Summary
The quantitative study of the cell growth [1-5] has led to many fundamental insights in our understanding of a wide range of subjects from cell cycle [6-9] to senescence [10]. Of particular importance is the growth rate, whose constancy represents a physiological steady state of an organism. Recent studies, however, suggest that the rate of elongation during exponential growth of bacterial cells decreases cumulatively with replicative age for both asymmetrically [11] and symmetrically [12,13] dividing organisms, implying that a “steady-state” population consists of individual cells that are never in a steady state of growth. To resolve this seeming paradoxical observation, we studied the long-term growth and division patterns of Escherichia coli cells by employing a microfluidic device designed to follow steady state growth and division of a large number of cells at a defined reproductive age. Our analysis of ~105 individual cells reveals a remarkable stability of growth of the mother cell inheriting the same pole for hundreds of generations. We further show that death of E. coli is not purely stochastic but is the result of accumulating damages. We conclude that E. coli, unlike all other aging model systems studied to date, has a robust mechanism of growth that is decoupled from cell death.
Results and Discussion
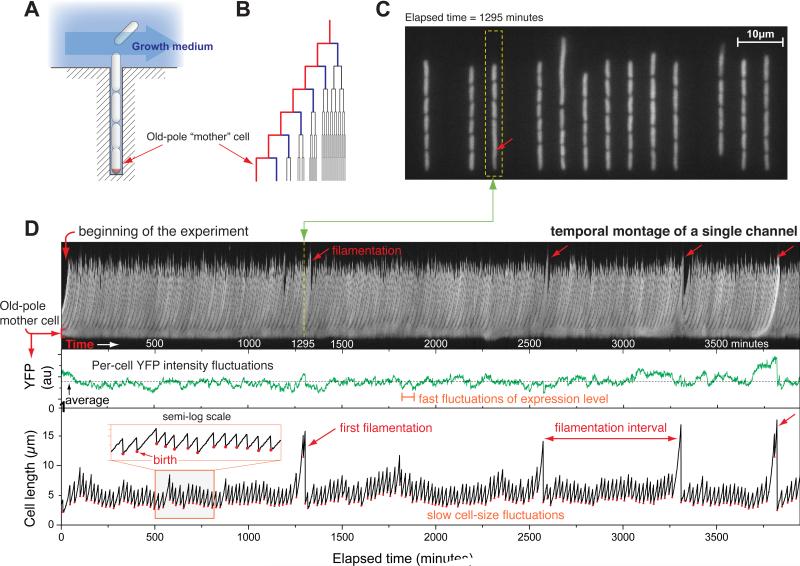
To follow a large number of cells inheriting the same pole and their progeny for many generations, we employed a high-throughput, continuous, microfluidic liquid culture device built using a standard soft-lithographic technique developed by others for cell biology studies [14-17]. Our device consists of a series of growth channels, oriented at right angles to a trench through which growth medium is passed at a constant rate (Fig. 1A). This constant flow results in diffusion of fresh medium into the growth channels as well as removal of cells as they emerge from the channels into the main trench (Fig. 1A). We measured the timescale of nutrient uptake by E. coli using the fluorescent glucose analog (2-NBDG) and found that diffusion into the channels is very much faster (~ 1 second) than the timescale of nutrient uptake (~2-3 minutes; Supplemental Experimental Procedures), ensuring steady-state conditions for all cells. The cell at the end of the growth channel, distal to the trench, is referred to as the “old-pole mother cell” (or mother cell) since one of its poles, abutting the end of the channel, is inherited from one generation to the next (Fig. 1A). The diameter of the growth channels prevents the mother cell from moving around. The replicative age of the mother cell, defined as the number of consecutive divisions from the young-pole daughter cell [12], increases by one generation at each cell division (Fig. 1B). It is noteworthy that this device, which we call the “mother machine,” allows us to follow orders of magnitude more generations of cells than has been possible with other single celled organisms including Saccharomyces cerevisiae [18] and E. coli [12] (see Experimental Procedures; Supplemental Movies S1A and S1B).
Fig. 1.
