Summary
Interphase cytogenetics are commonly used to identify clonal abnormalities in chronic lymphocytic leukemia (CLL) patients but fail to identify recurrent translocations that ultimately can direct more focused molecular characterization. Given the importance of del(17p13.1) in CLL outcome, we performed an extensive review of 1213 patients undergoing metaphase cytogenetics at our institution and identified 16 (1.3%) with a recurrent unbalanced translocation between the p arms of chromosomes 17 and 18 that results in a dicentric chromosome with loss of much of 17p and 18p. The dic(17;18)(p11.2;p11.2) was associated with a complex (three or more unrelated cytogenetic abnormalities) karyotype in 12 patients (75%) at the time that the abnormality was first identified, and eventually associated with a complex karyotype in 94% of patients. IGHV mutational analysis was un-mutated in 88% of cases where evaluation was possible. Except for one patient who was diagnosed with CLL incidentally during a workup for metastatic tonsillar cancer, all patients identified with dic(17;18)(p11.2;p11.2) met criteria for disease treatment, with a median time from diagnosis to first treatment of 15 months. Our data demonstrate that dic(17;18)(p11.2;p11.2) is a novel recurrent cytogenetic abnormality in CLL associated with early age at diagnosis and accelerated disease progression. Future efforts to identify genes disrupted by this translocation are warranted and ongoing.
Keywords: chronic lymphocytic leukemia, prognostic factors, cytogenetic abnormalities, FISH, dic(17;18)(p11.2;p11.2)
B-cell chronic lymphocytic leukemia (CLL), the clonal proliferation of mature B lymphocytes, is the most common form of adult leukemia. This disease has a variable clinical course; many patients do not require treatment for years and have survival equal to age-matched controls. Other patients, however, exhibit aggressive disease and have a poor prognosis despite appropriate therapy (Byrd et al, 2004).
Because of the variable course of CLL, much research has focused on the elucidation of characteristics that help predict clinical course. Staging systems by Binet et al (1981) and Rai et al (1975) have been used for many years to aid in disease prognosis. More recently, chromosomal aberrations have been identified that can help differentiate between patients who will have slowly progressive disease and those who will have an aggressive disease course. Clonal abnormalities have been found in 40–50% of patients with CLL by conventional cytogenetics (Juliusson et al, 1990; Juliusson et al, 1991), and in 80% by fluorescence in situ hybridization (FISH) analysis (Dohner et al, 2000). The most common and well-documented chromosomal abnormalities include del(13q14.3), trisomy 12, del(17p13.1), and del(11q22.3). Of these, deletions of 17p13.1 and 11q22.3 are associated with poor disease prognosis and deletion of 13q14.3 is associated with slower progression of disease (Dohner et al, 2000). Trisomy of chromosome 12 has a more variable course, but is also associated with more aggressive disease (Dohner et al, 2000; Athanasiadou et al, 2006). Because of the importance of these prognostic indicators, cytogenetic analysis has become routine in the evaluation of CLL patients. With knowledge of common abnormalities in CLL, FISH has proven useful because it allows rapid detection of these recurrent abnormalities. Conventional cytogenetics, on the other hand, allows analysis of the entire chromosome complement and the opportunity to discover previously unrecognized abnormalities and also to better localize areas for relevant genes disrupted in B-cell transformation. Most relevant to the identification of such genes are regions of clinical prognostic impact, such as del(17p13.1), for which no contributing gene has been identified. This abnormality is associated with a need for early therapy, poor response to conventional treatment (Dohner et al, 1995; Byrd et al, 2006; Grever et al, 2007; Van Den Neste et al, 2007), and shortened survival (Wattel et al, 1994; Dohner et al, 1995, 2000; Byrd et al, 2006).
Besides cytogenetic abnormalities, the mutational status of the variable region of the immunoglobulin heavy chains (IGHV) has been found to be predictive of disease progression, with unmutated IGHV associated with a poorer prognosis independent of disease stage or other cytogenetic markers (Hamblin et al, 1999). Unmutated IGHV has also been found to be highly associated with cytogenetic abnormalities (Karhu et al, 2003) including the poor prognostic indicators del(17p13.1) and del(11q22.3) (Krober et al, 2002).
