain occurs in all myosin crystal structures examined (Fig. 1A). Making this connection constant during the mechanochemical cycle still permits myosin to function in vitro.10 This was shown by cross-linking cysteines that were substituted for isoleucine 508 in the relay loop and arginine 759 in the converter domain of Dictyostelium myosin and demonstrating that actin binding and basal ATPase activity were unaffected; however, 29% reductions in actin-activated ATPase and Km were observed. Another study showed that disruption of this putative communication pathway severely affects myosin function. Mutation of Dictyostelium relay loop residue isoleucine 508 to alanine prevented myosin-based cell division in vivo and disabled in vitro motility, apparently as a result of reducing communication to the converter domain and inhibiting lever arm swing.9 Note that for the sake of consistency, we are using the chicken amino acid numbering system that we have typically employed, as chicken fast skeletal myosin was the first myosin described at atomic level resolution.2
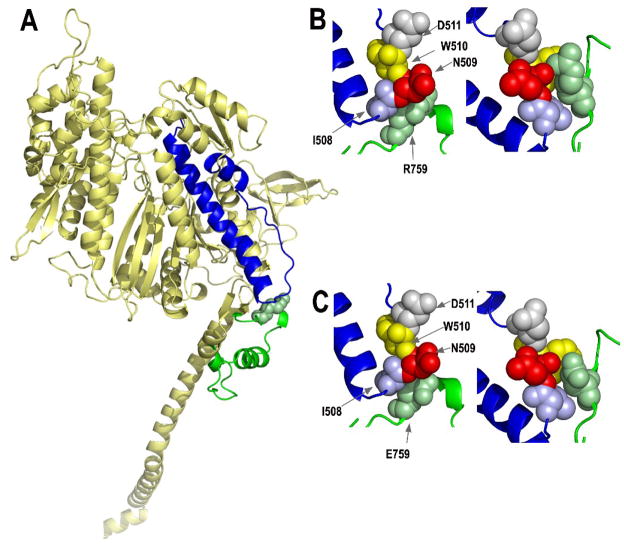
Fig. 1.
Locations of the converter and relay domains of myosin, and the effects of mutating the R759 converter residue. (A) Mapping of the amino acids of the Drosophila indirect flight muscle isoform (IFI) of myosin onto the scallop crystal structure in the pre-power stroke state (1qvi). The relay domain encoded by alternative exon 9a is highlighted in blue, whereas the central portion of the converter domain (residues 724–764) encoded by alternative exon 11e is shown in green. Converter domain residue R759 is shown as a space-filling model. The homology model was produced by fitting the Drosophila indirect flight muscle myosin S-1 amino acid sequence to the coordinates of scallop myosin S1 using the automated mode of the Swiss-Model homology modeling server (http://swissmodel.expasy.org/). PyMOL (http://www.pymol.org/, DeLano Scientific, Palo Alto, CA, USA) was used to visualize the output. (B) Interaction of converter domain residue R759 (green) with amino acid residues of the relay domain [I508 (blue), N509 (red), W510 (yellow), D511 (gray)] in the pre-power stroke state (left) and the post-power stroke state (1kk8; right). Space-filling models suggest the hydrophobic region of R759 near the peptide backbone interacts with I508 in the pre-power stroke state, while the polar terminal portion interacts with polar N509. In the post-power stroke state, interactions with I508 and N509 are retained, plus the changed orientation of the relay loop results in formation of a salt bridge between R759 and D511. See Bloemink et al.30 for further modeling and discussion. (C) Interaction of mutated converter domain residue E759 with amino acid residues of the relay domain in the pre-power stroke state (left) and the post-power stroke state (right). These models were produced as described above, except that E759 replaced R759 prior to modeling. The negatively charged region of E759 is located near hydrophobic I508, eliminating the hydrophobic interaction found in wild-type myosin. The mutation also reduces interaction of residue 759 with polar N509, particularly in the pre-power stroke state (left). Disruption of the relay loop/converter domain interface is exacerbated at the post-power stroke state (right), since E759 is unable to form a salt bridge with negatively charged D511. All residue numbers correspond to those of chicken skeletal muscle myosin.2
The transgenic approach we used involves mutagenesis of the codon encoding arginine 759 to glutamic acid in Drosophila muscle Mhc, followed by expression in Mhc-null indirect flight muscles. Molecular modeling at the pre- and post-power stroke states (Fig. 1B,C) suggests that replacing the hydrophobic portion of the arginine side chain (close to the protein backbone) with the more polar glutamic acid side chain would disrupt the hydrophobic interactions with relay loop residue isoleucine 508. Further, the shorter side-chain length in the mutant appears to reduce interaction with polar asparagine 509, particularly in the pre-power stroke state. Finally, the shorter side chain along with the charge change would eliminate interactions with negatively charged residue aspartate 511 in the post-power stroke state (Fig. 1B,C).
