Abstract
PARP-like zinc fingers are protein modules, initially described as nick-sensors of poly(ADP-ribosyl)-polymerases (PARPs), which are found at the N-terminus of different DNA repair enzymes. I chose to study the role of PARP-like fingers in AtZDP, a 3′ DNA phosphoesterase, which is the only known enzyme provided with three such finger domains. Here I show that PARP-like fingers can maintain AtZDP onto damaged DNA sites without interfering with its DNA end repair functions. Damage recognition by AtZDP fingers, in fact, relies on the presence of flexible joints within double-strand DNA and does not entail DNA ends. A single AtZDP finger is already capable of specific recognition. Two fingers strengthen the binding and extend the contacts on the bound DNA. A third finger further enhances the specific binding to damaged DNA sites. Unexpectedly, gaps but not nicks are bound by AtZDP fingers, suggesting that nicks on a naked DNA template do not provide enough flexibility for the recognition. Altogether these results indicate that AtZDP PARP-like fingers, might have a role in positioning the enzyme at sites of enhanced helical flexibility, where single-strand DNA breaks are present or are prone to occur.
INTRODUCTION
PARP-like zinc fingers are eukaryotic DNA binding domains that can function as strand-break sensors within eukaryotic cells (1–6). PARP-like fingers consist of unusually long zinc finger-like motifs of the form CX2C-X28/30-WHX2C, which can be present in one, two or three copies at the N-terminus of different DNA repair enzymes (3,5,7).
In poly(ADP-ribosyl)-polymerases (PARPs), the enzymes where PARP-like fingers were initially described, two such modules can be found, which dictate the strand-break dependent activation of these enzymes. Specifically, upon DNA-breaks recognition by the fingers, PARPs catalyse the reversible poly(ADP-ribosyl)ation of nuclear acceptor proteins, including PARPs themselves. As a consequence, only proteins confined at sites of damaged DNA are modified and lose their affinity for DNA, due to the negative charges of added ADPr units. Such PARPs-catalysed protein modification is required for efficient DNA repair and, according to the recent identification of new poly(ADP-ribosyl)ation enzymes, might be also involved in DNA replication as well as in DNA transcription processes (5,8–10).
The repair ligase III, the second identified PARP-like fingered enzyme, only has one finger, which is not required for the enzymatic activity (4,6). Indeed the role of ligase III finger in DNA repair mechanisms is not fully understood. It has been proposed to promote the displacement of PARPs from DNA strand breaks, thus allowing the entrance of the repair complex (3,11). Alternatively, it has also been proposed to facilitate the repair of strand breaks by binding to nearby DNA secondary structures (6).
The last identified PARP-like fingered enzyme is a repair protein from Arabidopsis named AtZDP, which comprises a DNA 3′-phosphoesterase domain preceded by three PARP-like finger modules. This plant protein has been shown to recognise single-strand and double-strand DNA breaks and to remove blocking groups from DNA 3′ ends. Similarly to the case of ligase III, the role of the fingers in AtZDP DNA repair activity is not clear, since the isolated catalytic domain is by itself functional, both in vivo and in vitro (7,12).
All three PARP-like fingered enzymes are implicated in the repair of single-strand DNA breaks. Such lesions are the most abundant in cellular DNA and can arise during the processes of DNA replication, repair or recombination as well as following exposure to endogenous and exogenous reactive oxygen species, and after exposure to ionising radiation (13). They thus represent a persistent challenge to genome integrity, and continuous sealing of DNA breaks must occur within the cell in order to prevent chromosome instability and/or cell death. Indeed, the association of single-strand break repair with neuro-degenerative disorders as well as with a cancer-prone phenotype has stimulated very intense studies on this subject in mammals (11,14–19). In particular, the scaffold protein XRCC1 has been clearly shown to orchestrate the efficient repair of single-strand DNA breaks, by organising and stimulating the enzymatic activity of DNA ligase III, DNA polymerase β and DNA kinase/phosphatase within a very efficient repair complex (11). It is worth noting that two different nick-sensors appear to be present within this complex, namely the ligase III PARP-like finger and a distinct domain, which is located in the XRCC1 N-terminal region (20). Interestingly, the plant homologue of XRCC1 lacks the N-terminal nick-sensing region, and there is no evidence for PARP-like fingered DNA ligases. In addition, DNA kinase/phosphatase activity in plants is not accomplished by the same polypeptide chain, as observed in the animal system. Instead, the 3′ DNA phosphatase activity in plants is associated with three PARP-like fingers in the AtZDP protein, which is assumed to provide the nick-sensing functions for a putative single-strand repair complex (7). On the contrary, PARPs with two finger modules are very well conserved throughout higher eukaryotes, including plants (8,10,21).
It is not known how the different DNA-break sensors interact at damaged sites, nor is it clear how they permit the entry of repair enzymes at the DNA ends. The present work aims to characterise the DNA-break sensor activity of AtZDP PARP-like fingers, and their specific interactions at damaged DNA sites.
MATERIALS AND METHODS
Sequence analyses
AtZDP PARP-like fingers were aligned with PARP-like finger domains retrieved from the PFAM database (http://www.sanger.ac.uk/Software/Pfam/) using ClustalW (22) and visualised with GeneDoc (23). Secondary structure predictions were obtained with the Predict Protein server (http://cubic.bioc.columbia.edu/predictprotein/). The phylogenetic tree was generated using the neighbour-joining algorithm of ClustalW, excluding positions with gaps and correcting for multiple substitutions. The resulting unrooted tree was rooted by the midpoint method using the Retree program of the Phylip package (http://evolution.genetics.washington.edu/phylip.html). Bootstrap percentages were calculated with 1000 replicates.
Expression and purification of recombinant AtZDP constructs
The cloning for the expression of recombinant AtZDP full-length protein, the three-fingers domain (3F) and the catalytic domain were described elsewhere (7). The histidine tagged two-fingered (2FCD) and the one-fingered (1FCD) AtZDP constructs, schematically represented in Figure 2A, were obtained after PCR amplification, CpoI restriction digestion and cloning in the pET-CpoI expression vector (7). Briefly, amplifications (20 cycles) were performed using a high-fidelity thermophilic DNA polymerase (VentR; New England Biolabs), 100 ng of the pET-AtZDP expression vector as template and the following CpoI-tailed primers: acgcggtccgattgctcataatgcgaaatcgag or acgcggtccgtctgagtatgcgaaatcgagcag as plus primers for 2FCD or 1FCD respectively, along with the ctccggaccgctaagtccctggcgatgtacttg CpoI-tailed minus primer. The pET-2FCD and the pET-1FCD plasmids thus obtained were sequenced and used for protein expression in BL21-Codon Plus (DE3)-RIL cells (Novagen). Recombinant proteins were affinity purified as previously described (7).