In this report, we describe a novel recurrent abnormality that is the result of an unbalanced translocation between chromosomes 17 and 18 whose identity has been appreciated previously in only 12 reported cases. We describe the clinical and cytogenetic features associated with this abnormality in a larger cohort of patients, review data available from previous cases, and identify directions for future research.
Materials and methods
Patients
Between January 2002 and February 2009, 1213 patients with CLL underwent cytogenetic and FISH analyses at our institution. Patients were enrolled on an Institutional Review Board approved prospective tissue bank study that allows assessment of prognostic factors and treatment outcome.
Diagnostic testing
Bone marrow or peripheral blood samples were cultured in RPMI 1640 medium (Gibco) by standard laboratory procedures From January, 2002 through September, 2007, all cultures were stimulated with pokeweed mitogen (Sigma-Aldrich, St Louis, MO, USA) at a final concentration of 10 ul/ml and phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich) at a final concentration of 40 ng/ml. From October 2007, CpG stimulation (20 μg/ml) was also added to the cultures. Cultures were for 72 h, and harvesting, fixation and banding were by standard laboratory techniques. Twenty metaphases per sample were completely analyzed. Chromosomes were classified according to the International System for Human Cytogenetic Nomenclature (ISCN 2005) (Schaffer & Tommerup, 2005). FISH was performed on all samples with commercially available probe sets (Abbott Molecular, Abbott Park, IL, USA) designed to detect the chromosome 12 centromere, MYC located at 8q24, MYB located at 6q23, D13S319 located at 13q14.3, ATM located at 11q22.3 and TP53 located at 17p13.1 as previously reported by our group (Byrd et al, 2006). IGHV mutational analysis was performed as previously described since 2006 and retrospectively in patients seen at Ohio State University since 2002 for whom stored samples were available.
Biostatistical analysis
Actuarial overall survival and treatment-free survival were estimated using the method of Kaplan–Meier. Treatment-free survival indicates the time from disease diagnosis until initiation of chemotherapy. Overall survival was calculated from the date of disease diagnosis until death.
Results
Cytogenetic description
Peripheral blood or bone marrow samples from total of 1213 patients with CLL were analyzed, and 16 (1.3%) of these patients were found, through banded metaphase cytogenetics, to have a dic(17;18)(p11.2;p11.2). This dicentric chromosome is the result of an unbalanced translocation between the p arms of chromosomes 17 and 18 and results in a dicentric chromosome with loss of much of 17p and 18p. The dicentric nature of the chromosome was proven with FISH analysis using chromosome 17 and chromosome 18 centromere probes in different colors (Abbott Molecular), which showed the presence and juxtaposition of both centromeres in each of the nine cases analyzed in this manner. An example image is provided in Fig 1. This chromosomal abnormality was associated with a complex karyotype in 12 patients (75%) at the time that dic(17;18)(p11.2;p11.2) was first identified; 31% of patients had three abnormalities, 19% had 4, 12.5% had five, and 12.5% had six or more cytogenetic abnormalities at this time. In one patient, dic(17;18)(p11.2;p11.2) was the sole abnormality seen on conventional cytogenetics at initial presentation, but FISH at that time was positive for trisomy 12 in 5.1% of cells, and the patient eventually developed a complex karyotype. At the time of this report, only one patient has not developed a complex karyotype. The dic(17;18)(p11.2;p11.2) was associated with trisomy 12 in seven patients (44%) and with del(13q) in five patients (31%) with no overlap between these two abnormalities. In 11 patients, this abnormality was seen at the time of first conventional cytogenetic analysis, whereas in two patients the abnormality was acquired following treatment. In patients who acquired the abnormality, the time from diagnosis to acquisition was 31 and 43 months respectively. In the remaining three patients, initial cytogenetics showed a normal karyotype, while FISH was positive for del(17p13.1) in 11.2%, 26%, and 36% (with 52.3% positive for trisomy 12) of cells, suggesting that the abnormality was present, but in such low copy number that it was not detected by conventional cytogenetic analysis. Of the two patients who are known to have acquired dic(17;18)(p11.2;p11.2) during the course of their disease, both also acquired other cytogenetic abnormalities.
Fig 1.

(A) Banded metaphase analysis showing 46,XX,del(13)(q14q22),dic(17;18)(p11.2;p11.2). (B) Metaphase FISH with CEP17 (SpecrumAqua) and CEP18 (SpectrumOrange) show the presence of both centromeres, indicating a dic(17;18).
