Abstract
We describe a computational approach to sequential resonance assignment in solid state NMR studies of uniformly 15N,13C-labeled proteins with magic-angle spinning. As input, the algorithm uses only the protein sequence and lists of 15N/13Cα crosspeaks from 2D NCACX and NCOCX spectra that include possible residue-type assignments of each crosspeak. Assignment of crosspeaks to specific residues is carried out by a Monte Carlo/simulated annealing algorithm, implemented in the program MC_ASSIGN1. The algorithm tolerates substantial ambiguity in residue-type assignments and coexistence of visible and invisible segments in the protein sequence. We use MC_ASSIGN1 and our own 2D spectra to replicate and extend the sequential assignments for uniformly labeled HET-s(218-289) fibrils previously determined manually by Siemer et al. (J. Biomolec. NMR, vol. 34, pp. 75-87, 2006) from a more extensive set of 2D and 3D spectra. Accurate assignments by MC_ASSIGN1 do not require data that are of exceptionally high quality. Use of MC_ASSIGN1 (and its extensions to other types of 2D and 3D data) is likely to alleviate many of the difficulties and uncertainties associated with manual resonance assignments in solid state NMR studies of uniformly labeled proteins, where spectral resolution and signal-to-noise are often sub-optimal.
Keywords: automated assignment, chemical shifts, amyloid, prion, HET-s, stochastic recoupling
1. Introduction
In solid state nuclear magnetic resonance (NMR) studies of uniformly 15N,13C-labeled proteins with magic-angle spinning (MAS), assignment of the observed 15N and 13C resonances to specific residues is a prerequisite for the determination of molecular structure or characterization of molecular dynamics. Resonance assignment typically proceeds in a sequential manner, by connecting 13C signals of residue k with 13C signals of residue k+1 through the 15N signals of backbone amide sites, often in two-dimensional (2D) or three-dimensional (3D) NCACX and NCOCX spectra [1-15]. Manual sequential assignment from 2D NCACX and NCOCX spectra is easy when most 15N chemical shifts are unique and well resolved and when most 13C chemical shifts can be assigned to unique residue types. When overlap and degeneracy of 15N resonances is severe and residue-type assignments are ambiguous, manual assignment becomes difficult because of the many possible candidates for k/k+1 residue pairs that must be explored and either proven or disproven. 3D spectroscopy helps, but still may not readily yield unique assignments. The situation becomes more complicated when only certain segments of the protein sequence contribute to the solid state NMR spectra [16-23], due to variations in rigidity and structural order, and when the identity of these segments is unknown. In the end, resonance assignment is a tedious and potentially error-prone process except when the solid state NMR data are of extremely high quality.
In this paper, we describe an alternative approach to resonance assignment in solid state MAS NMR of uniformly labeled proteins, in which residue-specific assignments are generated in an automated manner from lists of crosspeaks in 2D NCACX and NCOCX spectra by a Monte Carlo/simulated annealing (MC/SA) computational algorithm. Using our 2D spectra of uniformly-labeled HET-s(218-289) fibrils, for which resonance assignments from a manual analysis of 2D and 3D spectra have been reported previously by Siemer et al. [17], we show that the MC/SA algorithm leads to complete and correct assignments even when there is ambiguity in the residue-type assignments of many of the 2D crosspeaks. Unlike manual assignment procedures, the MC/SA algorithm provides a complete and objective picture of the information content of the solid state NMR data, allowing either full or partial assignments to be extracted from data that are not necessarily ideal.
2. Methods
2.1 Sample preparation
Uniformly 15N,13C-labeled HET-s(218-289) (sequence MKID AIVGRNSAKD IRTEERARVQ LGNVVTAAAL HGGIRISDQT TNSVETVVGK GESRVLIGNE YGGKGFWDN HHHHHH, representing residues 218-289 of the P. anserina HET-s protein with an additional N-terminal Met residue and a C-terminal hexa-His tag) was expressed and purified as previously described [17,21]. Fibrils were prepared by incubation at 0.4 mM protein concentration in Tris buffer at pH 8 and 4° C for 14 days. Fibril formation was confirmed by transmission electron microscopy (see Figure S1 of supplementary information). Fibrils were pelleted, lyophilized, and packed in a MAS rotor (1.8 mm outer diameter, 10.5 μl volume), then rehydrated in the rotor by addition of 5 μl of water. The sample contained approximately 5 mg of HET-s(218-289).
2.2 NMR measurements
Spectra were obtained with three-channel MAS probes constructed by the group of Dr. Ago Samoson (National Institute of Chemical Physics and Biophysics, Tallinn, Estonia). 2D NCACX and NCOCX spectra were acquired with a Varian Infinity spectrometer at 17.6 T (188.0 MHz 13C NMR frequency) and 17.0 kHz MAS, using 5.0 ms cross-polarization for 15N-13C polarization transfer to either Cα (NCACX) or CO (NCOCX) sites after the t1 period, followed by 2.82 ms finite-pulse radio-frequency-driven recoupling (fpRFDR) [24,25] for 13C-13C polarization transfer before the t2 period. 13C π pulses in the fpRFDR periods were 20.0 μs at 45 ppm carrier frequency (NCACX) or 10.0 μs at 105 ppm carrier frequency (NCOCX). Two-pulse phase-modulated (TPPM) proton decoupling [26] at 110 kHz was applied during t1 and t2. Continuous-wave decoupling at 110 kHz was applied during 15N-13C and 13C-13C polarization transfer periods. Maximum t1 and t2 periods were 9.10 ms and 7.68 ms, respectively. Total measurement times were 22 hr for each 2D spectrum, with 1.0 s recycle delays.
The 2D 13C-13C (CC) spectrum was acquired with a Varian InfinityPlus spectrometer at 14.1 T (150.7 MHz 13C NMR frequency) and 40.0 kHz MAS, using a novel zero-quantum stochastic dipolar recoupling (ZQ-SDR) pulse sequence for longitudinal 13C-13C polarization transfers between t1 and t2. The ZQ-SDR sequence consisted of four fpRFDR blocks with 7.5 μs 13C π pulses, 32 rotor periods in each block, separated by randomly-chosen delays that ranged from zero to three rotor periods in length. The delays were determined by a random number generator within the pulse program. This ZQ-SDR sequence is related conceptually to the previously-described double-quantum SDR technique [27,28], and similarly leads to 13C-13C polarization transfers that are unaffected by quantum mechanical interference between non-commuting pairwise dipole-dipole couplings (i.e., “dipolar truncation”). TPPM decoupling at 125 kHz was applied during the t1 and t2 periods. No decoupling was applied during the ZQ-SDR period. Maximum t1 and t2 periods were 7.97 ms and 15.36 ms, respectively. Total measurement time was 17 hr, with 1.5 s recycle delays.