Production of R759E transgenic lines
The Drosophila Mhc gene has five alternative versions of exon 11 that encode the central portion of the converter domain. We used in vitro mutagenesis of exon 11e to transform the codon encoding arginine 759 to glutamic acid. The expression of exon 11e has only been detected in the indirect flight muscle,19,20 so this mutation should not disrupt function in other muscle types or cause lethality. The mutant gene fragment was placed in the context of the Mhc gene and transgenic lines were obtained by P element-mediated germline transformation (see Table 1 legend). Transgenes were crossed into the Mhc10 line that is null for indirect flight muscle MHC,14 so that the product and effects of the transgene could be assessed in the absence of wild-type myosin. We characterized three independent lines that produced normal levels of MHC in their indirect flight muscles (Table 1).
Table 1.
Characteristics of transgenic lines expressing wild-type (PwMhc2) and mutant (PwMhcR759E) myosin
| Flight testing (at 2 days/7 days) | |||||||
|---|---|---|---|---|---|---|---|
| Line name (chromosome location) | Myosin level ± SE | Number tested | % Up | % Horizontal | % Down | % Not at all | Flight index ± SE |
| PwMhc2 (X) | 1.00±0.03 | 116/197 | 57.8/45.7 | 14.6/26.4 | 15.5/22.8 | 12.1/5.1 | 4.4±0.1a/4.2±0.1 |
| PwMhcR759E-V8 (X) | 0.98±0.04 | 98/114 | 0/0 | 27.5/0 | 40.9/1.8 | 31.6/98.2 | 1.3±0.2/0.03±0.01 |
| PwMhcR759E-7B (3) | 0.97±0.04 | 91/111 | 0/0 | 21.9/0 | 40.7/2.7 | 37.4/97.3 | 1.6±0.3/0.05±0.01 |
| PwMhcR759E-V5 (X) | 0.95±0.03 | 111/ND | 0/ND | 19/ND | 46.8/ND | 34.2/ND | 1.6±0.3/ND |
Flight index data for 2-day-old PwMhc2 are from Kronert et al.35
The R759E mutant Mhc gene was constructed using the wild-type genomic construct PwMhc2.36 The 12.5-kb Eag I restriction fragment was subcloned into pBluescriptKS (Stratagene, La Jolla, CA), which had been digested with Eag I. The resulting subclone, p3’Mhc was digested with Xba I and Spe I. The 160-bp fragment from this digest (containing the R759 coding region) was subcloned into pBluescriptKS that had been digested with Xba I and Spe I. The resulting subclone, pR759, was subjected to site-directed mutagenesis using the QuickChange II kit (Stratagene) and the exon specific-primer 5′-CCCGATATGTACGAAATTGGTCACACC-3′ containing the R759E nucleotide coding change (bold). Upon sequence confirmation of the R759E site-directed mutagenesis product, the pR759E subclone was digested with Xba I and Spe I. The resulting 160 bp fragment was used to replace the wild-type Xba I - Spe I fragment of p3’Mhc. The resulting clone was digested with Eag I, and the Eag I fragment was replaced back into the wild-type construct PwMhc2 at its Eag I site. Ligation sites were confirmed by DNA sequencing, as were all splice junctions and coding regions of the final pwMhcR759E plasmid. BestGene, Inc. (Chino Hills, CA) produced transgenic lines by P element-mediated transformation37 and mapped chromosome locations by standard genetic crosses using balancer chromosomes. Transgenes were crossed into the Mhc10 (null for myosin in the indirect flight muscle) background.14 To confirm the expression pattern of PwMhcR759E and to insure that no wild-type copy of Mhc