The “mother machine” and high-throughput observation of the mother cells. (A) Schematic illustration of the microfluidic mother machine. The old-pole mother cell is trapped at the end of the growth channel. (B) The outer-most branch of the lineage tree represents the old-pole mother cell and her progeny. (C) Snapshot of a typical field of view. (D) (top panel) Temporal montage of a single growth channel [within the dotted yellow box in (C)] from the beginning to the end of the experiment. The stable band on the bottom of the montage is the old-pole mother cell, whereas the “feather” of the montage show the growth and escape of her progeny; (mid panel) the average YFP intensity of the mother cell fast fluctuating around the mean without decay with time; (bottom panel) cell length vs. time of the old-pole mother cell, which shows occasional spikes (filamentations). In all three panels, the x-axis represents time in minutes.
We studied two distantly related strains of E. coli, B/r and MG1655, which constitutively express yellow fluorescent protein (YFP) from a chromosomal copy of the yfp gene, allowing visualization of the cells via live microscopy (Fig. 1C). A typical time series of a single growth channel from the beginning of the experiment until death of the mother cell is shown in the top panel of Fig. 1D (constructed from the time series images by following the growth channel indicated by the dotted yellow box in Fig. 1C; see Supplementary Movie S1B). This temporal montage shows the fluorescence level (YFP) of the mother cell and her progeny over time during the reproductive lifetime of the mother cell (Fig. 1D (mid panel)). A cell length vs. time curve was constructed for every cell in all of our experiments (e.g., Fig. 1D, bottom panel). This curve is well approximated by a straight line in a semi-log plot (see the inset of Fig. 1D). That is, each interval between birth and division can be fitted using a single exponential function to obtain the growth rate of the cell at that replicative age. The spikes which appear at random intervals in the size distribution are the result of limited filamentation, discussed in detail below.
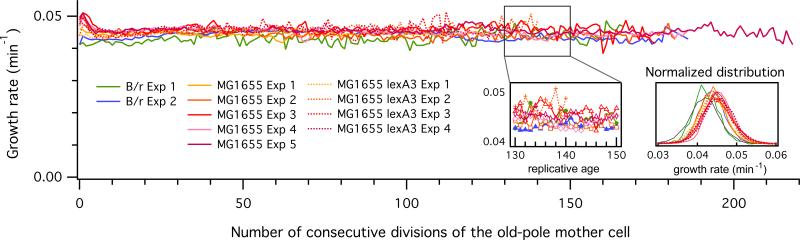
The growth rate of individual cells showed a striking long-term stability over hundreds of generations as indicated by the average growth rate vs. replicative age curves of the old-pole mother cells (Fig. 2). The growth rate remained constant under our experimental conditions, for both MG1655 and B/r. In contrast to this long-term stability, the growth rate of the old-pole mother cell showed only weak correlation between two consecutive cell cycles. Mother cells exhibited fast fluctuations with a timescale of less than one generation and following a Gaussian distribution (Fig. 2 inset; Fig. 3A). The daughter cells also showed the same growth-rate statistics as the mother cells as we summarize in Figures and Tables S1 and S2. In other words, the cell “forgets” immediately upon division how fast it was growing in the previous cell cycle.
Fig. 2.
Long-term stability of growth rate in E. coli. The elongation rate does not show any cumulative decrease over 200 generations in all three strains (B/r, MG1655, MG1655 lexA3). The growth rates show fast fluctuations with short-term correlation following Gaussian distribution. These eleven curves represent over ~105 individual mother cells, namely, ~107 measured cell length.
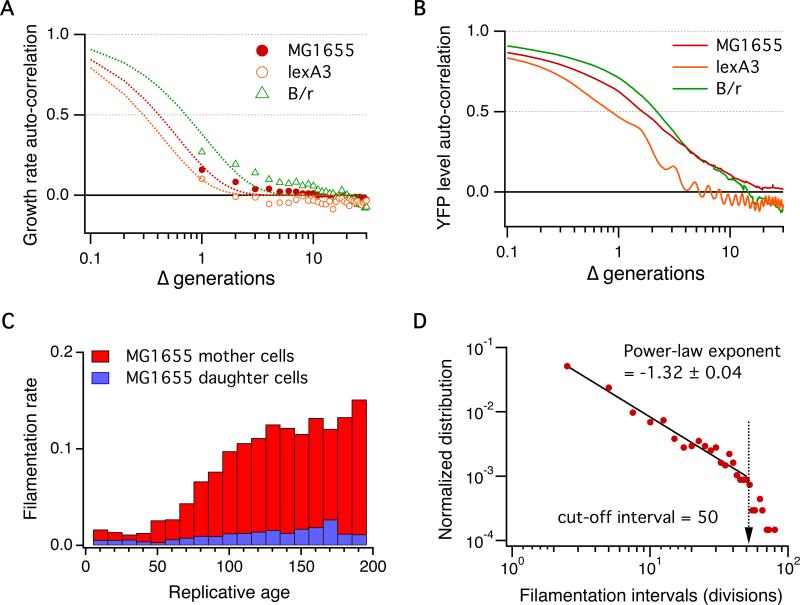
Fig. 3.