2D NMR spectra were processed with NMRPipe software [29]. Crosspeaks were picked manually using Sparky software (available at http://www.cgl.ucsf.edu/home/sparky/). Residue-type assignments of Cα chemical shifts were determined manually from crosspeak patterns (principally in the CC spectrum, but also from sets of 13C resonances with a common 15N chemical shift in the NCACX and NCOCX spectra). 13C and 15N chemical shifts are referred to tetramethylsilane and liquid ammonia, respectively, consistent with the work of Siemer et al. [17]
2.3 MC/SA algorithm
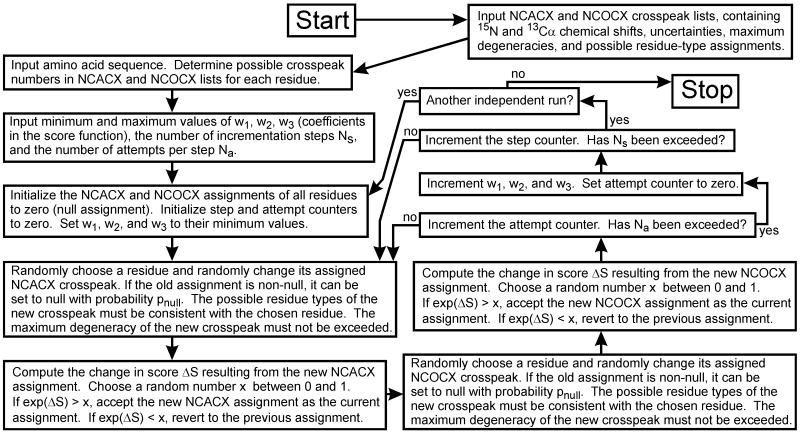
Residue-specific assignments were determined from lists of 15N/13Cα crosspeaks in the 2D NCACX and NCOCX spectra with the Fortran95 computer program MC_ASSIGN1 (see supplementary information), which implements the algorithm in Figure 1. The crosspeak lists include the 15N and 13C chemical shifts, the uncertainties in these shifts, the maximum degeneracies (in case more than one residue may contribute to a given crosspeak), and the possible residue-type assignments. Crosspeak lists are prepared by manual analyses of the 2D spectra. Residue-type assignments are typically determined from patterns of sidechain 13C chemical shifts in 2D CC spectra, supplemented by comparisons with the NCACX and NCOCX spectra. Each 15N/13Cα crosspeak can have multiple residue-type assignments. MC_ASSIGN1 attempts to assign one NCACX crosspeak and one NCOCX crosspeak to each residue in the protein sequence (or leave certain residues without assigned crosspeaks, called a “null assignment”, if the number of residues exceeds the number of crosspeaks) in such a way that the number of “good connections” (Ng) is maximized, the numbers of “bad connections” (Nb) and “edges” (Ne) are minimized, and the number of unused crosspeaks (Nu) is minimized. Ng is the number of residues with non-null NCACX and non-null NCOCX assignments for which the two 13Cα shifts agree to within the allowed uncertainty, plus the number of k/k+1 pairs for which the 15N shift in the NCOCX assignment of residue k and the NCACX assignment of residue k+1 agree to within the allowed uncertainty. Nb is the total number of these 13Cα shift pairs and 15N shift pairs that do not agree to within the allowed uncertainty. Ne is the number of residues that have a null NCACX assignment and a non-null NCOCX assignment or vice versa, plus the number of k/k+1 pairs for which residue k has a null NCOCX assignment and residue k+1 has a non-null NCACX assignment or vice versa. Nu is the number of crosspeaks in the NCACX and NCOCX lists that have been not been assigned to any residues. With these definitions, any assignment candidate has a score S, defined by
Figure 1.
Flow chart for the MC/SA algorithm implemented in the program MC_ASSIGN1. Input files contain the information in columns 1-7 of Tables 1 and 2. The scoring function S is defined in Equation (1).
| (1) |
where w1, w2, and w3 are weighting factors that are incremented gradually during execution of the algorithm. The factor of 1/4 in Equation (1) is somewhat arbitrary, motivated by the idea that Ne should be minimized (i.e., the lengths of fully assigned segments should be maximized if possible) but need not be zero, while Nb should be zero in the final, correct assignment. If the uncertainties in 15N or 13C chemical shifts in two spectra are εa and εb, a connection is considered good if the absolute value of the chemical shift difference is less than .
Starting with null assignments for all residues and with w1, w2, and w3 set to their minimum values, MC_ASSIGN1 chooses a residue at random and randomly changes its current NCACX assignment to another assignment (i.e., another NCACX crosspeak) that is allowed for the given residue type. If the current assignment is not null, the assignment can be changed to null with 40% probability or to another allowed assignment (if at least one exists) with 60% probability. If a given crosspeak has degeneracy nmax, then the same crosspeak can be assigned to as many as nmax residues. The change in score ΔS resulting from this random change in NCACX assignment of a single randomly-chosen residue this is then calculated. The quantity exp(ΔS) is then compared to a random number x from the interval (0,1). If exp(ΔS) ≥ x, the new assignment is accepted. If exp(ΔS) < x, the new assignment is rejected and the old assignment is restored.
In the same manner, MC_ASSIGN1 then attempts to change the NCOCX assignment of another randomly chosen residue. One attempted NCACX assignment change and one attempted NCOCX assignment change together constitute a single complete attempt. After Na attempts, the values of w1, w2, and w3 are incremented. Ns steps of incrementation are performed with Na attempts in each step (total of Ns×Na attempts in a single MC_ASSIGN1 run) to arrive at a final assignment. Multiple independent runs are performed to ensure that all assignments with high final values of S are identified. Typically, assignments with non-zero final values of Nb are discarded. If no assignments with non-zero Nb are found, one should re-examine the crosspeak lists to see whether the possible residue-type assignments or chemical shift uncertainties are unrealistically restrictive, or whether other errors in the manual analysis of the NMR spectra have been made.
The acceptance criterion described above is the standard Metropolis Monte Carlo criterion [30], leading to a probability that a given assignment is the current one proportional to exp(S) in the limit of many attempts. The gradual incrementation of w1, w2, and w3 represents a simulated annealing approach to optimization of the assignment.
The name MC_ASSIGN1 signifies the first version of this MC-based assignment algorithm. Subsequent versions may be developed to treat other data sets and higher-dimensional spectra.
3. Results
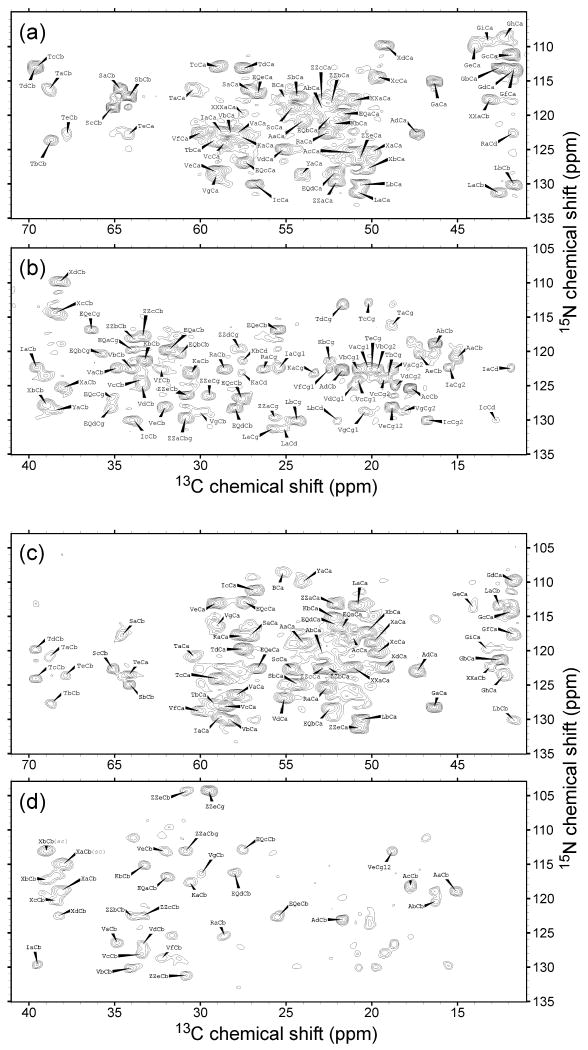
Figure 2 shows our 2D NCACX and NCOCX spectra of uniformly labeled HET-s(218-289) fibrils, with residue-type assignments determined by comparison of 13C chemical shifts of crosspeaks in these spectra with chemical shifts in the 2D CC spectrum in Figure 3. Importantly, residue-type assignments in Figures 2 and 3 were made without any reference to earlier work on HET-s(218-289) fibrils by Meier and coworkers [16-20]. For certain crosspeaks, the residue-type assignments were unambiguous, due to the characteristic Cα and sidechain chemical shift ranges of T, S, V, I, Y, A, and G residues. Some assignments to L, R, and K residues could also be made unambiguously, when correlations to sidechain signals were clear. In other cases, the residue-type assignments were partially ambiguous. For example, assignments to either E or Q could be made when both N-Cβ and N-Cγ crosspeaks were observed in the 2D NCACX and NCOCX spectra, but E and Q could not be distinguished from one another. Similarly, D and N crosspeaks were indistinguishable except when a crosspeak between the sidechain N and the Cβ were observed along with the backbone N crosspeaks in the 2D NCOCX spectra (peaks labeled Xa and Xb in Figures 2c and 2d). N/Cα crosspeak positions and residue-type assignments in the 2D NCACX and NCOCX spectra are summarized in columns 1-7 of Tables 1 and 2. The uncertainties in the 15N and 13C chemical shifts listed in Tables 1 and 2 are less than the full-width-at-half-maximum linewidths because of the high signal-to-noise ratio in the spectra. Degeneracies in Tables 1 and 2 are all equal to 1 because crosspeaks from different residues were generally fully or partially resolved. Overlapping signals in the N-Cα regions of the 2D NCACX and NCOCX spectra could be disentangled by comparisons with N-Cβ and other sidechain signals and with 2D CC crosspeaks.