Figure 2.
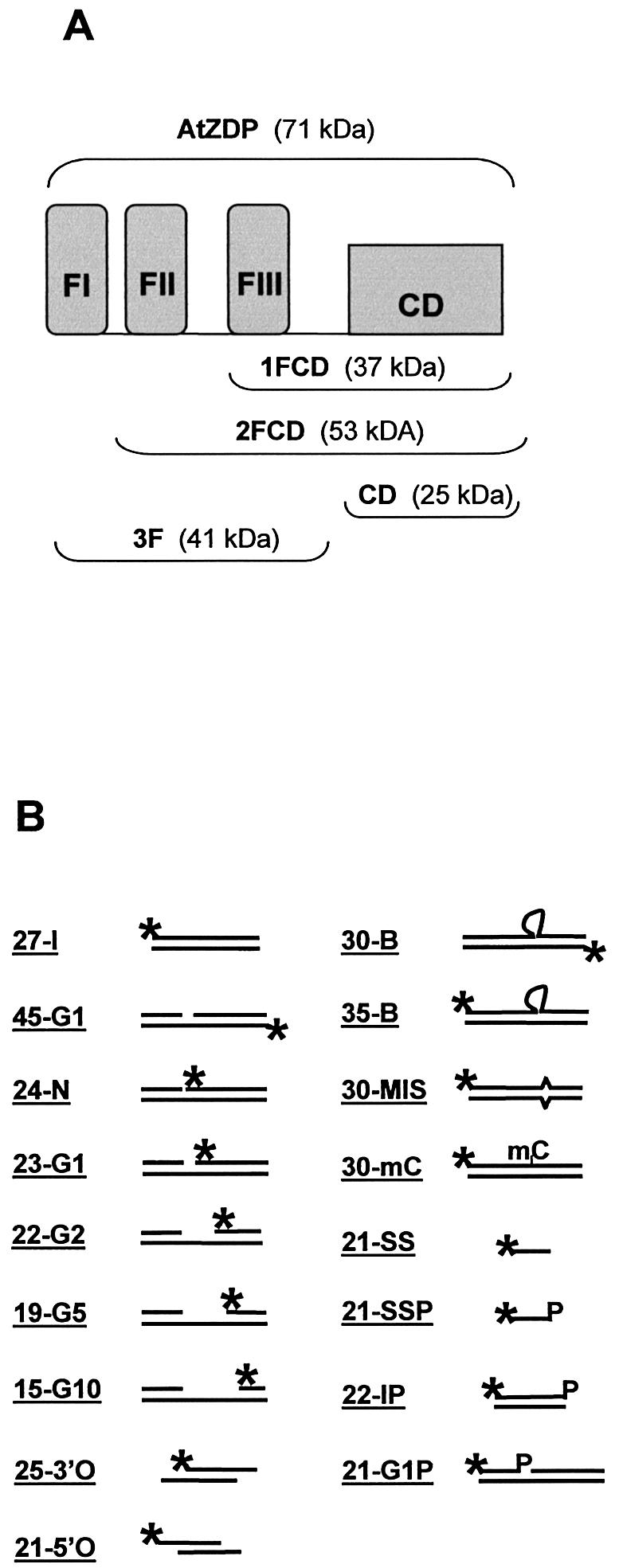
Structure of protein constructs and of oligo DNAs used in this work. (A) Schematic representation of the AtZDP deletion mutants. The three N-terminal PARP-like fingers (FI, FII, FIII, respectively) and the associated catalytic domain (CD) are shown as closed boxes. The modular composition of the wild type (above) and of the deletion mutants (below) are indicated with corresponding molecular weights enclosed in brackets. (B) Structure of the substrate oligonucleotides. The positions of 32P-labelled 5′-termini (asterisks) and of 3′-phosphate groups (P) are indicated. Heading numbers in oligo names refer to the length in bases of the labelled species. The 45-G1, 24-N, 23-G1, 22-G2, 19-G5, 21G1P are identical oligo- duplexes, only differing for the presence of a nick (24-N) or of a 1-base (45-G1, 23-G1, 21G1P), 2-base (22-G2), 5-base (19-G5), 10-base (15-G10) gap; the 25-3′O and the 21-5′O have overlapping double-strand domains with respectively 3′ or 5′ single-strand extensions on both strands; the 30-B, 35-B, 30-MIS, 30-mC only differ for the presence of a 5-nucleotide bulge (30-B and 35-B), a mismatched (30-MIS) or a methyl-cytosine (30-mC; see Materials and Methods for more details)
Oligo constructs
Substrates for the DNA binding and the phosphatase assays were prepared using the following oligonucleotides (Sigma): 21mer, 21p, 23mer, 45mer as previously described (24); 27mer, (catatactccgagcccgaacacgtcgc); 27-comp, (gcgacgtgtt cgggctcggagtatatg); 24-n, (ggcgcccaccaccactagctggcc); 23-comp, (ggccagctagtggtggtgggcgc); 25-3′, (gcccaccaccactag ctggccttcc); 21-5′, (ccttgcgcccaccaccactag); 22-g2, (cgcccaccaccactagctggcc); 19-g5, (ccaccaccactagctggcc); 15-g10, (caccactagctggcc); 35-b, (catatactccgagccgtctctgccgaacacgt cgc); 30mer, (catatactccgagccggccgaacacgtcgc); 30-mis, (catatactccgagccgggcgaacacgtcgc); 30-mC, (catatactccgagcc gg(methyl-c)cgaacacgtcgc); 30-comp, (gcgacgtgttcggccggctc ggagtatatg); 22p, (cgcccaccaccactagctggcc-P); 22-comp, (ggccagctagtggtggtgggcg).