Short-term correlation of growth rates vs. long-term inheritance of a “factor” in the mother cell. (A) Auto-correlation function of growth rates showing less than 1 generation of correlation time. The dashed lines are a fit to the data using a single exponential function (to guide the eye): exp(-0.7x), exp(-0.43x) and exp(-x) for MG1655, MG1655 lexA3 and B/r, respectively. (B) Auto-correlation function of YFP level also showing 1-2 generations of correlation time. (C) Filamentation rate of MG1655. At a critical replicative age of the first 50 generations, the filamentation rate of the mother cell starts to increase, in contrast to the daughter cells that continue to divide normally. (D) A power-law distribution of filamentation intervals characterized by a long tail. Together with the increasing filamentation rate in (C), this implies a long-term memory that is independent of the observed stable growth and protein synthesis shown in Figs. 1 and 2.
The observed stable growth is mirrored by the stable protein synthesis reflected in the long-term constant fluorescence level of the mother cell and its progeny (Fig. 1D, top two panels). Like the growth rate, the YFP fluorescence level also shows a short-time correlation of one to two generations, consistent with previous findings in E. coli and human cells [19,20] (Fig. 3B). Our results showing the long-term stability of growth and protein synthesis, accompanied by their short-term memory, argue strongly against a built-in aging mechanism in E. coli that is based on growth. In other words, in E. coli cultures, all cells will be in the same steady state of growth indistinguishable from one another regardless of their replicative age.
While our experiments unambiguously show that growth and protein synthesis are characterized by short-term correlations, surprisingly, further analysis revealed unexpected long-term correlation that spans dozens of generations. Specifically, at a critical replicative age of the first 50 generations, we noted a striking increase in filamentation of the mother cells of MG1655 (the “spikes” seen in Fig. 1D bottom panel; Fig. 3C; Fig. S3). Importantly, the filamentation rate of the daughter cells remained practically constant and, thus, the increased filamenation of the mother cell cannot be due to illumination. This means that the mother cell must inherit an unknown “factor” from one generation to the next, which serves as a long-term memory, causing filamentation independently of growth and protein synthesis. Indeed, filamentation in Fig. 1D occurs at intervals such that its distribution follows a power law characterized by a long tail (Fig. 3D). [Note that random events will produce an exponential distribution (“memory-less”).] Such a long-term effect could not have been detected by more conventional timelapse experiments of exponentially growing population, since it requires an observation of the mother cell inheriting the same old pole for hundreds of generations.
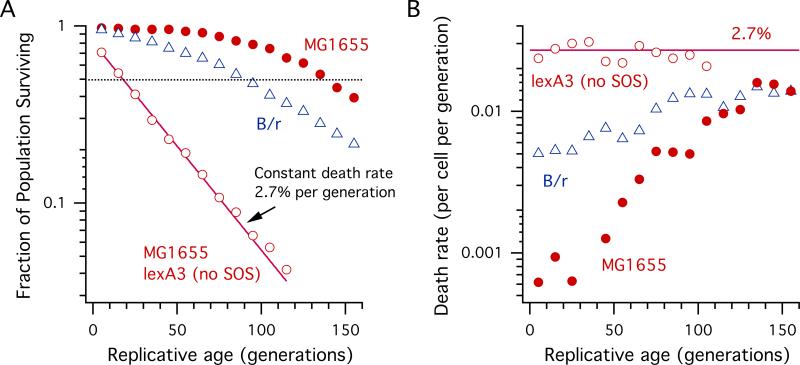
Since filamentation is a hallmark of the SOS response in bacteria, we asked how its suppression will affect our observation of filamentation. For this purpose we constructed a derivative of MG1655 carrying an allele of the lexA gene, lexA3, whose protein product constitutively represses SOS gene expression even under conditions of DNA damage. While the lexA3 mutant behaved virtually the same as MG1655 in terms of constant growth rate, its filamentation rate was significantly reduced, with a constant rate of ~ 1%, as expected. Note that B/r lacks sulA, a key SOS gene that inhibits cell division during the SOS response, also shows a similar low filamentatin rate (Fig. S3). A more important deviation lexA3 from MG1655 is that the population of the lexA3 mutant cells decayed exponentially with a constant death rate of 2.7% per cell per generation (Fig. 4).