was present in PwMhcR759E transgenic line, we used exon-specific primers and RT-PCR as previously described.35 RNA was isolated from upper thoraces of two-day-old female adult flies of wild type (yw) and PwMhcR759E. Restriction enzyme digests and sequence analysis of the RT-PCR products from PwMhc2R759E compared to yw confirmed there was no difference in alternative exon usage and that the mutagenized codon encoding R759E was present in PwMhcR759E but not in yw (data not shown). Myosin accumulation relative to actin was determined by SDS polyacrylamide gel electrophoresis. Dissected upper thoraces were homogenized in 50 μl SDS gel buffer from 5 two-day-old female flies. Eight μl of sample were loaded on a 9% polyacrylamide gel. Each transgenic line was analyzed five times, each time with a freshly prepared sample. Coomassie blue stained gels were digitally scanned and protein accumulation was determined using NIH image software. Mean values are compared to the wild-type transgenic control, PwMhc2. Flight assays were performed with two-day-old and one-week-old adult female flies at 22°C. Flight was assessed by the ability to fly up (U), horizontal (H), down (D) or not at all (N) when released in a plexiglass box with a light source at its top.38 The flight index is defined as: 6U/T + 4H/T + 2D/T + 0N/T; T is the total number of flies tested.39
In vitro properties of R759E transgenic myosin
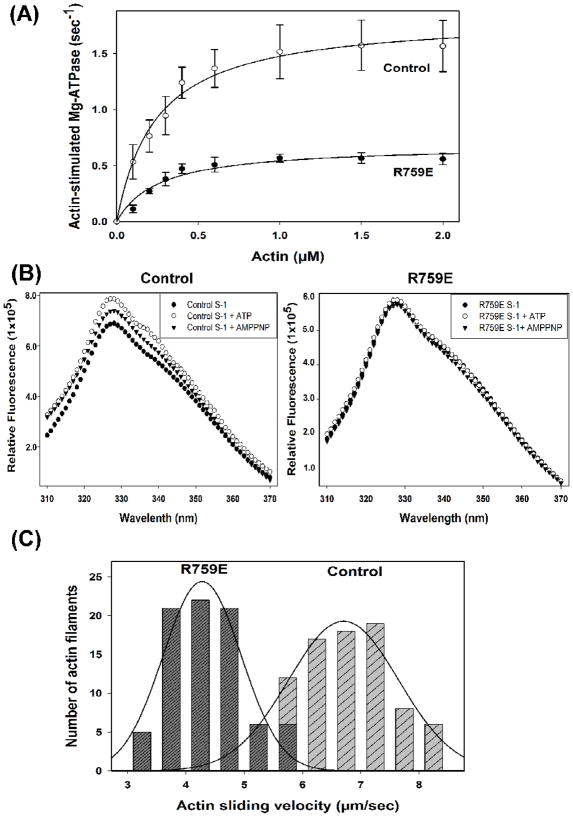
R759E myosin isolated from indirect flight muscles was tested as to ATPase activity, intrinsic tryptophan fluorescence and ability to drive in vitro sliding of actin filaments compared to wild-type control (PwMhc2) myosin. Ca and basal MgATPase activities of the mutant myosin were reduced ~60% compared to wild type (3.96 ± 0.99 s−1 vs. 10.34 ± 0.73 for CaATPase and 0.10 ± 0.04 s−1 vs. 0.26 ± 0.02 for basal MgATPase). Actin-activated MgATPase (Vmax) was reduced by 63% (0.69 ± 0.06 vs. 1.86 ± 0.33 s−1), although Km values were not affected by the mutation (Fig. 2A; Table 2). We analyzed intrinsic tryptophan fluorescence of the R759E S-1 fragment compared to the wild-type control. This is the first such analysis of Drosophila S-1. Consistent with findings for other myosins,9,21–23 wild-type Drosophila S-1 showed enhanced tryptophan fluorescence upon binding with ATP (15%) or AMP-PNP (8%). However, this enhancement did not occur in R759E S-1 (Fig. 2B). Assuming the main contributor of tryptophan fluorescence is W510 (Fig. 1B,C), as is the case in other myosins,21–24 and that the enhanced fluorescence mirrors ATP-induced relay movement and lever arm swing,9,25 this suggests that disruption of the relay loop/converter domain interface prevents efficient lever arm swing during the recovery stroke (see Conclusions). Further, it appears that W510 is less accessible to solvent in R759E S-1 in the absence of nucleotide, as the mutant protein yields a lower fluorescence intensity compared to an equal amount of wild-type S-1 (Fig 2B). In vitro motility assays showed that the ability of the converter mutant to drive actin filament sliding decreased by 36% (4.3 ± 0.6 vs. 6.7 ± 1.1 μm s−1; Fig. 2C). The ATPase, tryptophan fluorescence and in vitro motility data are summarized in Table 2.