3′-phosphorylated oligos were end-labelled as described in detail elsewhere (12). 3′-unphosphorylated oligos were end-labelled with polynucleotide kinase (Amersham) and [γ-32P]ATP. The specific activity of all labelled oligos ranged between 0.3 and 0.8 µCi/pmol. The intact oligo duplex 27-I was generated by annealing as previously described (12) the two complementary 27mer and 27-comp oligos, one of which was labelled at the 5′ end. The 45-G1 was generated by end-labelling of the 45mer, and annealing with the complementary, unlabelled 21mer and 23mer. Over-hanging 25-3′O and 21-5′O oligos were derived by the annealing of the end-labelled 25-3′ with the 23-comp oligo, and of labelled 21-5′ with the 22-comp oligo, respectively. Radioactive 24-n, 23mer, 22-g2, 19-g5 or 15-g10 oligos were each annealed with unlabelled 21mer and 45mer to generate 24-N, 23-G1, 22-G2, 19-G5 or 15-G10 oligo duplexes. 35-B, 30-B, 30-MIS or 30-mC was derived by the annealing of labelled 35-b, 30mer, 30-mis or 30-mC with the complementary 30-comp oligo. End-labelled 21p, herein defined as 21-SSP, was annealed with unlabelled 23mer and 45mer to give the 21-G1P oligo. Labelled 22p was used in the preparation of the 22-IP, by annealing with 22-comp.
Phosphatase assays
The 5′ labelled 3′-phosphorylated single-strand 21-SSP, the 3′-phosphrylated 22-IP double-strand or the 1-nucleotide gap 21-G1P oligo bearing a 3′-phosphate at the gap site, (as represented in Fig. 2B and described above) were used as reaction substrates. Assays were run at 30°C for 10 min in 15 µl reaction mixtures containing 100 mM Tris, pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol (DTT), 0.4 mM EDTA and 100 µg/ml bovine serum albumin plus the substrate and purified recombinant AtZDP proteins, as specified in the text. Reactions were stopped on ice by the addition of formamide-EDTA loading buffer and the products were analysed on 8% sequencing gels. Phosphorimages of dried gels were recorded with a Personal Imager FX (Bio-Rad) and analysed using the Multi-Analyst/PC software (Bio-Rad). The percent conversions were calculated on the basis of product appearance.
DNA binding assays
For electrophoretic mobility shift assays (EMSA) the 45-G1 gapped duplex and varying amounts of AtZDP recombinant proteins were incubated for 30 min on ice in 10 µl of EMSA buffer containing 20 mM Tris, pH 8.0, 100 mM KCl, 2 mM MgCl2, 3% glycerol, 30 ng of HaeIII restricted pBluescript plasmid DNA and 2 mM DTT (unless otherwise specified). Electrophoresis was carried at 4°C on 5–8% non-denaturing polyacrylamide gels at 150 V in 1× TBE. Gels were dried and subjected to phosphorimager analyses.
For southwestern blotting, 20–1000 pmol of recombinant AtZDP proteins were separated by 10% polyacrylamide– SDS gel electrophoresis and transferred onto nitrocellulose membranes (ECL, Amersham Pharmacia Biotech) according to the manufacturer’s instructions. Membrane bound proteins were renatured for 30 min at room temperature in RB (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% Nonidet P-40), and then equilibrated for 30 min at room temperature in DBB (20 mM Tris, pH 8.0, 100 mM KCl, 5 mM MgCl2, 2 mM DTT, 50 µM ZnCl2 and 0.1% Nonidet P-40). DNA binding was carried out in DBB for 1 h at 4°C in the presence of 32P-end-labelled oligos duplexes (1 nM and 0.5–0.8 µCi/pmol). After three washes (5 min each) at 4°C in DNA binding buffer, wet filters were directly subjected to phosphorimager analyses.
For the analyses of DNA binding to filter-bound proteins, and for the oligo-selection or footprint analyses, membrane-bound proteins were visualised immediately following the membrane transfer by staining of the filter with 0.2% Ponceau S in 3% trichloro-acetic acid at room temperature for 10 min. Filter-bound proteins, thus identified, were excised from the membrane and de-stained by soaking in a 1 M NaCl solution. Following 30 min renaturation in 500 µl RB and 30 min equilibration in 500 µl DBB at room temperature, DNA binding on filter-bound proteins was carried out for 30 min at 4°C in 100 µl DBB plus 0.1 pmol each (as evaluated by OD measurements at 260 nm) of the labelled probes (0.5–0.8 µCi/pmol), followed by 3× 500 µl washes with cold DBB. To quantitate the amount of radioactivity bound by each protein after southwestern blotting, samples were counted and background binding to the filter, as evaluated by counting of corresponding protein-free filter slices, was subtracted. The values thus obtained were normalised for the amount of protein loaded on southwestern gels, and expressed as the percentage of binding relative to the total amount of probe used in the binding reaction.
For the oligo-selection analyses, bound DNA was directly recovered in 100 µl of elution buffer (Tris 50 mM, pH 7.5, EDTA 5 mM, SDS 0.2%, carrier DNA 0.10 µg/ml), phenol/chloroform extracted and ethanol precipitated, along with aliquots of the oligo mixtures collected before and after the binding reactions. DNA samples were counted, directly resuspended in formamide-EDTA loading buffer, and same c.p.m. aliquots were analysed on 8–16% sequencing gels. Phosphorimages of dried gels were recorded and analysed. To calculate the relative oligo binding by each AtZDP protein in the oligo-selection experiments, signal intensities of bound DNAs were quantitated on phosphorimages of the denaturing gels, corrected for differences in the specific activity, and divided for the corresponding amount of the bound reference oligo.
For DNase I footprinting of AtZDP bound oligos, following the DNA binding and washing steps described above, filter-bound protein–DNA complexes were incubated for 5 min at room temperature in 100 μl DBB containing 2–3 ng/µl of DNase I (0.5–0.75 U). DNase I digestions were then stopped with 100 µl of 0.2 M NaCl, 20 mM EDTA, 1% SDS and 0.25 µg/ml carrier DNA, digestion products were recovered from the filter (as described above) and analysed by denaturing 16% polyacrylamide gel electrophoresis.