Figure 2.
2D NCACX (a,b) and NCOCX (c,d) spectra of uniformly-labeled HET-s(218-289) fibrils with residue-type assignments. For example, the label TaCb indicates the crosspeak for Thr residue “a” at the 13C shift of its β-carbon. 15N shifts are those of the same residue in the NCACX spectrum and of the subsequent residue in the NCOCX spectrum. Ambiguous residue-type labels are X (D or N), Z (E or Q), ZZ (E, Q, K, or R), XX (F, L, D, or N), XXX (I, F, L, D, or N), and B (anything other than A, G, or T). Contour levels increase by successive factors of 1.5, with the lowest contour being at approximately five (a,b) or three (c,d) times the root-mean-squared noise level.
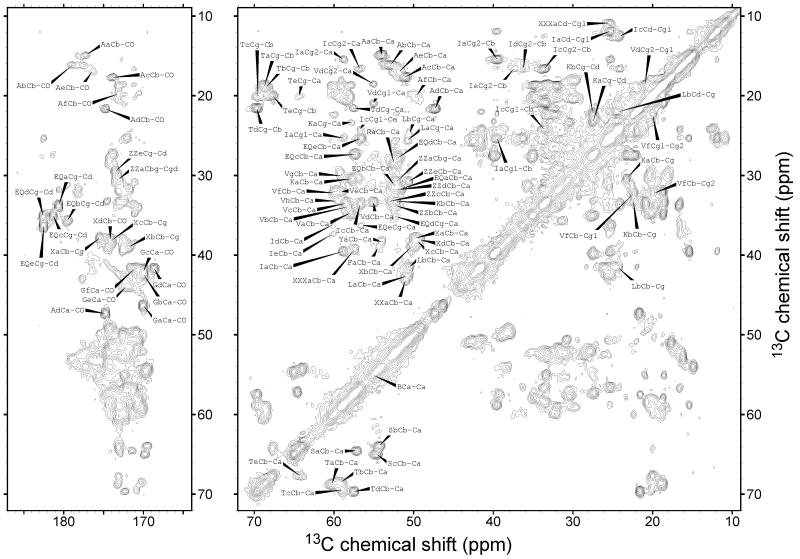
Figure 3.
2D CC spectrum of uniformly-labeled HET-s(218-289) fibrils with residue-type assignments. Contour levels increase by successive factors of 1.5, with the lowest contour being at approximately three times the root-mean-squared noise level.
Table 1.
Crosspeaks in 2D NCACX spectrum of HET-s(218) fibrils. Uncertainties in 15N and 13C shifts are ±εN and ±εC. Maximum degeneracy is nmax. Tentative assignments (present in the highest-scoring MC_ASSIGN1 assignment, but not in all assignments with Nb = 0) are indicated by **.
| NCACX crosspeak number | 15N shift (ppm) | 13C shift (ppm) | εN (ppm) | εC (ppm) | nmax | possible residue types | final assignment (1-72 numbering) | final assignment (standard numbering) |
|---|---|---|---|---|---|---|---|---|
| 1 | 120.7 | 54.0 | 0.3 | 0.15 | 1 | A | 30 | A247 |
| 2 | 119.1 | 52.9 | 0.3 | 0.15 | 1 | A | 31 | A248 |
| 3 | 125.5 | 51.1 | 0.3 | 0.15 | 1 | A | 20 | A237 |
| 4 | 122.7 | 47.3 | 0.3 | 0.15 | 1 | A | 11 | A228 |
| 5 | 125.3 | 50.0 | 0.3 | 0.15 | 1 | DN | 9 | N226 |
| 6 | 127.8 | 50.3 | 0.3 | 0.15 | 1 | DN | 45 | N262 |
| 7 | 114.4 | 49.7 | 0.3 | 0.15 | 1 | DN | 62 | N279 |
| 8 | 109.9 | 49.2 | 0.3 | 0.15 | 1 | DN | 26 | N243 |
| 9 | 119.8 | 51.7 | 0.3 | 0.15 | 1 | EQ | 17 | E234 |
| 10 | 120.1 | 52.4 | 0.3 | 0.15 | 1 | EQ | 63 | E280 |
| 11 | 126.9 | 57.4 | 0.3 | 0.15 | 1 | EQ | 48 | E265 |
| 12 | 128.3 | 52.1 | 0.3 | 0.15 | 1 | EQ | 55 | E272 |
| 13 | 116.8 | 56.7 | 0.3 | 0.15 | 1 | EQ | 18 | E235 |
| 14 | 126.5 | 50.7 | 0.3 | 0.15 | 1 | EQKR | 23 | Q240 |
| 15 | 115.2 | 46.4 | 0.3 | 0.15 | 1 | G | 54 | G271 |
| 16 | 113.1 | 42.6 | 0.3 | 0.15 | 1 | G | 52 | G269 |
| 17 | 111.3 | 41.9 | 0.3 | 0.15 | 1 | G | 61 | G278 |
| 18 | 113.4 | 41.7 | 0.3 | 0.15 | 1 | G | 66 | G283 |
| 19 | 110.0 | 44.0 | 0.3 | 0.15 | 1 | G | 65 | G282 |
| 20 | 113.1 | 41.7 | 0.3 | 0.15 | 1 | G | 25 | G242 |
| 21 | 108.7 | 42.3 | 0.3 | 0.15 | 1 | G | 68** | G285** |
| 22 | 109.2 | 43.2 | 0.3 | 0.15 | 1 | G | 36** | G253** |
| 23 | 122.4 | 58.8 | 0.3 | 0.15 | 1 | I | 14 | I231 |
| 24 | 130.0 | 56.8 | 0.3 | 0.15 | 1 | I | 60 | I277 |
| 25 | 119.8 | 57.5 | 0.3 | 0.15 | 1 | IFLDN | 37** | I254** |
| 26 | 123.1 | 57.6 | 0.3 | 0.15 | 1 | K | 12 | K229 |
| 27 | 121.3 | 52.2 | 0.3 | 0.15 | 1 | K | 53 | K270 |
| 28 | 131.2 | 50.9 | 0.3 | 0.15 | 1 | L | 24 | L241 |
| 29 | 130.1 | 50.6 | 0.3 | 0.15 | 1 | L | 59 | L276 |
| 30 | 117.8 | 51.1 | 0.3 | 0.15 | 1 | LDNF | 13 | D230 |
| 31 | 122.6 | 52.6 | 0.3 | 0.15 | 1 | R | 19 | R236 |
| 32 | 116.3 | 57.1 | 0.3 | 0.15 | 1 | S | 56 | S273 |
| 33 | 117.4 | 54.4 | 0.3 | 0.15 | 1 | S | 46 | S263 |
| 34 | 118.8 | 54.6 | 0.3 | 0.15 | 1 | S | 10 | S227 |
| 35 | 117.5 | 55.1 | 0.3 | 0.15 | 1 | not AGT | 67** | K284** |
| 36 | 116.2 | 60.4 | 0.3 | 0.15 | 1 | T | 29 | T246 |
| 37 | 123.6 | 59.2 | 0.3 | 0.15 | 1 | T | 44 | T261 |
| 38 | 112.9 | 59.0 | 0.3 | 0.15 | 1 | T | 49 | T266 |
| 39 | 113.2 | 57.5 | 0.3 | 0.15 | 1 | T | 16 | T233 |
| 40 | 122.8 | 64.2 | 0.3 | 0.15 | 1 | T | 43 | T260 |
| 41 | 122.6 | 57.8 | 0.3 | 0.15 | 1 | V | 22 | V239 |
| 42 | 122.6 | 58.2 | 0.3 | 0.15 | 1 | V | 58 | V275 |
| 43 | 124.0 | 58.6 | 0.3 | 0.15 | 1 | V | 50 | V267 |
| 44 | 125.0 | 55.1 | 0.3 | 0.15 | 1 | V | 47 | V264 |
| 45 | 128.3 | 59.1 | 0.3 | 0.15 | 1 | V | 51 | V268 |
| 46 | 122.6 | 60.0 | 0.3 | 0.15 | 1 | V | 27 | V244 |
| 47 | 128.8 | 59.1 | 0.3 | 0.15 | 1 | V | 28 | V245 |
| 48 | 129.7 | 52.1 | 0.3 | 0.15 | 1 | EQKR | 15 | R232 |
| 49 | 118.3 | 52.4 | 0.3 | 0.15 | 1 | EQKR | 21 | R238 |
| 50 | 117.9 | 53.0 | 0.3 | 0.15 | 1 | EQKR | 57 | R274 |
| 51 | 128.6 | 54.1 | 0.3 | 0.15 | 1 | Y | 64 | Y281 |
Table 2.