Fig. 2.
Actin-activated ATPase activity, tryptophan fluorescence and in vitro motility of wild-type control and R759E Drosophila myosin. (A) ATPase activities of myosins isolated from the transgenic lines expressing wild-type (PwMhc2) and R759E mutant myosin were determined as reported previously31,32 and described in the legend to Table 2. Chicken actin concentration ranged from 0–2 μM. Basal Mg-ATPase activities obtained in the absence of actin were subtracted from data points, which represent duplicate samples from six separate preparations of myosin for each genotype. Data were fitted with the Michaelis-Menten equation (rectangular hyperbola, y = ax/(b + x)). Vmax of R759E myosin was reduced by ~60% compared to wild-type myosin, but Km for actin was not affected (see Table 2). (B) Tryptophan fluorescence of wild-type control myosin (left) and R759E myosin (right) in the absence and presence of 0.1 mM ATP or AMP-PNP. Myosin sub- fragment-1 (S-1) was prepared after chymotrypsin digestion as previously reported.30,33 Intrinsic tryptophan fluorescence of control and R759E S-1 (0.02 mg/ml, final concentration) was recorded at 25°C in the presence or absence of ATP or AMP-PNP (0.1 mM each, pH 7.0) in 20 mM MOPS, pH 7.0, containing 100 mM KCl and 5 mM MgCl2 as previously described.9,22,23 The excitation wavelength for tryptophan fluorescence was 295 nm and fluorescence emission was recorded at 310–370 nm, using a PTI spectrofluorometer (Photon Technology International). The figure represents uncorrected tryptophan fluorescence of control S-1 and R759E S-1 in the absence (closed circles) or in the presence of ATP (open circles) or AMP-PNP (triangles). The fluorescence peak maxima (330 nm) in wild-type S-1 increased ~15% and ~8% upon incubation with ATP or AMP-PNP, respectively. However, R759E S-1 tryptophan fluorescence remained unchanged upon addition of either nucleotide. (C) Histograms comparing rates of in vitro actin sliding for wild-type control and R759E myosin. Actin sliding velocities are shown from all continuously moving actin filaments assessed from six preparations of each myosin type. The mutation decreases filament motility by ~35% (see Table 2).
Table 2.
Ca+2, Mg+2, actin-stimulated Mg+2 ATPase kinetics, tryptophan fluorescence and actin-sliding velocities of wild-type (PwMhc2) and R759E indirect flight muscle myosins
| Myosin | Basal Ca-ATPase ± SD (s−1) | Basal Mg-ATPase± SD(s−1) | Actin-stimulated Vmax ± SD(s−1) | Km (actin) ± SD (μM) | Peak tryptophan fluorescence (% compared to apo state ± SD) | Actin sliding velocity ± SD (μm s−1) |
|---|---|---|---|---|---|---|
| Wild-type (PwMhc2) | 10.34 ± 0.73 (n = 6) | 0.26 ± 0.02 (n = 6) | 1.86 ± 0.33 (n = 6) | 0.26 ± 0.10 (n = 6) | 115.0 ± 0.4 (ATP) 108.2 ± 0.3 (AMP-PNP) (n=3) | 6.7 ± 1.1 (n = 6) |
| R759E | 3.96 ± 0.99 (n = 6)* | 0.10 ± 0.04 (n = 6)* | 0.69 ± 0.06 (n = 6)* | 0.27 ± 0.08 (n = 6) | 99.9 ± 1.1 (ATP) 97.8 ± 1.6 (AMP-PNP) (n=3) | 4.3 ± 0.6 (n = 6)* |
Statistically different from PwMhc2 myosin for ATPase or actin sliding velocity data (p < 0.001, Student’s t test).