RESULTS
Sequence features of the PARP-like finger domains
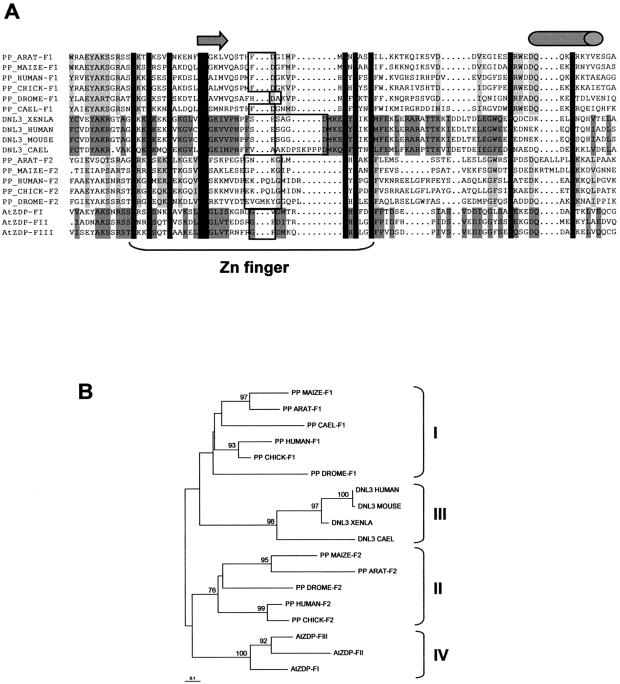
The PARP-like finger regions from different species and enzymes were compared by sequence alignment and the predicted secondary structures that are conserved were annotated. As shown in Figure 1A, sequence conservation is not confined to the putative zinc binding residues, but it extends upstream and downstream with respect to them, and terminates in a highly conserved alpha-helical structure at the carboxy-terminal side of all these domains. Possibly, fingertips lie in the conserved turn located between the putative zinc-coordinating residues. In a phylogenetic tree generated from this alignment (Fig. 1B), four types of PARP-like fingers can be identified. Type I corresponds to the N-terminal F1 finger of all PARP enzymes, whereas type II comprises all F2 fingers of PARPs. Type III, which is more related to the F1 finger of PARPs, belongs to DNA ligases III. Finally, all three fingers of AtZDP cluster in a fourth type, which appears to be more related to the PARPs’ F2 fingers. These phylogenetic relationships suggest the existence of a correlation between finger types and functional properties of PARP-like finger modules.
Figure 1.
Sequence comparison of PARP-like zinc fingers. (A) Alignment of known PARP-like finger modules. Amino acid residues conserved between all PARP-like finger domains are in black, and amino acids conserved within the clusters identified by the phylogenetic analysis shown in (B) are shaded. Gaps introduced to optimise the alignment are indicated by dots. Sequences within the zinc-coordinating residues are indicated as Zn finger. The arrow and the cylinder indicate respectively, the β-sheet and the α-helical secondary structures, which are predicted to be conserved between all the PARP-like finger domains. The amino acid residues included in a conserved turn, which might indicate the fingertip, are boxed. (B) Phylogenetic relationship among PARP-like finger domains. The tree was generated from the alignment shown in (A) using the neighbour-joining algorithm, and it was rooted with the mid-point distance method. Branches are drawn to scale as indicated by the scale bar, which corresponds to 0.1 changes per site. When >70%, bootstrap percentages are indicated at the nodes. Sequences clusters corresponding to distinct PARP-like finger types are included in brackets.
Functional interactions between PARP-like fingers and the catalytic domain in AtZDP
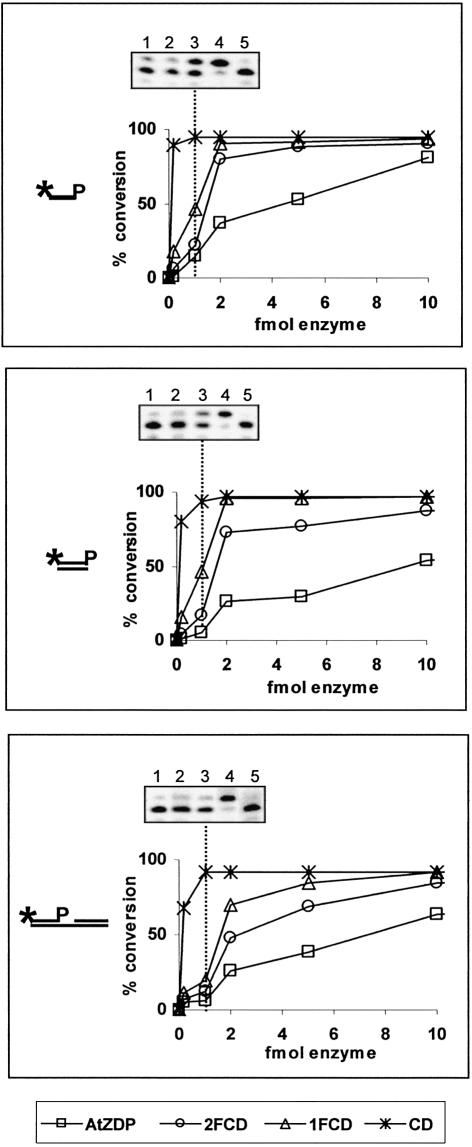
In order to assign a role to each AtZDP PARP-like finger, various AtZDP protein constructs were over-expressed and purified. These included proteins with no (CD), one (1FCD) or two (2FCD) PARP-like fingers located upstream of the AtZDP catalytic domain, as well as the three-fingers domain alone (3F) and the full-length enzyme that included the three PARP-like fingers and the catalytic domain (AtZDP; schematised in Fig. 2A). The purified proteins were then tested for their 3′-phosphatase activity on different DNA substrates (21-SSP, 22-IP and 21-G1P as represented in Fig. 2B). As expected, AtZDP proteins containing the catalytic domain catalysed the removal of a 3′-phosphate group from DNA strand-breaks, either located on single- or double-strand DNA ends, or at the gap site of a double-strand DNA (Fig. 3). When the 3F construct, which lacks the catalytic domain, was employed in the assay no activity was observed, excluding the possibility that the observed phosphatase activity was due to contaminating bacterial enzymes. Notably, none of the AtZDP proteins exhibited a clear preference between these different substrates, and in all cases the isolated catalytic domain had maximal enzymatic activity. These results clearly prove that AtZDP fingers are not required to direct substrate recognition by AtZDP, when a purified substrate is used. In fact, when comparing the activity of corresponding amounts of enzymes, it appears that increasing the number of PARP-like fingers associated with the catalytic domain results in a decreased percentage of substrate conversion. This observation is suggestive of an interaction between the fingers and the AtZDP catalytic domain, which might limit the catalytic properties of the enzyme.
Figure 3.