Fig. 4.
Survival and mortality rate analysis showing aging of E. coli. (A) Survival curves of B/r, MG1655 and MG1655 lexA3 mutant (no SOS response). The pure exponential decay of the MG1655 lexA3 population with time allows us to directly extract 2.7% of constant death rate per generation in the absence of the SOS response. The higher survival rate of MG1655 is due to the SOS response. The dotted horizontal line represents 50% decay of the initial populations. (B) Death rate computed by numerically differentiating the survival curves in (A). Both B/r and MG1655 show increasing mortality rates, whereas MG1655 lexA3 shows a constant 2.7% rate of cell death.
These findings have important implications for the cause of cell death. That is, the death of the lexA3 mutant is random, a process requiring the SOS response for survival. The much slower death rate of wild type MG1655 cannot simply be due to a purely stochastic, age-independent, DNA damage or metabolic fluctuation, otherwise we would have observed an exponential decay like that of the lexA3 mutant (Fig. 4). Instead, death of MG1655 is likely caused by a growth-independent inheritance and accumulation of a lethal “factor” as indicated by the long-term correlation observed in the mother cell described above. It is possible that this “factor” corresponds to the protein aggregates described in recent work [13,21], which are asymmetrically distributed in E. coli. An alternative but not mutually exclusive idea is that the physically aging cell wall at the pole accumulates defects due to its metabolic inertness [22], which also could be linked to the lethal element.
In previous work by Stewart et al. [12], it was found that the growth rate of the mother cell decreased cumulatively with replicative age, about 2% per generation. While our results show otherwise, this could be due to the differences in the experimental conditions, e.g., two-dimensional surface on an agar pad (Stewart et al.) vs. one-dimensional growth channel where fresh liquid medium is constantly supplied (current study). In addition, we excluded the data from the first ten generations of replicative age to ensure that our results reflect steady-state growth conditions. Nevertheless, we note that the average generation times of the mother cells of B/r, MG1655 and lexA3 mutant are in precise agreement with the generation time measured from the growth curves of the liquid culture (see Experimental Procedures). This strongly argues that, in our study, it is unlikely there is a decrease of the growth rate of the mother cell regardless of its replicative age, i.e., all cells are in the same steady state of growth.
In summary, using the microfluidic mother machine, we have shown a striking constant growth rate of the mother cells of E. coli and their immediate sister cells for hundreds of generations. Also, from the qualitative difference in death rate between MG1655 and its lexA3 mutant derivative, we concluded that death of E. coli cells cannot be due to random events such as DNA damage but must be a consequence of growth-independent accumulation of lethal element. These observations have been made possible because the mother machine allows us to follow a large number of cells for a long time at the single-cell level in a precisely controlled environment, and produces amounts of data comparable to studies at a population level (e.g., FACS). Our experimental approach can be directly and immediately applied to a wide range of problems.
In our view, the most important lesson from our study is that E. coli must have a very robust mechanism of growth. The next major challenge, then, is to understand the origin of the stability of bacterial growth. Understanding it at the systems level may shed a new light on an important related question, i.e., how replication and division are coupled via growth and coordinated robustly in bacteria in the absence of eukaryotic-like checkpoints [23].