Clinical and laboratory characteristics
The demographics of the patients with dic(17;18)(p11.2;p11.2) are summarized in Table I. Of 16 patients with this cytogenetic abnormality, nine were male (56%), and the median age at diagnosis was 57 years (range 37–68). At diagnosis, 62% of patients were Rai stage 0 or 1; three patients were stage 4 at diagnosis. The mean WBC at the time of diagnosis in the 13 patients where the information was available was 24 × 109/l. IGHV mutational status was known for 48% of patients, and of these eight patients, seven (88%) were found to have unmutated IGHV status.
Table I.
Patient characteristics.
| Characteristic | Number (%) |
|---|---|
| Age (years) at diagnosis (median) | 57 (range 37–68) |
| Sex | |
| Male | 9 (56) |
| Female | 7 (44) |
| Rai stage at diagnosis | |
| 0 | 5 (31) |
| 1 | 5 (31) |
| 2 | 3 (19) |
| 3 | 0 |
| 4 | 3 (19) |
| WBC (×109/l) at diagnosis (median) | 24.0 (n = 13) |
| Time from diagnosis to first treatment | 15 months (n = 14) |
| Fludarabine refractory | |
| Yes | 7 (44) |
| No | 5 (31) |
| Unknown | 4 (24) |
| Cytogenetics | |
| Complex (three or more unrelated abnormalities) | 12 (75) |
| Trisomy 12 | 7 (44) |
| Del 11q | 1 (6) |
| Del 13q | 5 (31) |
| IGHV mutational status | |
| Mutated | 1 (12) |
| Unmutated | 7 (88) |
| Unknown | 8 |
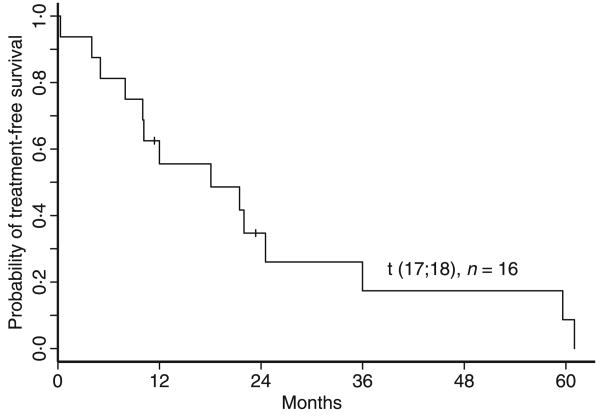
Treatment data are available for 14 of the 16 patients identified with this abnormality, and survival data in all patients. Except in the case of one patient who was diagnosed with CLL incidentally during a workup for metastatic tonsillar cancer, all patients identified with dic(17;18)(p11.2;p11.2) met criteria for disease treatment, with a median time from diagnosis to first treatment of 15 months, as outlined in Fig 2. In the two patients where follow-up data are not available, both were advised to begin treatment at their visit to our institution. Of the 12 patients who received fludarabine-based therapy, seven (58%) were refractory. Three patients have received stem cell transplant for recurrent/refractory disease, and four are currently undergoing chemotherapy. With a mean follow-up of 54 months, four patients have died, 21, 42, 49 and 92 months after diagnosis. Kaplan–Meier estimates of overall survival are shown in Fig 3. All of the deceased patients had a complex karyotype.
Fig 2.
Treatment free survival. Median time from diagnosis to first treatment is 15 months.
Fig 3.
Kaplan–Meier estimates of overall survival.