Crosspeaks in 2D NCOCX spectrum of HET-s(218) fibrils.
| NCOCX crosspeak number | 15N shift (ppm) | 13C shift (ppm) | εN (ppm) | εC (ppm) | nmax | possible residue types | final assignment (1-72 numbering) | final assignment (standard numbering) |
|---|---|---|---|---|---|---|---|---|
| 1 | 119.0 | 54.0 | 0.3 | 0.15 | 1 | A | 30 | A247 |
| 2 | 120.5 | 52.9 | 0.3 | 0.15 | 1 | A | 31 | A248 |
| 3 | 118.1 | 51.1 | 0.3 | 0.15 | 1 | A | 20 | A237 |
| 4 | 123.1 | 47.4 | 0.3 | 0.15 | 1 | A | 11 | A228 |
| 5 | 118.8 | 49.9 | 0.3 | 0.15 | 1 | N | 9 | N226 |
| 6 | 117.3 | 50.3 | 0.3 | 0.15 | 1 | N | 45 | N262 |
| 7 | 120.4 | 49.7 | 0.3 | 0.15 | 1 | DN | 62 | N279 |
| 8 | 122.5 | 49.3 | 0.3 | 0.15 | 1 | DN | 26 | N243 |
| 9 | 116.7 | 51.7 | 0.3 | 0.15 | 1 | EQ | 17 | E234 |
| 10 | 128.4 | 52.4 | 0.3 | 0.15 | 1 | EQ | 63 | E280 |
| 11 | 112.9 | 57.4 | 0.3 | 0.15 | 1 | EQ | 48 | E265 |
| 12 | 122.6 | 56.7 | 0.3 | 0.15 | 1 | EQ | 18 | E235 |
| 13 | 116.2 | 52.1 | 0.3 | 0.15 | 1 | EQ | 55 | E272 |
| 14 | 131.3 | 50.8 | 0.3 | 0.15 | 1 | EQKR | 23 | Q240 |
| 15 | 128.3 | 46.4 | 0.3 | 0.15 | 1 | G | 54 | G271 |
| 16 | 121.3 | 42.5 | 0.3 | 0.15 | 1 | G | 52 | G269 |
| 17 | 114.3 | 41.9 | 0.3 | 0.15 | 1 | G | 61 | G278 |
| 18 | 109.9 | 41.7 | 0.3 | 0.15 | 1 | G | 25 | G242 |
| 19 | 113.7 | 44.1 | 0.3 | 0.15 | 1 | G | 65 | G282 |
| 20 | 117.6 | 41.7 | 0.3 | 0.15 | 1 | G | 66 | G283 |
| 21 | 123.5 | 42.3 | 0.3 | 0.15 | 1 | G | 68** | G285** |
| 22 | 119.8 | 43.3 | 0.3 | 0.15 | 1 | G | 36** | G253** |
| 23 | 129.6 | 58.9 | 0.3 | 0.15 | 1 | I | 14 | I231 |
| 24 | 111.2 | 56.8 | 0.3 | 0.15 | 1 | I | 60 | I277 |
| 25 | 117.9 | 57.5 | 0.3 | 0.15 | 1 | K | 12 | K229 |
| 26 | 115.2 | 52.2 | 0.3 | 0.15 | 1 | K | 53 | K270 |
| 27 | 113.4 | 50.8 | 0.3 | 0.15 | 1 | L | 24 | L241 |
| 28 | 130.0 | 50.6 | 0.3 | 0.15 | 1 | L | 59 | L276 |
| 29 | 122.4 | 51.2 | 0.3 | 0.15 | 1 | LDNF | 13 | D230 |
| 30 | 125.4 | 52.6 | 0.3 | 0.15 | 1 | R | 19 | R236 |
| 31 | 117.8 | 57.1 | 0.3 | 0.15 | 1 | S | 56 | S273 |
| 32 | 124.9 | 54.3 | 0.3 | 0.15 | 1 | S | 46 | S263 |
| 33 | 122.7 | 54.7 | 0.3 | 0.15 | 1 | S | 10 | S227 |
| 34 | 108.6 | 55.2 | 0.3 | 0.15 | 1 | not AGT | 67** | K284** |
| 35 | 120.9 | 60.4 | 0.3 | 0.15 | 1 | T | 29 | T246 |
| 36 | 127.8 | 59.2 | 0.3 | 0.15 | 1 | T | 44 | T261 |
| 37 | 124.1 | 58.9 | 0.3 | 0.15 | 1 | T | 49 | T266 |
| 38 | 119.8 | 57.5 | 0.3 | 0.15 | 1 | T | 16 | T233 |
| 39 | 123.6 | 64.2 | 0.3 | 0.15 | 1 | T | 43 | T260 |
| 40 | 126.4 | 57.9 | 0.3 | 0.15 | 1 | V | 22 | V239 |
| 41 | 130.1 | 58.3 | 0.3 | 0.15 | 1 | V | 58 | V275 |
| 42 | 128.1 | 58.7 | 0.3 | 0.15 | 1 | V | 50 | V267 |
| 43 | 126.9 | 55.1 | 0.3 | 0.15 | 1 | V | 47 | V264 |
| 44 | 113.1 | 59.1 | 0.3 | 0.15 | 1 | V | 51 | V268 |
| 45 | 128.8 | 60.0 | 0.3 | 0.15 | 1 | V | 27 | V244 |
| 46 | 116.3 | 59.0 | 0.3 | 0.15 | 1 | V | 28 | V245 |
| 47 | 113.2 | 52.2 | 0.3 | 0.15 | 1 | EQKR | 15 | R232 |
| 48 | 122.5 | 52.5 | 0.3 | 0.15 | 1 | EQKR | 21 | R238 |
| 49 | 122.6 | 52.9 | 0.3 | 0.15 | 1 | EQKR | 57 | R274 |
| 50 | 110.0 | 54.0 | 0.3 | 0.15 | 1 | Y | 64 | Y281 |
Information in columns 1-7 of Tables 1 and 2 was used as input to MC_ASSIGN1. No other information was used, aside from the amino acid sequence of HET-s(218-289). Twenty independent MC_ASSIGN1 runs were performed, starting with the null assignment and using different random number sequences in each run. The parameters w1, w2, and w3 were incremented simultaneously and linearly from 0 to 10, 0 to 10, and 0 to 5, respectively, in 30 steps with 106 assignment change attempts in each step. As these weighting parameters increased, the acceptance rate (i.e., the fraction of attempts that were accepted) decreased from 1.0 to 0.0, with acceptance rates generally being below 0.001 in the second half of each run. Total execution time for each run (3 × 107 attempts) was 20 s on an Acer TravelMate 6292 computer with 2.20 GHz processor speed.