DNA 3′-phosphatase activities of AtZDP proteins. The percentages of single-stranded (21-SSP, upper panel), double-stranded (22-IP, middle panel) or of a gapped duplex (21-G1P, lower panel) substrate conversion are plotted against enzyme quantities. All reactions contained 100 fmol of the 5′ labelled-substrate. Unphosphorylated reaction products were separated from phosphorylated DNA substrates on denaturing 8% polyacrylamide gels, quantitated on phosphorimages, and used to calculate the percent of substrate conversion. Data are the average of two independent experiments that differed by <10% of the mean. Symbols are as follows: wild type AtZDP, squares; 2FCD, circles; 1FCD, triangles; CD, asterisks. Phosphorimages of representative phosphatase assays using 1 fmol of AtZDP (lane 1), 2FCD (lane 2), 1FCD (lane 3), CD (lane 4) or 100 fmol of 3F (lane 5) are reported above each graphic. Note that the phosphate-terminated oligonucleotide migrates faster than the hydroxyl-terminated oligonucleotide of the same size.
The DNA binding activities of the different AtZDP constructs were next compared by electrophoresis mobility shift assay (EMSA). Figure 4A shows that full-length AtZDP completely shifted the gapped 45-G1 DNA template in a well-resolved protein–DNA complex (lanes 2 and 3), while corresponding amounts of the other AtZDP constructs recruited a much smaller fraction of the labelled oligo duplex (lanes 4–9). Higher protein concentrations led to the accumulation of aggregates not resolved under these gel conditions (visible at the top of lanes 2–9), and a very weak binding activity was observed using a large excess of the isolated catalytic domain (lanes 10 and 11). It thus appears that AtZDP fingers must cooperate with the catalytic domain for binding, since the absence of either domain results in reduced DNA binding capacity. It is worth noticing that in the absence of DTT specific binding by the fingered proteins AtZDP, 2FCD and 3F, was lost (Fig. 4B).
Figure 4.

Interactions between the PARP-like fingers and the DNA 3′-phosphesterase domain modulate AtZDP DNA binding. (A) The DNA binding activity of AtZDP constructs. Phosphorimages of protein–DNA complexes analysed by non-denaturing polyacrylamide gel electrophoresis. 100 fmol of the 32P-labelled 45-G1 gapped duplex (see Fig. 2B) were incubated with the indicated amounts of AtZDP (lanes 2 and 3), 3F (lanes 4 and 5), 2FCD (lanes 6 and 7), 1FCD (lanes 8 and 9) or CD (lanes 10 and 11). The labelled DNA ligand incubated without proteins was run in lane 1. (B) DTT-dependence of AtZDP DNA binding. Phosphorimages of protein–DNA complexes were analysed as in (A) after incubation of 1 pmol of AtZDP (lanes 2 and 3), 3FD (lanes 4 and 5) or 2FCD (lanes 6 and 7) with 100 fmol of the 32P-labelled 45-G1 gapped duplex, either in the absence (lanes 2, 4 and 6) or in the presence (lanes 3, 5 and 7) of DTT. The labelled DNA ligand incubated without protein was run in lane 1.
Altogether these results show that functional interactions occur between the catalytic and the PARP-like finger domains of AtZDP, which can modulate the enzyme activity and increase DNA binding. Possibly, unpaired PARP-like fingers in the AtZDP deletion constructs may provide independent DNA binding activity, which results in the observed high molecular weight aggregates.
DNA binding preferences by AtZDP fingers
In order to understand the contribution of single AtZDP domains to DNA damage recognition, I next compared the relative affinity of AtZDP proteins for oligo duplexes bearing different types of DNA strand breaks. To this purpose, the functional properties of filter-bound AtZDP proteins were initially tested by southwestern analysis (Fig. 5A and Table 1). In agreement with EMSA analysis, full length AtZDP showed stronger DNA binding activity than the deleted AtZDP forms. Binding was totally lost in the absence of DTT or by using single-stranded DNA as a probe. Only a weak binding was observed in the presence of large amounts of the isolated catalytic domain. Filter-bound proteins were catalytically active, as revealed by the analysis of the bound oligo, recovered after southwestern blotting with the 21-G1P probe. Indeed DNA bound to AtZDP, 2FCD and 1FCD, but not to 3F, was completely converted to the corresponding 3′-unphosphorylated species by filter-bound proteins (Table 1). The relative affinity of AtZDP fingers for different DNA strand-breaks was then evaluated by analysing the DNA binding preferences of filter-bound proteins. For such an assay, double-strand oligos were designed, each identifiable for the different DNA damage on the basis of the labelled single-strand component (see Fig. 2B and Materials and Methods). Equimolar amounts of the radiolabelled double-strand oligos were then incubated with the filter-bound AtZDP proteins, bound and unbound oligo fractions were recovered and directly analysed on sequencing gels. Figure 5B reveals the binding preferences of the AtZDP constructs, when incubated with an equimolar mixture of an intact (27-I), a nicked (24-N) and a gapped (22-G2) oligo template. In order to quantitate this oligo-selection experiment, the signal intensity of the intact 27-I duplex bound to each protein species was taken as a reference and compared to the corresponding signals derived from the differently modified DNA templates (Fig. 5C). A binding ratio of 1 indicates that the three oligos were bound at similar affinity, implying that, presumably, only end binding has occurred on all templates. On the other hand, a binding ratio higher or lower than 1 respectively indicates that the analysed DNA template was, or was not, selected for binding. It is evident from the oligo-selection experiment presented in Figure 5 that the fingered (AtZDP, 3F, 2FCD, 1FCD) constructs have a clear preference for gapped DNA. None of the AtZDP protein constructs made a specific selection for the 24-N oligo DNA. The catalytic domain equally bound at a very low level all three templates, and background binding to the unblotted filter did not exhibit template preference either (not shown).
Figure 5.