Discussion
Here we describe the largest series of dic(17;18)(p11.2;p11.2) CLL patients reported in the literature and firmly establish this as a recurring abnormality in CLL. Our data demonstrated that dic(17;18)(p11.2;p11.2) is a novel recurrent cytogenetic abnormality in CLL associated with early age at diagnosis, accelerated disease progression, and a trend toward more refractory disease. These features are similar to previously published reports of patients with loss of p53 or del(17p13.1) alone, which found these patients to require early therapy, to have poor response to standard purine analog therapy, and have shortened survival (Fenaux et al, 1992; Wattel et al, 1994; Dohner et al, 1995; Cordone et al, 1998; Oscier et al, 2002). Within this subset of del(17p.13.) patients are individuals that can have an indolent natural history for an extended period of time. It is possible that this subset of patients having dic(17;18)(p11.2;p11.2) will represent the most aggressive subset of del(17p13.1) CLL patients identified by FISH analysis. Indeed, it is interesting that, besides the obvious association with loss of the p53 allele, dic(17;18)(p11.2;p11.2) also frequently accompanied other negative prognostic markers. Sixty-four percent of patients were found to have this abnormality as part of a complex karyotype, and 88% of those tested had unmutated IGHV, both of which have been shown to be markers of more aggressive disease (Damle et al, 1999; Hamblin et al, 1999; Maloum et al, 2000; Oscier et al, 2002; Vasconcelos et al, 2003). These patients described with dic(17;18)(p11.2;p11.2) exhibited characteristics in common with patients with del(17p13.1), including short time from diagnosis to need for treatment (Dohner et al, 1995) and short overall survival (Dohner et al, 1995). Our patients also showed an association between dic(17;18)(p11.2;p11.2) and trisomy 12, with 44% of patients exhibiting both abnormalities. This association was not seen in any of the previously reported cases. It is unclear whether this is a true association or whether trisomy 12 is a common abnormality and these patients are predisposed to the development of further cytogenetic abnormalities. An extensive review of the literature identified 12 reported cases of dic(17;18)(p11.2;p11.2) (Dohner et al, 1995; Callet-Bauchu et al, 1999; Espinet et al, 2000; Chena et al, 2002; Adeyinka et al, 2007). Our study adds to the clinical relevance of this abnormality as, to date, very little clinical data have been associated with this abnormality. Of the reported cases, the median age at diagnosis was 59 (Espinet et al, 2000; Chena et al, 2002; Adeyinka et al, 2007) (n = 5), and 67% of patients were male. Complex karyotypes were found in 50% of these previously reported patients with this cytogenetic abnormality, and 33% had this as the sole abnormality (Dohner et al, 1995; Callet-Bauchu et al, 1999; Espinet et al, 2000; Chena et al, 2002; Adeyinka et al, 2007). Three patients are known to have been treated with fludarabine (Callet-Bauchu et al, 1999; Chena et al, 2002), and all were refractory; three patients were refractory to anthracycline-based regimens (Callet-Bauchu et al, 1999; Espinet et al, 2000; Chena et al, 2002). These anecdotal findings are consistent with our larger series of patients.
It has been shown previously that 17p10–17p12 is a common breakpoint in CLL (Fink et al, 2006) and other malignancies. This region is genetically unstable due to the presence of multiple low copy repeats of DNA sequences (LCR) (Stankiewicz et al, 2003; Fink et al, 2006). With the identification of dic(17;18)(p11.2;p11.2) as a recurrent abnormality, it is possible that chromosome band 18p11.2 is another unstable area predisposed to breakage, and may be an area worthy of further investigation.
Because del(17p13.1) is often seen in advanced stages of CLL, it is thought to be a late event in pathogenesis. In our patients, dic(17;18)(p11.2;p11.2) appeared frequently at the time of diagnosis or shortly thereafter, making it likely to be an earlier event in leukemogenesis. Although our patients all had dic(17;18)(p11.2;p11.2) in association with other chromosomal abnormalities, in previous reports 33% of patients had this as the sole abnormality, so it is possible that this abnormality disrupts DNA repair in a way that predisposes to further genetic abnormalities.
Future study of this cytogenetic abnormality could more fully characterize the genes lost in this translocation. It is difficult to compare our data with previously published reports of patients with del(17p13.1) because of significant differences in diagnostic patterns and advances in treatment, so future study could focus on comparison among patients with dic(17;18)(p11.2;p11.2), del(17p13.1), and average risk cytogenetics. It will be important to evaluate whether there are differences in prognosis or response between patients with dic(17;18)(p11.2;p11.2) and those with del(17p13.1) to determine whether routine differentiation by conventional cytogenetics or FISH probes to detect a dicentric chromosome would be beneficial.
Acknowledgments
This project was supported by K23 CA102276-05, P30 CA 16058, The Leukemia and Lymphoma Society, and the D. Warren Brown Foundation.