Results of the twenty runs are summarized in Table S1 of supplementary information. Nineteen runs produced identical assignments with final values Ng = 98, Nb = 0, Nu = 0, and Ne = 6 (final S = 965). One run produced final values Ng = 97, Nb = 0, Nu = 0, and Ne = 8 (final S = 950) All runs produced identical, non-null assignments for residues 226-248 and 260-283 (residues 9-31, and 43-66 if the sequence is numbered 1-72) and null assignments for residues 218-225, 249-250, 255-259, and 287-289. The only variable assignments were at residues 251-254 and 285-287. These variations involve assignments of crosspeaks 21, 22, 25, and 35 in the 2D NCACX spectrum and crosspeaks 21, 22, and 34 in the 2D NCOCX spectrum, all of which are relatively weak signals that may arise from partially mobile residues. We tentatively assign these signals to G253, I254, K284, and G285, in accordance with the higher-scoring and more frequent MC_ASSIGN1 result.
Table 3 shows the final 15N and 13C chemical shifts for HET-s(218-289) fibrils derived solely from the 2D spectra in Figures 2 and 3, the information in columns 1-7 of Tables 1 and 2, and the MC_ASSIGN1 program. Figure 4 shows the 2D NCACX and NCOCX spectra, now labeled with site-specific assignments. It is important to note that MC_ASSIGN1 does not directly use any 13C chemical shifts other than those of Cα sites, except to the extent that these shifts lead to residue-type assignments. Also, although different crosspeaks with the same residue-type assignments are labeled differently in Figures 2 and 3 (e.g., EQa, EQb, EQc, etc.), MC_ASSIGN1 does not use these labels.
Table 3.
15N and 13C chemical shifts in HET-s(218-289) fibrils, assigned to specific residues by the program MC_ASSIGN1. Shifts are in ppm relative to liquid ammonia (15N) and tetramethylsilane (13C).
| residue | backbone N | CO | Cα | Cβ | Cγ | Cδ | sidechain N |
|---|---|---|---|---|---|---|---|
| N226 | 125.3 | 49.9 | 38.0 | 175.5 | 115.0 | ||
| S227 | 118.8 | 169.8 | 54.6 | 65.1 | |||
| A228 | 122.7 | 174.8 | 47.3 | 21.7 | |||
| K229 | 123.1 | 57.7 | 30.5 | 23.3 | 27.5 | ||
| D230 | 117.8 | 51.1 | 431. | ||||
| I231 | 122.4 | 58.8 | 39.5 | 25.3, 15.4 | 11.9 | ||
| R232 | 129.7 | 52.1 | 30.8 | 25.7 | |||
| T233 | 113.2 | 172.4 | 57.5 | 69.7 | 21.6 | ||
| E234 | 119.8 | 51.7 | 31.8 | 33.9 | 180.6 | ||
| E235 | 116.9 | 56.6 | 25.4 | 36.3 | 182.5 | ||
| R236 | 122.6 | 52.6 | 28.5 | 26.3 | 41.8 | ||
| A237 | 125.4 | 173.9 | 51.1 | 17.7 | |||
| R238 | 118.3 | 52.4 | 33.9 | ||||
| V239 | 122.5 | 57.9 | 34.8 | 20.2, 18.7 | |||
| Q240 | 126.5 | 171.2 | 50.7 | 30.8 | 29.5 | 173.5 | 104.3 |
| L241 | 131.3 | 50.8 | 42.6 | 25.7 | 25.1 | ||
| G242 | 113.1 | 168.7 | 41.7 | ||||
| N243 | 109.9 | 49.2 | 38.2 | 174.2 | |||
| V244 | 122.6 | 60.0 | 32.2 | 22.4, 19.9 | |||
| V245 | 128.8 | 59.0 | 30.0 | 20.4, 18.2 | |||
| T246 | 116.2 | 60.3 | 68.8 | 18.8 | |||
| A247 | 120.8 | 177.2 | 54.0 | 15.0 | |||
| A248 | 119.0 | 178.9 | 52.9 | 16.2 | |||
| G253** | 109.2 | 172.5 | 43.2 | ||||
| I254** | 119.8 | 57.4 | 39.3 | 25.4 | 10.8 | ||
| T260 | 122.7 | 173.1 | 64.2 | 67.7 | 20.0 | ||
| T261 | 123.6 | 170.3 | 59.2 | 68.6 | 19.0 | ||
| N262 | 127.8 | 50.2 | 38.9 | 171.9 | 113.1 | ||
| S263 | 117.3 | 169.5 | 54.3 | 64.1 | |||
| V264 | 125.0 | 55.1 | 33.4 | 21.0, 18.6 | |||
| E265 | 126.8 | 57.4 | 27.5 | 35.1 | 181.3 | ||
| T266 | 112.9 | 170.8 | 58.9 | 69.5 | 20.1 | ||
| V267 | 124.1 | 58.6 | 33.5 | 20.8, 19.3 | |||
| V268 | 128.2 | 59.1 | 32.1 | 18.8 | |||
| G269 | 113.1 | 170.0 | 42.5 | ||||
| K270 | 121.3 | 52.2 | 33.2 | 22.4 | 27.5 | ||
| G271 | 115.2 | 170.0 | 46.3 | ||||
| E272 | 128.3 | 52.1 | 27.9 | 35.2 | 182.6 | ||
| S273 | 116.2 | 171.3 | 57.1 | 64.6 | |||
| R274 | 117.9 | 53.0 | 33.3 | ||||
| V275 | 122.6 | 58.2 | 34.0 | 20.8, 19.7 | |||
| L276 | 130.1 | 50.6 | 41.6 | 24.5 | 22.0 | ||
| I277 | 130.0 | 56.8 | 33.7 | 24.2, 16.7 | 12.6 | ||
| G278 | 111.2 | 170.0 | 41.9 | ||||
| N279 | 114.4 | 49.6 | 38.4 | 174.2 | |||
| E280 | 120.3 | 52.4 | 31.2 | 35.7 | 179.5 | ||
| Y281 | 128.6 | 54.0 | 38.4 | 125.5 | |||
| G282 | 110.0 | 172.2 | 44.0 | ||||
| G283 | 113.4 | 171.1 | 41.7 | ||||
| K284** | 117.5 | 55.1 | |||||
| G285** | 108.7 | 170.0 | 42.2 | ||||
| F286 | 54.5 | 39.1 | 134.3 |
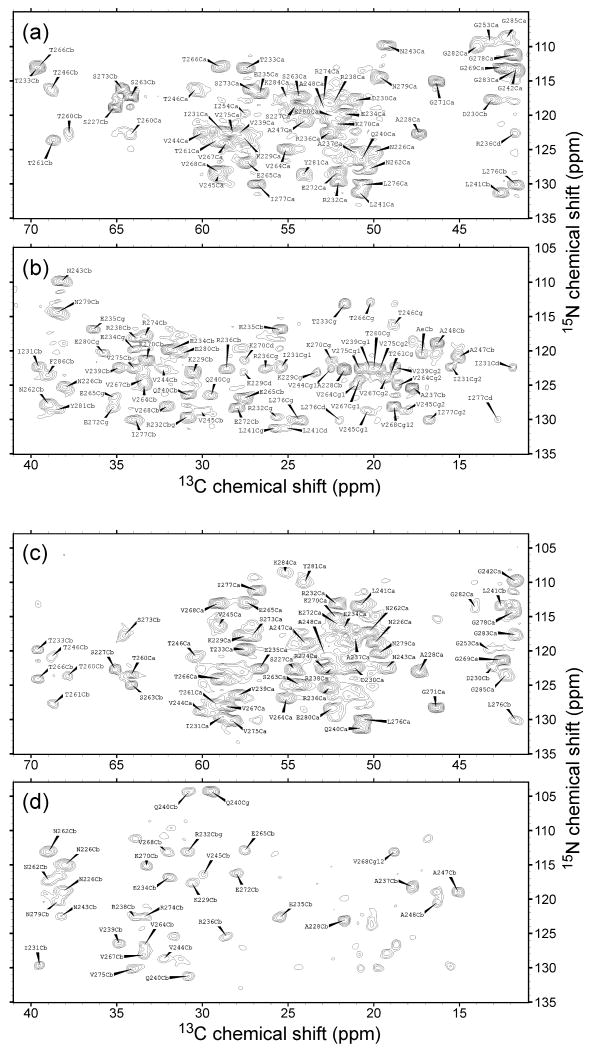
Figure 4.