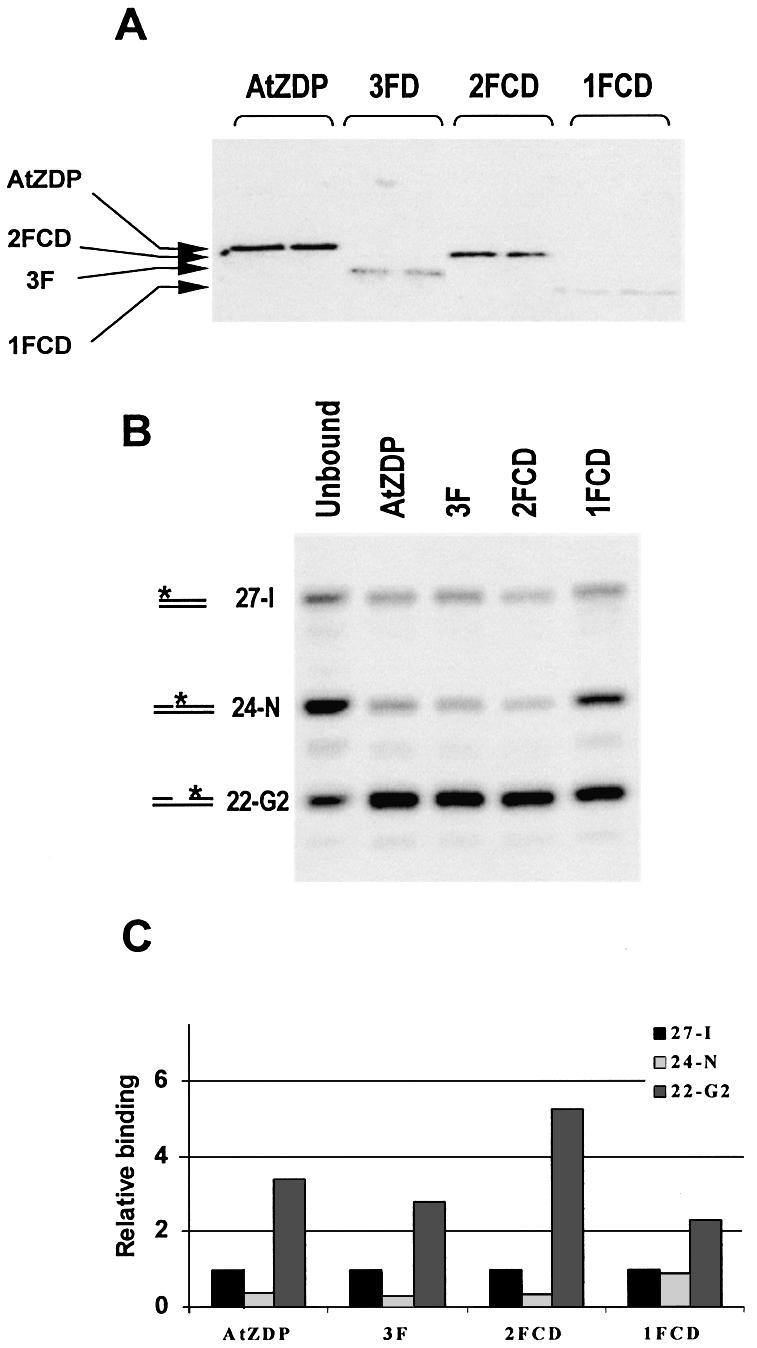
Oligo-selection analysis in membrane-bound protein–DNA complexes. (A) Phosphorimage of southwestern blotting of fingered AtZDP proteins. Purified proteins were run on an SDS–polyacrylamide gel, transferred to a nitrocellulose membrane and incubated with the 32P-labelled gapped 45-G1 oligo duplex. The migration positions of purified AtZDP proteins loaded on the gel and visualised on the filter after gel transfer are indicated. (B) Phosphorimage of oligo selected by AtZDP proteins. Filter-bound AtZDP proteins were incubated with an equimolar mixture of the 32P-labelled intact 27-I, nicked 24-N and gapped 22-G2 oligo duplexes. After elution from the filter, same amounts of bound radioactivity were analysed by denaturing polyacrylamide gel electrophoresis, as indicated on top of the figure. A sample of the unbound oligo-mixture, recovered after the hybridisation, was run alongside for comparison (Unbound). The migration positions of the labelled species corresponding to the 27-I, the 24-N and the 22-G2 oligo duplexes are indicated on the left. (C) DNA binding preferences by AtZDP constructs. The phosphorimage presented in (B) was used to calculate the relative amount of templates selected by each of the AtZDP constructs. The radioactivity associated with bound oligo 27-I was taken as a reference and given an arbitrary value of 1.
Table 1. The DNA binding properties of filter-bound AtZDP proteins.
| Probe | AtZDP | 3F | 2FCD | 1FCD | CD | |
|---|---|---|---|---|---|---|
| Gap binding | 45-G1 | 11.5a | 7.3 | 2.6 | 1.4 | 0.02 |
| DTT dependence | 45-G1 | ndb | ||||
| Gap binding-3′P oligo | 21-G1P | 7.9 | 5.2 | |||
| Phosphatase activityc | 21-G1P | + | – | + | + | + |
| Single strand binding | 21-SS | nd | nd |
Filter-bound protein–DNA complexes were analysed after incubations with the 32P-labeled probes (1 nM).
aValues are expressed as the percentage of filter-bound probe relative to the total amount of probe used in the incubation, after normalization for the amount of protein loaded onto western gels. Data are the average of at least two independent experiments that differ by <10% of the mean.
bnd, not detected.
cPhosphatase activity was monitored by gel electrophoresis analysis of the probe (as described in the legend of Fig. 3), after recovery from the filter-bound proteins.
It can be inferred from this oligo-selection experiment that AtZDP finger FIII is by itself capable of recognising gapped DNA. The presence of the FII finger largely augments DNA binding and increases the preference for gapped DNA templates (compare 1FCD and 2FCD in Fig. 5B and C and Table 1). Finally, the FI finger can further increase DNA binding to both gap sites and double-strand DNA ends (compare 2FCD and AtZDP in Fig. 5B and C and Table 1). Surprisingly, AtZDP PARP-like fingers do not selectively recognise nicked DNA.