References
- Adeyinka A, Wei S, Sanchez J. Loss of 17p is a major consequence of whole-arm chromosome translocations in hematologic malignancies. Cancer Genetics and Cytogenetics. 2007;173:136–143. doi: 10.1016/j.cancergencyto.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Athanasiadou A, Stamatopoulos K, Tsompanakou A, Gaitatzi M, Kalogiannidis P, Anagnostopoulos A, Fassas A, Tsezou A. Clinical, immunophenotypic, and molecular profiling of trisomy 12 in chronic lymphocytic leukemia and comparison with other karyotypic subgroups defined by cytogenetic analysis. Cancer Genetics and Cytogenetics. 2006;168:109–119. doi: 10.1016/j.cancergencyto.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Binet JL, Auquier A, Dighiero G, Chastang C, Piguet H, Goasguen J, Vaugier G, Potron G, Colona P, Oberling F, Thomas M, Tchernia G, Jacquillat C, Boivin P, Lesty C, Duault MT, Monconduit M, Belabbes S, Gremy F. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer. 1981;48:198–206. doi: 10.1002/1097-0142(19810701)48:1<198::aid-cncr2820480131>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Byrd JC, Stilgenbauer S, Flinn IW. Chronic lymphocytic leukemia. Hematology. American Society of Hematology Educational Program Book. 2004;2004:163–183. doi: 10.1182/asheducation-2004.1.163. [DOI] [PubMed] [Google Scholar]
- Byrd JC, Gribben JG, Peterson BL, Grever MR, Lozanski G, Lucas DM, Lampson B, Larson RA, Caligiuri MA, Heerema NA. Select high-risk genetic features predict earlier progression following chemoimmunotherapy with fludarabine and rituximab in chronic lymphocytic leukemia: justification for risk-adapted therapy. Journal of Clinical Oncology. 2006;24:437–443. doi: 10.1200/JCO.2005.03.1021. [DOI] [PubMed] [Google Scholar]
- Callet-Bauchu E, Salles G, Gazzo S, Poncet C, Morel D, Pages J, Coiffier B, Coeur P, Felman P. Translocations involving the short arm of chromosome 17 in chronic B-lymphoid disorders: frequent occurrence of dicentric rearrangements and possible association with adverse outcome. Leukemia. 1999;13:460–468. doi: 10.1038/sj.leu.2401272. [DOI] [PubMed] [Google Scholar]
- Chena C, Cerretini R, Noriega MF, Narbaitz M, Scolnik M, Palacios MF, Neme D, Bruno S, Slavutsky I. Cytogenetic, FISH, and molecular studies in a case of B-cell chronic lymphocytic leukemia with karyotypic evolution. European Journal of Haematology. 2002;69:309–314. doi: 10.1034/j.1600-0609.2002.02793.x. [DOI] [PubMed] [Google Scholar]
- Cordone I, Masi S, Mauro FR, Soddu S, Morsilli O, Valentini T, Vegna ML, Guglielmi C, Mancini F, Giuliacci S, Sacchi A, Mandelli F, Foa R. p53 expression in B-cell chronic lymphocytic leukemia: a marker of disease progression and poor prognosis. Blood. 1998;91:4342–4349. [PubMed] [Google Scholar]
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- Dohner H, Fischer K, Bentz M, Hansen K, Benner A, Cabot G, Diehl D, Schlenk R, Coy J, Stilgenbauer S. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood. 1995;85:1580–1589. [PubMed] [Google Scholar]
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, Dohner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. New England Journal of Medicine. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- Espinet B, Sole F, Lloveras E, Abella E, Besses C, Woessner S, Florensa L. Dicentric (17;18) in a case of atypical B-cell chronic lymphocytic leukemia. Cancer Genetics and Cytogenetics. 2000;121:194–197. doi: 10.1016/s0165-4608(00)00255-7. [DOI] [PubMed] [Google Scholar]
- Fenaux P, Preudhomme C, Lai JL, Quiquandon I, Jonveaux P, Vanrumbeke M, Sartiaux C, Morel P, Loucheux-Lefebvre MH, Bauters F. Mutations of the p53 gene in B-cell chronic lymphocytic leukemia: a report on 39 cases with cytogenetic analysis. Leukemia. 1992;6:246–250. [PubMed] [Google Scholar]