2D NCACX (a,b) and NCOCX (c,d) spectra of uniformly-labeled HET-s(218-289) fibrils with site-specific assignments.
4. Discussion
4.1 Comparison with previous HET-s(218-289) assignments
After making assignments as described above, we compared our results with the assignments reported by Siemer et al., which were derived from a larger set of 2D and 3D spectra by manual methods [17]. Agreement is generally good, apart from chemical shift differences that are within the linewidths. Where differences greater than 0.3 ppm exist, we have verified that the assigned spectral features are the same in our work and in that of Siemer et al., so that these differences may be due to variations in signal-to-noise and chemical shift references. The signal-to-noise ratios in our spectra appear to be higher than those in the spectra of Siemer et al., probably because our HET-s(218-289) fibril sample was lyophilized, packed in the MAS rotor as a dry powder, and then rehydrated. Compared with the alternative method of centrifuging a hydrated fibril pellet into the rotor, our method results in a higher protein density in the MAS rotor.
Several differences are noted: (a) Siemer et al. report assignments for R238 but not R274. R238 and R274 have similar 15N and 13C shifts, but we clearly see separate signals in our 2D NCACX and NCOCX spectra (see Figures 4b and 4d); (b) Assignments reported by Siemer et al. span residues 226-248 and 262-282, which they interpret to be the segments that form the immobilized fibril core. Our definite assignments span residues 226-248 and 260-284. Signals that we assign to T260, T261, G283, K284 are present, but apparently weak, in the published spectra of Siemer et al. (see Figures 3 and 4 of ref. [17]). Signals we assign tentatively to G285 are clear in our spectra, but not in the spectra of Siemer et al. Signals from T260, G283, K284, and G285 signals are weaker than most other signals in our spectra, consistent with larger amplitudes or longer correlation times for local motions at these residues; (c) Signals we assign tentatively to G253 and I254 are weak, but clearly present, in our spectra. These signals are apparently absent from the spectra of Siemer et al., leading to their conclusion that residues 249-261 comprise a flexible loop in HET-s(218-289) fibrils. Our data suggest that this segment is not entirely a flexible loop.
All strong signals in our spectra are assigned definitively. Several weak signals in the 2D CC spectrum are not observed or assigned in the 2D NCACX and NCOCX spectra. These include two Ala signals (labeled Ae and Af in Figure 3), two Ile signals (labeled Id and Ie in Figure 3), one Val signal (labeled Vh in Figure 3), and one Phe signal (labeled Fa in Figure 3). By default, Fa must be F286. Ae and Af must be A249 and A221, with the more intense Ae signal most likely being A249, which is at the end of the first assigned segment. Id and Ie could be I219, I222, or I256. Vh could only be V223.
Siemer et al. report that signals attributed to Ala, Arg, Asn/Asp, Gln, Glu, His, Ile, Lys, and Val residues can be observed in “solution NMR” spectra of HET-s(218-289) fibrils, i.e., spectra that are recorded under conditions that select for highly mobile sites [18]. Observation of Ala, Glu, and Val signals under these conditions is somewhat surprising, given that all residues of these types contribute to our solid state NMR spectra, which were recorded under conditions that select against highly mobile sites. It is possible that differences in sample preparation (lyophilization followed by rehydration vs. uninterrupted hydration) or sample temperatures during NMR measurements produce real differences in mobility in the loop segments of HET-s(218-289) fibrils. It is also possible that the loop segments have variable mobility within all samples, due to inherent variations in lateral association or “bundling” of the fibrils (see Figure S1 of supplementary information). We see no evidence that lyophilization perturbs the structure of the immobilized segments, as our solid state 15N and 13C NMR chemical shifts are not significantly different from those reported by Siemer et al.
4.2 Effects of lower data quality
As originally discovered by Meier and coworkers [16,17,19,20], solid state NMR spectra of HET-s(218-289) are exceptionally well resolved when compared with spectra of other protein and peptide fibrils and other noncrystalline samples [21,31-33]. In addition to the sharp lines, relatively long T2 and T1ρ relaxation times contribute to the high quality of these spectra, especially the high signal-to-noise ratio for multiple-bond crosspeaks, reflecting favorable time scales and amplitudes of local molecular motions at temperatures near 20-30° C. In solid state NMR studies of other uniformly labeled proteins in noncrystalline states, the quality of the data is typically lower, resulting in greater ambiguity of residue-type assignments. To test the effect of greater ambiguity, we repeated the MC/SA analysis of our 2D NCACX and NCOCX spectra, increasing the residue-type ambiguities in column 7 of Tables 1 and 2 so that all E, Q, K, R, H, or W assignments became EQKRHW and all L, D, N, F, or Y assignments became LDNFY. This grouping of residue types places all residues with similar random-coil Cα and Cβ shifts together. Assignments to A, G, T, S, V, and I residues (with the one exception in Table 1) remained unambiguous, as these residue types are generally distinguishable due to their unique chemical shift patterns. Twenty runs of MC_ASSIGN1, with the same parameters described above, resulted in 18 assignments with Ng = 98, Nb = 0, Nu = 0, and Ne = 6 (final S = 965). Two other assignments had lower final scores. The 18 high-scoring assignments were identical to the final assignments in Tables 1 and 2 (assignment #1 in Table S1). Thus, increasing the ambiguity of residue-type assignments to a level that should be readily achievable in many solid state NMR studies does not prevent the determination of unique resonance assignments.
Next, we increased all 15N and 13C chemical shift uncertainties by a factor of two (to ±0.6 ppm and ±0.3 ppm, respectively) while keeping the greater residue-type ambiguities discussed above, to mimic the larger 15N and 13C linewidths that commonly result from minor static structural disorder in noncrystalline samples. Twenty MC_ASSIGN1 runs, with the same parameters described above, resulted in a larger diversity of assignments, with final scores ranging from 875 to 965. The highest score was obtained in 8 of the 20 runs. These runs were unanimous in their non-null assignments for residues 228-233, 235-237, 239-241, 243, 244, 246-248, 260, 261, 264-271, 273, 275-282, 284, and 285, all of which agreed with the final assignments in Tables 1 and 2. Moreover, all highest-scoring runs produced only null assignments for residues 218-225, 249-251, 255-259, and 287-289, and only non-null assignments for residues 226-248 and 260-285. Thus, even in the presence of highly ambiguous residue-type assignments and inhomogeneously broadened MAS NMR lines, the MC_ASSIGN1 algorithm provides partial assignments and reliable information concerning the segments within the protein sequence that contribute to solid state NMR signals. This information can be used to guide additional measurements that distinguish among the possible alternative assignments.
4.3 Comparison with previous automated assignment approaches
Automation of the resonance assignment process has been an active area of research in protein NMR for many years [34-44]. Monte Carlo-based approaches have been described in several previous publications [38,39,42,43]. An approach quite similar to ours is used in the MONTE program of Hitchens et al. [42], which was developed to analyze solution NMR data. MONTE begins with a random assignment of chemical shift correlations from multidimensional spectra (arising from polarization transfers driven by either scalar couplings or nuclear Overhauser effects) and other information (such as residue-type assignments) to backbone nitrogen-proton sites. MONTE then swaps the assignments among sites, evaluates changes in a scoring function, accepts or rejects the swap according to a Metropolis criterion, and performs simulated annealing to arrive at optimal assignments. MONTE includes a “cache” of extra sites, outside the real protein sequence, that serves a purpose similar to that of the null assignments described above. In contrast to other previous automated assignment programs, our MC_ASSIGN1 program is designed specifically for solid state NMR studies of proteins and peptides. In particular, the scoring function in Equation (1) and the choice of inputs to MC_ASSIGN1 are motivated by common solid state NMR measurements and scenarios. Our inclusion of edges in the scoring function is intended to treat situations in which the number of NMR signals is less than the number of amino acids, due to coexistence of mobile and immobile segments within the protein sequence. Our treatment of residue-type ambiguity is also motivated specifically by a common situation in solid state NMR studies.