Myosin was isolated from dissected dorsolongitudinal indirect flight muscle31 and actin was prepared from chicken skeletal muscle.40 ATPase measurements were performed using [γ-32P] as described.31,32 Tryptophan fluorescence was measured as described in the legend to Figure 2. The percentage of peak fluorescence (330 nm) compared to apo state (in the absence of nucleotide) is reported. In vitro motility was performed as previously detailed,31 with some modifications.35
Effects of the R759E mutation on muscle ultrastructure and locomotion
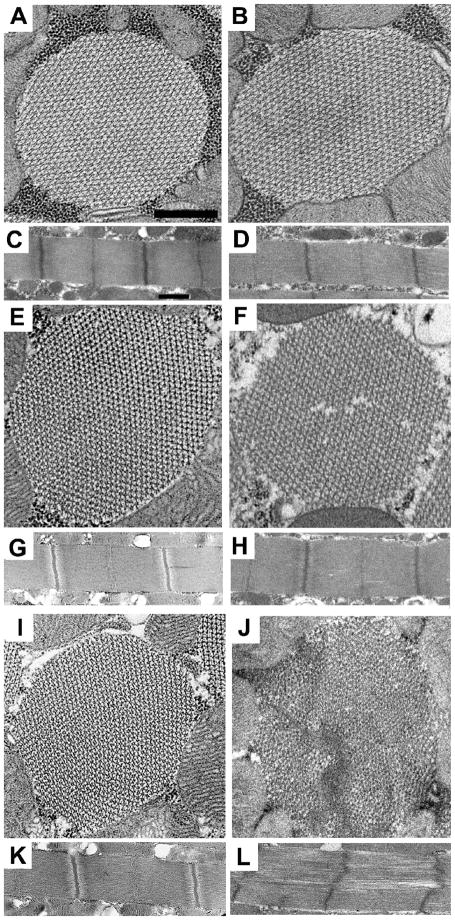
We used transmission electron microscopy to examine how the R759E mutation affects indirect flight muscle myofibril assembly and maintenance. At the late pupal stage, myofibril structure appears normal (not shown), with normal thick and thin filament arrangements and intact sarcomeres. This is true for adults at two hours post eclosion (Figs. 3A–D). At two days of age, sarcomere structure is generally normal (Figs. 3E–H), but occasional cracks in the mutant myofilament lattice occur (Fig. 3F). At one-week post eclosion (Figs. 3I–L), mutant myofibrils show moderate to severe disruption. Hexagonal packing of filaments is not as regular (Fig. 3J), with gaps in the myofilament lattice (Fig. 3L). Thus while mutant muscle assembly appears normal, myofibrils degenerate as flies age.
Fig. 3.
Muscle structure of the dorsal longitudinal indirect flight muscle in the R759E myosin converter mutant. Transmission electron microscopy, as previously described,34 was used to determine the effects of transgene expression on muscle structure of the dorsal longitudinal indirect flight muscles at three stages of adult development (two hours, two days and one week). Cross-sections and longitudinal sections were obtained from females for each transgenic line, with at least three different samples examined for each. Representative images are shown. (A) Transverse section from PwMhc2 two-hour-old adults. (B) Transverse section from R759E two-hour-old adults. Myofibril structure of R759E shows normal hexagonal packing of thick and think filaments compared to PwMhc2. (C) Longitudinal section from PwMhc2 two-hour-old adults. (D) Longitudinal section from R759E two-hour-old adults. R759E sarcomere structure is normal compared to PwMhc2. (E) Transverse section from PwMhc2 two-day-old adults. (F) Transverse section from R759E two-day-old adults. Myofibril structure of R759E shows subtle cracking with slight disruption in the hexagonal packing of thick and thin filaments compared to PwMhc2. (G) Longitudinal section from PwMhc2 two-day-old adults. (H) Longitudinal section from R759E two-day-old adults. R759E sarcomere structure appears essentially normal compared to PwMhc2. (I) Transverse section from PwMhc2 one-week-old adults. (J) Transverse section from R759E one-week-old adults. R759E shows further cracking and more severe disruption of the normal hexagonal packing of thick and thin filaments compared to PwMhc2 and R759E two-day-old adults. (K) Longitudinal section from PwMhc2 one-week-old adults. (L) Longitudinal section from R759E one-week-old adults. The sarcomere structure of R759E is cracked and Z-bands are slightly disrupted compared to PwMhc2 sarcomeres. Scale bars, 0.5 μm.
We assessed the effects of the R759E mutation on locomotion by flight testing at two days and at one week post-eclosion (Table 1). Flight ability is severely impaired in young flies, with a flight index of ~1.5 compared to 4.4 for the transgenic control (the maximum index is 6.0 for 100% upward flight). While nearly 60% of the control flies flew upward, none of the mutant flies were capable of doing so. At one week, the flight index of the mutant line was reduced to ~0.4, while that of the control did not change significantly (4.2). While 72% of the control flies few upward or horizontally at one week, none of the R759E mutant flies could do so. All of the mutants glided downward or were incapable of flight. This reduction in flight ability mirrors the degeneration in myofibril structure noted above.