Experimental Procedures
Growth condition, microscopy, and microfluidics
Cultures were grown overnight in Luria-Bertani (LB) at 37°C. The next day, 30 μl of the overnight culture was back-diluted in 3ml of fresh LB at 37°C. At OD450 = 0.2 ~ 0.3 (or OD600 = 0.1 ~ 0.15), the cell culture was 10x concentrated by centrifugation for injection into the mother machine. The cells were loaded by diffusion until over 80% of the channels were filled with the cells. Fresh LB medium was infused by a syringe pump. For each experiment, images were acquired from 10 – 12 fields at one-minute intervals using NIS-Elements software and a Nikon Eclipse Ti fluorescence microscope equipped with a motorized stage and a CCD camera (Photometrics CoolSnap HQ2). We tested a wide range of dosage by varying the exposure time (1.5s – 4.0s) and the intensity of the illumination light using neutral density filters (ND32 to ND128; ND128 means that only 1/128 of the illumination light is used for imaging). Above ND32 and up to 2 seconds of exposure time, the growth rate measured in the mother machine precisely agreed with the growth rate measured from the growth curve for both E. coli strains B/r and MG1655 thus used in our studies as MG1655 = 20.9 ± 0.3 (minutes), MG1655 lexA3 = 20.7 ± 0.8 (minutes), B/r D = 22.6 ± 0.4 (minutes).
Our device consists of 4000 growth channels and an automated microscope stage, which we use to continuously scan 10-12 fields of view for 72 hours at one minute intervals (SI Movie 1). Since each field of view contains ~102 cells at 100x magnification (Fig. 1c), we acquire and process images of ~106 – 107 individual cells per experiment. See Supplementary Experimental Procedures for more details.
Image analysis and data acquisition
Custom software was developed using C++ to analyze time-lapse images. Our software has a user-friendly interface similar to ImageJ as shown below. Since each experiment generated about 50,000 to 100,000 images with each image containing ~ 102 cells, we analyzed typically ~107 cells per each time-lapse experiment. The current version of software can complete analysis in about 3~5 hours on a standard desktop PC, which is significantly faster than other software developed using more high-level programming languages such as Matlab. Segmentation of cells was directly performed on fluorescent images by finding basins of intensity profile along channel direction.
Bacterial Strains
The wild type strains of Escherichia coli MG1655 and B/r D were modified to express the gene encoding the yellow fluorescent protein (YFP) under the control of the constitutive promoter λPR [gene construct from M. Elowitz (Elowitz 2002)]. The YFP gene was inserted in the intC locus, along with a chloramphenicol resistance gene by P1 transduction from M. Elowitz's MRR strain (Elowitz 2002) and selection for chloramphenicol resistance. The lexA3 allele, present in strain RB258 [Braun RE and Wright A., Mol Gen Genet. 202(2):246-50, 1986], was introduced into MG1655, along with the closely linked marker malE::Tn5, by P1 transduction, with selection for kanamycin resistance. The MG1655 strain used is the poorly motile strain that does not contain an IS1 insertion sequence element in the regulatory region of the flhD promoter (Barker 2004) (CGSC 6300 of the E. coli Genetic Stock Center).
Autocorrelation function
The normalized auto-correlation function A(Δt) has been calculated as follows: , where I(t) denotes the quantity of interest at time t (e.g., yfp intensity, cell length). 〈〉t and 〈〉i denote average over time and cells, respectively. From the typical timescale of decay of the (normalized) auto-correlation function, we can estimate how long the memory of the process lasts, for example, by measuring how fast it drops from 1 to 0.5. If A(Δt) decays exponentially as exp(-t/τ), the inverse decay constant τ is defined as the correlation time of the process. However, if A(Δt) decays slowly, e.g, as a power-law t-α, where α is the exponent, there is no intrinsic timescale associated with the process. In this case, the process is said to have a long-term memory.
Filamentation
We considered a cell to be filamentous if its new-born cell size is larger than the population average by more than 2σ, where σ is the standard deviation of the cell-size distribution. In our experiments, filaments were visually obvious (Fig. 1C and 1D), well-separated from the normal cells in the size distributions (Fig. S2B), and our results are insensitive to the choice of the threshold. To compute the filamentation intervals, we only counted intervals between new filamentations developed from a normal sized cell. For example, the double spikes around 1250 minutes in Fig. 1D are counted as a single filamentation event.