Automation of site-specific resonance assignments is distinct from automation of the assignment of crosspeaks that contain information about internuclear distances or inter-residue contacts [45-47].
4.4 Concluding remarks
Resonance assignment is a major hurdle in solid state NMR studies of uniformly labeled proteins. Unless the NMR lines are very sharp and the signal-to-noise ratio is very high, the manual assignment process is tedious, subjective, and potentially error-prone. When a set of assignments that are consistent with the available spectra is finally obtained, one can not be sure that all alternative assignments would be inconsistent with the same spectra. The MC/SA algorithm demonstrated above has the potential to alleviate these problems. This algorithm (and its extensions to other types of 2D and 3D solid state NMR spectra) minimizes the subjectivity of the assignment process and allows all assignments that are consistent with the available spectra to be identified. In essence, this algorithm displays the full information content of the solid state NMR data, including not only the resonance assignments in segments where unique assignments can be determined, but also the identity of segments that are definitely contributing to the solid state NMR signals even if unique assignments can not be determined and the identity of segments that are definitely not contributing to the solid state NMR signals. By repeating the MC/SA algorithm many times, one can check that the information content of the data is not being overestimated.
Subjectivity plays a role only in the residue-type assignments, the chemical shift uncertainties, and the degeneracies. As demonstrated above, residue-type assignments can be highly ambiguous without compromising the ability of the MC/SA algorithm to find unique assignments, provided that the data are otherwise of good quality. Thus, users of this algorithm are encouraged to err on the side of generosity when developing residue-type assignments. It may also be possible to automate the determination of residue types in solid state NMR spectra, but residue-type assignments are not the primary obstacle to sequential assignment as long as ambiguous residue-type assignments are acceptable.
Chemical shift uncertainties are readily estimated from linewidths and signal-to-noise ratios. Values of nmax greater than 1 should be used only when certain crosspeak volumes are obviously larger than expected for a single site. When any of these factors are unclear, it is a simple matter to modify the input files and repeat the runs.
The execution time of the MC_ASSIGN1 program is proportional to the number of runs and the number of attempts in each run, independent of the amino acid sequence length and the number of entries in the crosspeak tables. We have not performed a systematic study to determine how many attempts and how many runs are required to identify the correct assignment, or how these numbers depend on the sequence length or other factors. However, we find that correct assignments are found in roughly 90% of the runs when we artificially double the HET-s(218-298) sequence, double the number of entries in each crosspeak table, and quadruple the number of attempts in each run.
In general terms, the existence of a unique set of resonance assignments depends on several intertwined factors that are difficult to quantify manually. One factor is obviously the NMR linewidths. If all 15N lines are fully resolved in one dimension, then unique sequential assignments can be determined from 2D NCACX and NCOCX data even without residue-type assignments and without using sidechain 13C shifts. Another factor is the complexity and length of the protein sequence. If the sequence contains at most one copy of each amino acid, then unique sequential assignments can obviously be determined from residue-type assignments alone, without any resolution of 15N shifts. When residue-type assignments are not completely unambiguous or the sequence contains multiple copies of various amino acids (which is necessarily true for real proteins unless they are selectively labeled), then some 15N resolution is required, but the required 15N resolution depends on the order of amino acids in the sequence, the 13C resolution, and the details of the residue-type ambiguities. In our view, the complexity of these factors makes an objective, computational approach to resonance assignments especially valuable.
Several extensions to the MC_ASSIGN1 program are obvious and will be pursued in future work. These include direct use of sidechain 13C chemical shifts in the evaluation of good and bad connections and generalization of the MC/SA algorithm to larger sets of 2D and 3D NMR data. The generalized algorithm may prove useful in solution NMR studies as well.
Supplementary Material
Acknowledgments
We thank Reed B. Wickner for his assistance with the production of HET-s(218-289) fibrils. This work was supported by the Intramural Research Program of the National Insitute of Diabetes and Digestive and Kidney Diseases, part of the National Institutes of Health. Additional support was provided by the Intramural AIDS Targeted Antiviral Program of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McDermott A, Polenova T, Bockmann A, Zilm KW, Paulsen EK, Martin RW, Montelione GT. Partial NMR assignments for uniformly 13C,15N-enriched BPTI in the solid state. J Biomol NMR. 2000;16:209–219. doi: 10.1023/a:1008391625633. [DOI] [PubMed] [Google Scholar]
- 2.Pauli J, Baldus M, van Rossum B, de Groot H, Oschkinat H. Backbone and side-chain 13C and 15N signal assignments of the alpha-spectrin SH3 domain by magic angle spinning solid state NMR at 17.6 tesla. ChemBioChem. 2001;2:272–281. doi: 10.1002/1439-7633(20010401)2:4<272::AID-CBIC272>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Petkova AT, Baldus M, Belenky M, Hong M, Griffin RG, Herzfeld J. Backbone and side chain assignment strategies for multiply labeled membrane peptides and proteins in the solid state. J Magn Reson. 2003;160:1–12. doi: 10.1016/s1090-7807(02)00137-4. [DOI] [PubMed] [Google Scholar]
- 4.Igumenova TI, Wand AJ, McDermott AE. Assignment of the backbone resonances for microcrystalline ubiquitin. J Am Chem Soc. 2004;126:5323–5331. doi: 10.1021/ja030546w. [DOI] [PubMed] [Google Scholar]
- 5.Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM. Magic-angle spinning solid state NMR spectroscopy of the beta 1 immunoglobulin binding domain of protein G (GB1): 15N and 13C chemical shift assignments and conformational analysis. J Am Chem Soc. 2005;127:12291–12305. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- 6.Heise H, Seidel K, Etzkorn M, Becker S, Baldus M. 3D NMR spectroscopy for resonance assignment and structure elucidation of proteins under MAS: Novel pulse schemes and sensitivity considerations. J Magn Reson. 2005;173:64–74. doi: 10.1016/j.jmr.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Kaiser JM, Polenova T, Yang J, Rienstra CM, Mueller LJ. Backbone assignments in solid state proteins using J-based 3D heteronuclear correlation spectroscopy. J Am Chem Soc. 2007;129:10650–10651. doi: 10.1021/ja073498e. [DOI] [PubMed] [Google Scholar]
- 8.Franks WT, Kloepper KD, Wylie BJ, Rienstra CM. Four-dimensional heteronuclear correlation experiments for chemical shift assignment of solid proteins. J Biomol NMR. 2007;39:107–131. doi: 10.1007/s10858-007-9179-1. [DOI] [PubMed] [Google Scholar]
- 9.Goldbourt A, Gross BJ, Day LA, McDermott AE. Filamentous phage studied by magic-angle spinning NMR: Resonance assignment and secondary structure of the coat protein in Pf1. J Am Chem Soc. 2007;129:2338–2344. doi: 10.1021/ja066928u. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Berthold DA, Frericks HL, Gennis RB, Rienstra CM. Partial 13C and 15N chemical-shift assignments of the disulfide-bond-forming enzyme DSBb by 3D magic-angle spinning NMR spectroscopy. ChemBioChem. 2007;8:434–442. doi: 10.1002/cbic.200600484. [DOI] [PubMed] [Google Scholar]