GAP recognition by AtZDP PARP-like fingers
I next wanted to establish whether binding of AtZDP fingers requires close proximity between the 3′ and the 5′ ends of a broken strand at gapped DNA sites. To elucidate this point a new oligo-selection experiment was performed, using 5′ over-hanging (21-5′O) and 3′ over-hanging (25-3′O) oligo duplexes, along with the blunt-ended oligo 27-I, used as a reference. As shown in Figure 6A, full-length AtZDP and 3F both display a clear preference for blunt-end versus over-hang DNA. The CD domain clearly participates in the binding to the recessed DNA ends, since AtZDP shows higher affinity for the over-hang templates than 3F. By comparing the data presented in Figures 5 and 6 it appears that, while AtZDP PARP-like fingers select gapped over intact oligo duplex, DNAs with 3′ or 5′ over-hanging strands are very poorly selected relative to the blunt end DNA. It thus follows that double-stranded DNA structures flanking both sides of the gap must be present to allow recognition by the PARP-like fingers. This finding further raised the question about the gap length that is tolerated for binding. Oligo duplexes containing gaps of various size (23-G1, 19-G5, 15-G10, Fig. 2B) were thus prepared and analysed. Figure 6B shows that AtZDP fingers, either associated or not with AtZDP catalytic domain, can recognise with similar affinity DNAs bearing from 1 to 10 nucleotide-long gaps.
Figure 6.

Gap sensing by AtZDP PARP-like fingers. Phosphorimage of labelled oligos, following AtZDP or 3F oligo-selection analyses. DNA templates recovered from filter-bound proteins or from the unbound fraction, as reported in the figure, were identified according to their migration position on denaturing acrylammide gels, and are indicated on the left. (A) Binding to overhanging DNA templates. Overhanging 25-3′O and 21-5′O oligo duplexes, along with the blunt 27-I oligo duplex, were used for the oligo-selection analysis. A quantification of protein-bound oligos relative to bound 27-I duplex is given on the right. (B) AtZDP finger binding to variously gapped DNAs. An equimolar mixture of a 1 (23-G1), 5 (19-G5) and 10 (15-G10) nucleotides-gap oligo duplexes was analysed. Binding ratios are given relative to the amount of bound 23-G1 gapped oligo duplex.
It was concluded from these experiments that AtZDP fingers sense double-stranded DNA structures flanking both sides of a gap, and that a variable gap length does not interfere with such recognition.
DNA ends are dispensable for gap sensing by AtZDP fingers
Previous studies have demonstrated that PARP-like fingered enzymes bend the DNA upon binding to gap sites, and it has been postulated that the flexible joint at a strand break might provide the target for binding by PARP-like fingers (7,8,25). If this is indeed the case, in an oligo-selection experiment, isolated PARP-like fingers should be able to specifically select an intact oligo duplex that can yield V-shaped structures. The 35-B oligo-duplex fulfils these requirements since it bears an internal five-base bulge, which can produce a V-like DNA conformation (25,26). Figure 7 demonstrates that AtZDP fingers specifically sense the flexible junction within the 35-B oligo (upper panel). By contrast, corresponding oligo duplexes bearing internal modifications like a mismatch (middle panel) or a methylated residue (lower panel), which are not expected to induce V-shaped secondary structures, are not preferentially selected by 3F for binding.
Figure 7.

PARP-like fingers sense flexible joints on double-stranded DNA. Phosphorimages of labelled oligos left unbound or recovered from filter-bound 3F, after the incubation with equimolar mixtures of 35-B and 27-I (upper panel), 30-MIS and 27-I (middle panel) or 30-mC and 27-I (lower panel). The migration position of labelled species, and the amounts of 3F-bound oligos relative to the 27-I duplex are indicated on the left and right side, respectively.
AtZDP finger interaction with flexible DNA sites
Little is known about PARP-like finger-DNA contacts due to the lack of crystallographic data and due to the difficulties of footprinting nicked or gapped DNA strands. I tried to elucidate the interactions of AtZDP PARP-like fingers with DNA by taking advantage of their specific binding to the 35-B DNA template, which does not bear internal strand breaks and can therefore be analysed on both strands. The results of DNase I footprint generated by AtZDP proteins on the 35-B duplex as well as on the continuous strand of gapped oligos are presented in Figure 8. As expected from the oligo-selection experiments, protected regions span the flexible joints corresponding to the gap or the bulge sites present on these templates. Protection patterns obtained with the three-fingered (AtZDP and 3F) and the two-fingered proteins (2FCD) were in all cases very similar. Therefore it was not possible to define specific contact sites for the FI finger (by comparing AtZDP and 2FCD) nor for the CD domain (by comparing AtZDP and 3F) from these footprint data. On the other hand, footprints produced by 2FCD (and AtZDP or 3F) over the flexible joints of the bulged or of the gapped templates extended those observed with the one-fingered 1FCD protein (compare 1FCD and 2FCD in Fig. 8). These data therefore suggest that the FIII finger contacts double-stranded DNA flanking both sides of the bulge or of the gap. 1FCD binding is however weak (see Table 1) and, as summarised in Figure 8D, it is not symmetric with respect to the flexible joints on the DNA. The addition of a second finger strengthens gap binding, expands the protein–DNA contacts and appears to restore the symmetry of binding relative to the flexible joints. In agreement with these interpretations, by enlarging the gap size, a corresponding shift of AtZDP finger footprints over the double-stranded DNA flanking the gap was observed (Fig. 8D and data not shown).
Figure 8.
AtZDP finger-DNA contact sites. Phosphorimages (A), (B) and (C) and a schematic summary (D) of the DNase I footprint analyses using the 35-B (A), the 30-B (B) or the 45-G1 (C) labelled probe and the indicated AtZDP proteins. A digestion of the protein-free probes (Free) was run alongside for comparison. Nucleotide-numbers are reported relative to the 5′-labelled ends of the corresponding probe. DNA regions protected by AtZDP, 3F and 2FCD are indicated within longer brackets, while shorter brackets delimit the protection by 1FCD. The bulge or gap site is indicated by parentheses on the right side of the phosphorimages. Dashed lines in (D) indicate the footprint extension, which is observed upon a 5-nucleotide extension of the gap.
DISCUSSION
This work aimed at understanding the role of AtZDP PARP-like finger modules in the context of DNA repair functions. It is indeed very intriguing that PARP-like fingers can be found in different numbers and in association with different enzymatic functions of the single-strand DNA repair process. In this regard, AtZDP, a unique 3′ DNA phosphoesterase, is particularly well suited for such investigation because it is the only enzyme bearing three copies of PARP-like finger modules (7).