Conclusions
Our work presents the first mutational evidence that the linkage between the relay loop and converter domain is necessary for normal muscle function. At the molecular level, the R759E mutation substantially reduces basal and actin-activated ATPase activity as well as in vitro actin filament sliding. The failure to observe enhanced tryptophan fluorescence upon ATP addition suggests reduced ability of the lever arm to move into the pre-power stroke configuration, perhaps as a result of inhibition of the 25° rotation proposed to arise from interaction of the relay helix with the converter.18 This assumes that the fluorescence increase in wild-type myosin is largely due to tryptophan 510, which has been shown to be the case in both muscle and non-muscle myosins.21–24 Molecular modeling (Figure 1) suggests that the relay loop/converter interface is significantly disrupted in the post-power stroke configuration, and this may inhibit the ability of the relay loop repositioning to efficiently move the converter domain and thereby trigger lever arm cocking and the subsequent power stroke. Disrupting the relay loop-converter interface thus appears to feed forward to affect lever arm repositioning as well as to feed back to the ATP binding pocket to reduce hydrolysis rate.
It is interesting that the R759E mutation does not grossly affect myofibril assembly. Indirect flight muscles at both the late pupal and young adult stages appear structurally normal. This contrasts with some myosin mutants in C. elegans and Drosophila wherein motor function defects prevented thick filament accumulation.26,27 Minor myofibrillar disarray is detected in R759E indirect flight muscle at 2 days of age, and more severe defects are seen at one week. Hence the degeneration in structure, as well as further degeneration in function, appears to be use related. It is important to note that at 2 days post-eclosion R759E muscle is essentially normal in structure, yet is barely able to support flight. Thus the functional defects in ATPase activity, lever arm swing and ability to translocate actin filaments are likely to give rise directly to the poor flight ability observed in 2 day old flies, rather than causing defects in myofibril assembly that secondarily result in defects in muscle function. We hypothesize that the mutation re-tunes the motor so that its duty ratio is increased, resulting in the S-1 domain remaining bound to the thin filament longer per cycle. Stretch by the opposing muscle set while some of the heads remain attached to the thin filament could result in progressive structural degeneration of the stretched myofibrils. We have found that chimeric myosins with slowed kinetics and an increased duty ratio typically show this sort of degenerative phenotype.28,29 Overall we conclude that relay to converter communication is essential to the biochemical properties of muscle myosin and the ability of muscle to function.
Acknowledgments
We appreciate comments on the manuscript from Dr. Douglas Swank (Rensselaer Polytechnic Institute) and Drs. Michael Geeves and Marieke Bloemink (University of Kent, Canterbury). Funds to support this research were provided by NIH grants R01 GM32443 to S.I.B. An NSF equipment grant (0308029) to Dr. Steven Barlow of the SDSU Electron Microscope Facility supported the purchase of the electron microscope.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gourinath S, Himmel DM, Brown JH, Reshetnikova L, Szent-Gyorgyi AG, Cohen C. Crystal structure of scallop Myosin S1 in the pre-power stroke state to 2.6 Å resolution: flexibility and function in the head. Structure. 2003;11:1621–1627. doi: 10.1016/j.str.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 3.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 4.Fisher AJ, Smith CA, Thoden JB, Smith R, Sutoh K, Holden HM, Rayment I. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP. BeFx and MgADP.AlF4. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 5.Houdusse A, Szent-Gyorgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc Natl Acad Sci USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Himmel DM, Gourinath S, Reshetnikova L, Shen Y, Szent-Gyorgyi AG, Cohen C. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc Natl Acad Sci USA. 2002;99:12645–12650. doi: 10.1073/pnas.202476799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gulick AM, Bauer CB, Thoden JB, Rayment I. X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain. Biochemistry. 1997;36:11619–11628. doi: 10.1021/bi9712596. [DOI] [PubMed] [Google Scholar]