- Fink SR, Smoley SA, Stockero KJ, Paternoster SF, Thorland EC, Van Dyke DL, Shanafelt TD, Zent CS, Call TG, Kay NE, Dewald GW. Loss of TP53 is due to rearrangements involving chromosome region 17p10 approximately p12 in chronic lymphocytic leukemia. Cancer Genetics and Cytogenetics. 2006;167:177–181. doi: 10.1016/j.cancergencyto.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Grever MR, Lucas DM, Dewald GW, Neuberg DS, Reed JC, Kitada S, Flinn IW, Tallman MS, Appelbaum FR, Larson RA, Paietta E, Jelinek DF, Gribben JG, Byrd JC. Comprehensive assessment of genetic and molecular features predicting outcome in patients with chronic lymphocytic leukemia: results from the US Intergroup Phase III Trial E2997. Journal of Clinical Oncology. 2007;25:799–804. doi: 10.1200/JCO.2006.08.3089. [DOI] [PubMed] [Google Scholar]
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- Juliusson G, Oscier DG, Fitchett M, Ross FM, Stockdill G, Mackie MJ, Parker AC, Castoldi GL, Guneo A, Knuutila S. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. New England Journal of Medicine. 1990;323:720–724. doi: 10.1056/NEJM199009133231105. [DOI] [PubMed] [Google Scholar]
- Juliusson G, Gahrton G, for the International Working Party on Chromosomes in CLL (IWCCLL) Cytogenetic findings and survival in B-cell chronic lymphocytic leukemia. Second IWCCLL compliation of data on 662 patients. Leukemia & Lymphoma. 1991;5:21–25. doi: 10.3109/10428199109103374. [DOI] [PubMed] [Google Scholar]
- Karhu R, Tobin G, Thunberg U, Vilpo L, Sundstrom C, Knuutila S, Rosenquist R, Vilpo J. More extensive genetic alterations in unmutated than in hypermutated cases of chronic lymphocytic leukemia. Genes, Chromosomes and Cancer. 2003;37:417–420. doi: 10.1002/gcc.10227. [DOI] [PubMed] [Google Scholar]
- Krober A, Seiler T, Benner A, Bullinger L, Bruckle E, Lichter P, Dohner H, Stilgenbauer S. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416. [PubMed] [Google Scholar]
- Maloum K, Davi F, Merle-Beral H, Pritsch O, Magnac C, Vuillier F, Dighiero G, Troussard X, Mauro FF, Benichou J. Expression of unmutated VH genes is a detrimental prognostic factor in chronic lymphocytic leukemia. Blood. 2000;96:377–379. [PubMed] [Google Scholar]
- Oscier DG, Gardiner AC, Mould SJ, Glide S, Davis ZA, Ibbotson RE, Corcoran MM, Chapman RM, Thomas PW, Copplestone JA, Orchard JA, Hamblin TJ. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100:1177–1184. [PubMed] [Google Scholar]
- Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46:219–234. [Google Scholar]
- Schaffer LG, Tommerup N. ISCN (2005): An International System for Human Cytogenetic Nomenclature. S. Karger; Basel: 2005. [Google Scholar]
- Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, Shaffer LG, Withers M, Elizondo L, Park SS, Lupski JR. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. American Journal of Human Genetics. 2003;72:1101–1116. doi: 10.1086/374385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Neste E, Robin V, Francart J, Hagemeijer A, Stul M, Vandenberghe P, Delannoy A, Sonet A, Deneys V, Costantini S, Ferrant A, Robert A, Michaux L. Chromosomal translocations independently predict treatment failure, treatment-free survival and overall survival in B-cell chronic lymphocytic leukemia patients treated with cladribine. Leukemia. 2007;21:1715–1722. doi: 10.1038/sj.leu.2404764. [DOI] [PubMed] [Google Scholar]
- Vasconcelos Y, Davi F, Levy V, Oppezzo P, Magnac C, Michel A, Yamamoto M, Pritsch O, Merle-Beral H, Maloum K, Ajchenbaum-Cymbalista F, Dighiero G. Binet's staging system and VH genes are independent but complementary prognostic indicators in chronic lymphocytic leukemia. Journal of Clinical Oncology. 2003;21:3928–3932. doi: 10.1200/JCO.2003.02.134. [DOI] [PubMed] [Google Scholar]
- Wattel E, Preudhomme C, Hecquet B, Vanrumbeke M, Quesnel B, Dervite I, Morel P, Fenaux P. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood. 1994;84:3148–3157. [PubMed] [Google Scholar]