Supplementary Material
Acknowledgments
Author contributions: SJ conceived and supervised the project. WLD, SJ and LR developed the experimental protocol. WLD and SJ designed the microfluidic “mother machine” and jointly constructed the device with PW. LR and AW constructed the strains. SJ and JP initiated image analysis method, and PW wrote CellTracker software used in the study. WLD, SJ, JP, LR and PW performed experiments and SJ, JP and PW analyzed the data. SJ wrote the paper with AW. All authors discussed the results and implications and commented on the manuscript.
We thank Irene Chen, Philippe Cluzel, Petra Levin, Steve Sandler, Bodo Stern for critical reading. We are particularly grateful to Andrew Murray for numerous stimulating and invaluable discussions on our manuscript. We also thank JD Deng (Harvard cleanroom), Peter Galajda, Daan Kiviet, Ariel Linder and Sander Tans for helpful discussions on the development of microfluidic device in the early stage of this work, and Robert Rioux (Whitesides lab) for help with chemical treatment of PDMS device.
This work was supported by Bauer Fellowship from Harvard University and the National Institutes of Health grant P50GM068763 (SJ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- The mother machine can follow thousands of cells for hundreds of generations.
- The mother cell of E. coli can divide hundreds of generations.
- The growth rate of the mother cell does not decrease until the cell dies.
- Death of E. coli is likely caused by accumulation of damage, independent of growth.
References
- 1.Monod J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949;3:371–394. [Google Scholar]
- 2.Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on Medium and Temperature of Cell Size and Chemical Composition during Balanced Growth of Salmonella typhimurium. J Gen Microbiol. 1958;19(3):592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 3.Kjeldgaard NO, Maaloe O, Schaechter M. The transition between different physiological states during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19(3):607–616. doi: 10.1099/00221287-19-3-607. [DOI] [PubMed] [Google Scholar]
- 4.Helmstetter C, Cooper S. DNA synthesis during the division cycle of rapidly growing Escherichia coli B/r. J. Mol. Biol. 1968;31:507–518. doi: 10.1016/0022-2836(68)90424-5. [DOI] [PubMed] [Google Scholar]
- 5.Donachie WD. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- 6.Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- 7.Neidhardt F. Bacterial growth: Constant Obsession with dN/dt. J. Bacteriol. 1999;181:7405–7408. doi: 10.1128/jb.181.24.7405-7408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weart RB, Lee AH, Chien A, Haeusser DP, Hill NS, Levin PA. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzur A, Kafri R, LeBleu VS, Lahav G, Kirschner MW. Cell Growth and Size Homeostasis in Proliferating Animal Cells. Science. 2009;325:167–171. doi: 10.1126/science.1174294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson KA, Gottschling DE. A mother's sacrifice: what is she keeping for herself? Curr. Op. Cell Biol. 2008;20:723–728. doi: 10.1016/j.ceb.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ackermann M, Stearns SC, Jenal U. Senescence in a bacterium with asymmetric division. Science. 2003;300:1920. doi: 10.1126/science.1083532. [DOI] [PubMed] [Google Scholar]
- 12.Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3(2):e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkler J, Seybert A, Konig L, Pruggnaller S, Haselmann U, et al. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 2010;29:910–923. doi: 10.1038/emboj.2009.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 15.Balagadde FK, You L, Hansen CL, Arnold FH, Quake SR. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science. 2005;309:137–140. doi: 10.1126/science.1109173. [DOI] [PubMed] [Google Scholar]
- 16.Lu T, Shen T, Bennett MR, Wolynes PG, Hasty J. Phenotypic variability of growing cellular populations. Proc. Nat. Acad. Sci. USA. 2007;104:18982–18987. doi: 10.1073/pnas.0706115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weibel DB, DiLuzio WR, Whitesides GM. Microfabrication meets microbiology. Nat. Rev. Microbiol. 2007;5:209–218. doi: 10.1038/nrmicro1616. [DOI] [PubMed] [Google Scholar]
- 18.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 19.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 20.Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, Rosenfeld N, Danon T, Perzov N, Alon U. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- 21.Linder AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Nat. Acad. Sci. USA. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang JD, Levin PA. Metabolism, cell growth and the bacterial cell cycle. Nat. Rev. Microbiol. 2009;7:822–827. doi: 10.1038/nrmicro2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.