- 8.Geeves MA, Holmes KC. The molecular mechanism of muscle contraction. Adv Protein Chem. 2005;71:161–193. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki N, Ohkura R, Sutoh K. Dictyostelium myosin II mutations that uncouple the converter swing and ATP hydrolysis cycle. Biochemistry. 2003;42:90–95. doi: 10.1021/bi026051l. [DOI] [PubMed] [Google Scholar]
- 10.Shih WM, Spudich JA. The myosin relay helix to converter interface remains intact throughout the actomyosin ATPase cycle. J Biol Chem. 2001;276:19491–19494. doi: 10.1074/jbc.M010887200. [DOI] [PubMed] [Google Scholar]
- 11.Rozek CE, Davidson N. Drosophila has one myosin heavy-chain gene with three developmentally regulated transcripts. Cell. 1983;32:23–34. doi: 10.1016/0092-8674(83)90493-2. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein SI, Mogami K, Donady JJ, Emerson CP., Jr Drosophila muscle myosin heavy chain encoded by a single gene in a cluster of muscle mutations. Nature. 1983;302:393–397. doi: 10.1038/302393a0. [DOI] [PubMed] [Google Scholar]
- 13.Chun M, Falkenthal S. Ifm(2)2 is a myosin heavy chain allele that disrupts myofibrillar assembly only in the indirect flight muscle of Drosophila melanogaster. J Cell Biol. 1988;107:2613–2621. doi: 10.1083/jcb.107.6.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collier VL, Kronert WA, O’Donnell PT, Edwards KA, Bernstein SI. Alternative myosin hinge regions are utilized in a tissue-specific fashion that correlates with muscle contraction speed. Genes Dev. 1990;4:885–895. doi: 10.1101/gad.4.6.885. [DOI] [PubMed] [Google Scholar]
- 15.O’Donnell PT, Collier VL, Mogami K, Bernstein SI. Ultrastructural and molecular analyses of homozygous-viable Drosophila melanogaster muscle mutants indicate there is a complex pattern of myosin heavy-chain isoform distribution. Genes Dev. 1989;3:1233–1246. doi: 10.1101/gad.3.8.1233. [DOI] [PubMed] [Google Scholar]
- 16.Gyimesi M, Kintses B, Bodor A, Perczel A, Fischer S, Bagshaw CR, Malnasi-Csizmadia A. The mechanism of the reverse recovery step, phosphate release, and actin activation of Dictyostelium myosin II. J Biol Chem. 2008;283:8153–8163. doi: 10.1074/jbc.M708863200. [DOI] [PubMed] [Google Scholar]
- 17.Mesentean S, Koppole S, Smith JC, Fischer S. The principal motions involved in the coupling mechanism of the recovery stroke of the myosin motor. J Mol Biol. 2007;367:591–602. doi: 10.1016/j.jmb.2006.12.058. [DOI] [PubMed] [Google Scholar]
- 18.Koppole S, Smith JC, Fischer S. The structural coupling between ATPase activation and recovery stroke in the myosin II motor. Structure. 2007;15:825–837. doi: 10.1016/j.str.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Hastings GA, Emerson CP., Jr Myosin functional domains encoded by alternative exons are expressed in specific thoracic muscles of Drosophila. J Cell Biol. 1991;114:263–276. doi: 10.1083/jcb.114.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Bernstein SI. Spatially and temporally regulated expression of myosin heavy chain alternative exons during Drosophila embryogenesis. Mech Dev. 2001;101:35–45. doi: 10.1016/s0925-4773(00)00549-9. [DOI] [PubMed] [Google Scholar]
- 21.Batra R, Manstein DJ. Functional characterisation of Dictyostelium myosin II with conserved tryptophanyl residue 501 mutated to tyrosine. Biol Chem. 1999;380:1017–1023. doi: 10.1515/BC.1999.126. [DOI] [PubMed] [Google Scholar]
- 22.Yengo CM, Chrin LR, Rovner AS, Berger CL. Tryptophan 512 is sensitive to conformational changes in the rigid relay loop of smooth muscle myosin during the MgATPase cycle. J Biol Chem. 2000;275:25481–25487. doi: 10.1074/jbc.M002910200. [DOI] [PubMed] [Google Scholar]
- 23.Malnasi-Csizmadia A, Kovacs M, Woolley RJ, Botchway SW, Bagshaw CR. The dynamics of the relay loop tryptophan residue in the Dictyostelium myosin motor domain and the origin of spectroscopic signals. J Biol Chem. 2001;276:19483–19490. doi: 10.1074/jbc.M010886200. [DOI] [PubMed] [Google Scholar]