- 11.Pintacuda G, Giraud N, Pierattelli R, Bockmann A, Bertini I, Emsley L. Solid state NMR spectroscopy of a paramagnetic protein: Assignment and study of human dimeric oxidized Cu(II)-Zn(II) superoxide dismutase (SOD) Angew Chem-Int Edit. 2007;46:1079–1082. doi: 10.1002/anie.200603093. [DOI] [PubMed] [Google Scholar]
- 12.Huang L, McDermott AE. Partial site-specific assignment of a uniformly 13C, 15N enriched membrane protein, light-harvesting complex 1 (LH1), by solid state NMR. Biochim Biophys Acta-Bioenerg. 2008;1777:1098–1108. doi: 10.1016/j.bbabio.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Berthold DA, Gennis RB, Rienstra CM. Chemical shift assignment of the transmembrane helices of DSBb, a 20-kDa integral membrane enzyme, by 3D magic-angle spinning NMR spectroscopy. Protein Sci. 2008;17:199–204. doi: 10.1110/ps.073225008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higman VA, Flinders J, Hiller M, Jehle S, Markovic S, Fiedler S, van Rossum BJ, Oschkinat H. Assigning large proteins in the solid state: A MAS NMR resonance assignment strategy using selectively and extensively 13C-labeled proteins. J Biomol NMR. 2009;44:245–260. doi: 10.1007/s10858-009-9338-7. [DOI] [PubMed] [Google Scholar]
- 15.Han Y, Ahn J, Concel J, Byeon IJL, Gronenborn AM, Yang J, Polenova T. Solid state NMR studies of HIV-1 capsid protein assemblies. J Am Chem Soc. 2010;132:1976–1987. doi: 10.1021/ja908687k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 17.Siemer AB, Ritter C, Steinmetz MO, Ernst M, Riek R, Meier BH. 13C, 15N resonance assignment of parts of the HET-s prion protein in its amyloid form. J Biomol NMR. 2006;34:75–87. doi: 10.1007/s10858-005-5582-7. [DOI] [PubMed] [Google Scholar]
- 18.Siemer AB, Arnold AA, Ritter C, Westfeld T, Ernst M, Riek R, Meier BH. Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc. 2006;128:13224–13228. doi: 10.1021/ja063639x. [DOI] [PubMed] [Google Scholar]
- 19.Siemer AB, Ritter C, Ernst M, Riek R, Meier BH. High-resolution solid state NMR spectroscopy of the prion protein HET-s in its amyloid conformation. Angew Chem-Int Edit. 2005;44:2441–2444. doi: 10.1002/anie.200462952. [DOI] [PubMed] [Google Scholar]
- 20.Ritter C, Maddelein ML, Siemer AB, Luhrs T, Ernst M, Meier BH, Saupe SJ, Riek R. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxa U, Wickner RB, Steven AC, Anderson DE, Marekov LN, Yau WM, Tycko R. Characterization of beta-sheet structure in Ure2p1-89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 22.Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP. Molecular conformation and dynamics of the Y145Stop variant of human prion protein. Proc Natl Acad Sci U S A. 2008;105:6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helmus JJ, Surewicz K, Surewicz WK, Jaroniec CP. Conformational flexibility of Y145Stop human prion protein amyloid fibrils probed by solid state nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2010;132:2393–2403. doi: 10.1021/ja909827v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett AE, Rienstra CM, Griffiths JM, Zhen WG, Lansbury PT, Griffin RG. Homonuclear radio frequency-driven recoupling in rotating solids. J Chem Phys. 1998;108:9463–9479. [Google Scholar]
- 25.Ishii Y. 13C-13C dipolar recoupling under very fast magic angle spinning in solid state nuclear magnetic resonance: Applications to distance measurements, spectral assignments, and high-throughput secondary-structure determination. J Chem Phys. 2001;114:8473–8483. [Google Scholar]
- 26.Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 27.Tycko R. Stochastic dipolar recoupling in nuclear magnetic resonance of solids. Phys Rev Lett. 2007;99:187601. doi: 10.1103/PhysRevLett.99.187601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tycko R. Theory of stochastic dipolar recoupling in solid state nuclear magnetic resonance. J Phys Chem B. 2008;112:6114–6121. doi: 10.1021/jp076808o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe: A multidimensional spectral processing system based on Unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 30.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. Equation of state calculations by fast computing machines. J Chem Phys. 1953;21:1087–1092. [Google Scholar]
- 31.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc Natl Acad Sci U S A. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharpe S, Kessler N, Anglister JA, Yau WM, Tycko R. Solid state NMR yields structural constraints on the V3 loop from HIV-1 gp120 bound to the 447-52d antibody Fv fragment. J Am Chem Soc. 2004;126:4979–4990. doi: 10.1021/ja0392162. [DOI] [PubMed] [Google Scholar]
- 34.Nelson SJ, Schneider DM, Wand AJ. Implementation of the main chain directed assignment strategy: Computer-assisted approach. Biophys J. 1991;59:1113–1122. doi: 10.1016/S0006-3495(91)82326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartels C, Guntert P, Billeter M, Wuthrich K. GARANT: A general algorithm for resonance assignment of multidimensional nuclear magnetic resonance spectra. J Comput Chem. 1997;18:139–149. [Google Scholar]
- 36.Buchler NEG, Zuiderweg ERP, Wang H, Goldstein RA. Protein heteronuclear NMR assignments using mean-field simulated annealing. J Magn Reson. 1997;125:34–42. doi: 10.1006/jmre.1997.1106. [DOI] [PubMed] [Google Scholar]
- 37.Li KB, Sanctuary BC. Automated resonance assignment of proteins using heteronuclear 3D NMR. 2. Side chain and sequence-specific assignment. J Chem Inf Comput Sci. 1997;37:467–477. doi: 10.1021/ci960372k. [DOI] [PubMed] [Google Scholar]
- 38.Lukin JA, Gove AP, Talukdar SN, Ho C. Automated probabilistic method for assigning backbone resonances of 13C,15N-labeled proteins. J Biomol NMR. 1997;9:151–166. doi: 10.1023/a:1018602220061. [DOI] [PubMed] [Google Scholar]
- 39.Leutner M, Gschwind RM, Liermann J, Schwarz C, Gemmecker G, Kessler H. Automated backbone assignment of labeled proteins using the threshold accepting algorithm. J Biomol NMR. 1998;11:31–43. doi: 10.1023/a:1008298226961. [DOI] [PubMed] [Google Scholar]
- 40.Bailey-Kellogg C, Widge A, Kelley JJ, Berardi MJ, Bushweller JH, Donald BR. The NOESY jigsaw: Automated protein secondary structure and main-chain assignment from sparse, unassigned NMR data. J Comput Biol. 2000;7:537–558. doi: 10.1089/106652700750050934. [DOI] [PubMed] [Google Scholar]
- 41.Moseley HNB, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Method Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- 42.Hitchens TK, Lukin JA, Zhan YP, McCallum SA, Rule GS. MONTE: An automated Monte Carlo based approach to nuclear magnetic resonance assignment of proteins. J Biomol NMR. 2003;25:1–9. doi: 10.1023/a:1021975923026. [DOI] [PubMed] [Google Scholar]
- 43.Lemak A, Steren CA, Arrowsmith CH, Llinas M. Sequence specific resonance assignment via multicanonical Monte Carlo search using an ABACUS approach. J Biomol NMR. 2008;41:29–41. doi: 10.1007/s10858-008-9238-2. [DOI] [PubMed] [Google Scholar]
- 44.Tycko R. Prospects for resonance assignments in multidimensional solid state NMR spectra of uniformly labeled proteins. J Biomol NMR. 1996;8:239–251. doi: 10.1007/BF00410323. [DOI] [PubMed] [Google Scholar]
- 45.Meiler J, Baker D. Rapid protein fold determination using unassigned NMR data. Proc Natl Acad Sci U S A. 2003;100:15404–15409. doi: 10.1073/pnas.2434121100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korukottu J, Schneider R, Vijayan V, Lange A, Pongs O, Becker S, Baldus M, Zweckstetter M. High-resolution 3D structure determination of kaliotoxin by solid state NMR spectroscopy. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loquet A, Bardiaux B, Gardiennet C, Blanchet C, Baldus M, Nilges M, Malliavin T, Bockmann A. 3D structure determination of the Crh protein from highly ambiguous solid state NMR restraints. J Am Chem Soc. 2008;130:3579–3589. doi: 10.1021/ja078014t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.