- 24.Park S, Burghardt TP. Isolating and localizing ATP-sensitive tryptophan emission in skeletal myosin subfragment 1. Biochemistry. 2000;39:11732–11741. doi: 10.1021/bi000945t. [DOI] [PubMed] [Google Scholar]
- 25.Malnasi-Csizmadia A, Toth J, Pearson DS, Hetenyi C, Nyitray L, Geeves MA, Bagshaw CR, Kovacs M. Selective perturbation of the myosin recovery stroke by point mutations at the base of the lever arm affects ATP hydrolysis and phosphate release. J Biol Chem. 2007;282:17658–17664. doi: 10.1074/jbc.M701447200. [DOI] [PubMed] [Google Scholar]
- 26.Bejsovec A, Anderson P. Functions of the myosin ATP and actin binding sites are required for C. elegans thick filament assembly. Cell. 1990;60:133–140. doi: 10.1016/0092-8674(90)90723-r. [DOI] [PubMed] [Google Scholar]
- 27.Kronert WA, O’Donnell PT, Bernstein SI. A charge change in an evolutionarily-conserved region of the myosin globular head prevents myosin and thick filament accumulation in Drosophila. J Mol Biol. 1994;236:697–702. doi: 10.1006/jmbi.1994.1182. [DOI] [PubMed] [Google Scholar]
- 28.Swank DM, Knowles AF, Suggs JA, Sarsoza F, Lee A, Maughan DW, Bernstein SI. The myosin converter domain modulates muscle performance. Nat Cell Biol. 2002;4:312–316. doi: 10.1038/ncb776. [DOI] [PubMed] [Google Scholar]
- 29.Swank DM, Braddock J, Brown W, Lesage H, Bernstein SI, Maughan DW. An alternative domain near the ATP binding pocket of Drosophila myosin affects muscle fiber kinetics. Biophys J. 2006;90:2427–2435. doi: 10.1529/biophysj.105.075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloemink MJ, Dambacher CM, Knowles AF, Melkani GC, Geeves MA, Bernstein SI. Alternative exon 9-encoded relay domains affect more than one communication pathway in the Drosophila myosin head. J Mol Biol. 2009;389:707–721. doi: 10.1016/j.jmb.2009.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swank DM, Bartoo ML, Knowles AF, Iliffe C, Bernstein SI, Molloy JE, Sparrow JC. Alternative exon-encoded regions of Drosophila myosin heavy chain modulate ATPase rates and actin sliding velocity. J Biol Chem. 2001;276:15117–15124. doi: 10.1074/jbc.M008379200. [DOI] [PubMed] [Google Scholar]
- 32.Swank DM, Knowles AF, Kronert WA, Suggs JA, Morrill GE, Nikkhoy M, Manipon GG, Bernstein SI. Variable N-terminal regions of muscle myosin heavy chain modulate ATPase rate and actin sliding velocity. J Biol Chem. 2003;278:17475–17482. doi: 10.1074/jbc.M212727200. [DOI] [PubMed] [Google Scholar]
- 33.Miller BM, Nyitrai M, Bernstein SI, Geeves MA. Kinetic analysis of Drosophila muscle myosin isoforms suggests a novel mode of mechanochemical coupling. J Biol Chem. 2003;278:50293–50300. doi: 10.1074/jbc.M308318200. [DOI] [PubMed] [Google Scholar]
- 34.O’Donnell PT, Bernstein SI. Molecular and ultrastructural defects in a Drosophila myosin heavy chain mutant: differential effects on muscle function produced by similar thick filament abnormalities. J Cell Biol. 1988;107:2601–2612. doi: 10.1083/jcb.107.6.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kronert WA, Dambacher CM, Knowles AF, Swank DM, Bernstein SI. Alternative relay domains of Drosophila melanogaster myosin differentially affect ATPase activity, in vitro motility, myofibril structure and muscle function. J Mol Biol. 2008;379:443–456. doi: 10.1016/j.jmb.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swank DM, Wells L, Kronert WA, Morrill GE, Bernstein SI. Determining structure/function relationships for sarcomeric myosin heavy chain by genetic and transgenic manipulation of Drosophila. Microsc Res Tech. 2000;50:430–442. doi: 10.1002/1097-0029(20000915)50:6<430::AID-JEMT2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 37.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 38.Drummond DR, Hennessey ES, Sparrow JC. Characterisation of missense mutations in the Act88F gene of Drosophila melanogaster. Mol Gen Genet. 1991;226:70–80. doi: 10.1007/BF00273589. [DOI] [PubMed] [Google Scholar]
- 39.Tohtong R, Yamashita H, Graham M, Haeberle J, Simcox A, Maughan D. Impairment of muscle function caused by mutations of phosphorylation sites in myosin regulatory light chain. Nature. 1995;374:650–653. doi: 10.1038/374650a0. [DOI] [PubMed] [Google Scholar]
- 40.Pardee J, Spudich J. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]