PARP-like fingers as sensors of flexible DNA structures
The DNA binding properties of AtZDP PARP-like fingers were evaluated by oligo-selection and footprinting analyses, and a schematic view of the obtained results is presented in Figure 9. Strand-break recognition by isolated AtZDP fingers does not imply direct contacts with the DNA ends, since they recognise double-stranded structures flanking gaps or other flexible sites within the DNA. Recent studies using atomic force microscopy also revealed that finger-containing PARPs can bind to hairpins in DNA heteroduplexes, and that these enzymes can bind to a promoter bearing dyad symmetry elements (27). It thus appears that PARP-like fingers can confer additional properties, beyond strand-break binding, to fingered enzymes. It is worth noting, however, that DNA binding by PARPs is modulated by the enzyme auto-poly(ADP-ribosyl)lation, while neither DNA ligase nor AtZDP enzymatic activity can directly modulate its own DNA binding capacity.
Figure 9.
Schematic view of AtZDP binding to a flexible joint on the DNA. DNA filaments are represented as a ladder, with arrowheads representing the 3′ DNA ends. The bulged or gapped strand is in black. Atoms of the nucleotides contacted by the FIII and the FII fingers are presented using the ball and stick format and are coloured in yellow and pink, respectively. Putative positions of the CD and of the FI finger domains are also suggested.
A single AtZDP finger seems already capable of making contacts on both sides of the flexible joint. A second finger, however, produces a much stronger binding and it extends the contacts on the double-strand DNA flanking the flexible joint. The presence of a third finger further reinforces DNA binding and hence, it presumably interacts with the DNA. Specific FI finger-DNA contacts, however, could not be observed in our footprint experiments. Differences in the size of the footprints over the gapped and the bulged templates suggest that the precise location of each finger along the flanking double-stranded DNA depends upon the degree of DNA flexibility.
Surprisingly, AtZDP PARP-like fingers seem to function as gap- but not as nick-sensors, as if a nick would not provide enough flexibility to allow for recognition. In fact, a close inspection of the DNA binding properties of ligase III would also suggest that the ligase PARP-like finger is a gap- more than a nick-sensor, since mutations in the finger domain abolishes gap but not nick binding by this enzyme (6). This could be an important feature in the case of the DNA ligase III, considering that nick binding by the PARP-like finger could actually compete with the sealing activity of the enzyme. Similarly, in the case of PARPs, the fingers are not the unique DNA binding domains within these enzymes, and two helix–turn–helix motifs located in the proximity of the fingers have been shown to be substantial to DNA binding (28).
From the above observations it would follow that the nick-sensing activity usually attributed to PARP-like fingers may result from intramolecular and/or intermolecular interactions between the fingers and the associated domains. Likewise, the phylogenetic classification of PARP-like fingers in four different types may reflect their different abilities to mediate such interactions in different protein contexts. There is indeed clear evidence that PARP-like fingers can communicate their DNA binding status to the associated catalytic domains. In PARPs, the enzymatic activity is strongly activated upon DNA-break recognition (2). In the case of DNA ligase III, the binding of the PARP-like finger to damaged DNA has been postulated to modify the repair properties of the enzyme (6). Interestingly, in the case of AtZDP, the association of the PARP-like fingers with the catalytic domain results in a decreased enzyme activity. Such a decrease, however, is not dependent on the DNA binding preferences of AtZDP fingers, since it occurs to similar extents in the presence of gapped substrates, which are bound by the fingers, and of single-stranded DNAs, which are not stably bound. Considering that the AtZDP catalytic domain displays a potent phosphatase activity that can also dephosphorylate 3′-phosphorylated mononucleotides (12), it is conceivable that the AtZDP fingers play a role in retaining the protein at damaged DNA sites and in limiting unspecific 3′-phosphate nucleotide dephosphorylation.
PARP-like fingers and single-strand DNA break repair
The results presented in this work clearly show that the AtZDP-catalysed removal of 3′ blocking groups from naked DNA is independent from DNA binding by PARP-like-fingers, and that AtZDP fingers can bind to both substrates and products of the AtZDP catalysed reaction. It thus appears that the enzyme can sense and repair damaged DNA independently from the PARP-like fingers and that the fingers might serve additional functions. Possibly, they can compete for nucleosome assembly onto damaged DNA until the completion of the repair process, and/or they might prevent the erroneous access of exonucleases and of recombination proteins. In agreement with that, in vitro and in vivo experiments have shown that the damage recognition by fingered PARPs does not provide a necessary step in DNA repair, but PARPs activity does increase the efficiency of the repair process, while decreasing the frequency of sister chromatid exchange and of homologous recombination (5,9,29). In fact, the finding that bulges within double-strand DNAs can be efficiently recognised by PARP-like fingers opens up the possibility that they can also play a role during non-homologous recombination processes, as well as at other DNA sites where DNA rearrangements are taking place.
Within a putative plant repair complex, we could envisage that, following 3′ end repair, the three-fingered plant AtZDP remains at gapped DNA sites where it favours the access of other repair enzymes. Indeed, the putative plant homologue of the scaffold protein XRCC1 does not contain a strand-break sensor, nor are there PARP-like fingered ligases encoded in the plant genome. In a similar scenario in animal cells, XRCC1 and/or ligase III, bearing strand-break sensors, could anchor the repair complex at damaged DNA sites. Repair complexes could then bind either before or after poly(ADP-ribosyl)ation by PARPs, depending on their availability and/or relative affinity for the damaged DNAs. Indeed, in vivo data using animal cells show that the over-expression of isolated PARPs’ fingers is inhibitory for DNA repair processes (30,31), and that XRCC1 and ligase III can inhibit the activity of PARPs when present in excess (3,32). Possibly, PARPs function by a ‘hit and run’-like mechanism, with PARPs clearing out damaged sites by protein poly(ADP-ribosyl)ation, but not being necessarily bound in order to direct the entry of a repair complex. Very recent evidence about spatial and temporal responses to single-strand breaks in human cells is consistent with such a mechanism (33). Moreover, similarly to the nucleotide excision repair systems (34), also in the case of the single-strand repair complex a flexible assembly of repair proteins could be preferred over a rigidly pre-assembled repairosome.
An extensive comparison between the in vivo binding properties of the different DNA-damage sensors is obviously required to provide final experimental support to these hypotheses.
Acknowledgments
ACKNOWLEDGEMENTS
I gratefully acknowledge Riccardo Percudani for his skilled assistance with sequence analyses, Alessio Peracchi, Simone Ottonello and Giorgio Dieci for critical reading of the manuscript, and Roberto Tirindelli for helpful discussions and continuous support. I am grateful to Gian Luigi Rossi for encouragement and assistance. This work was supported by the National Research Council of Italy (CNR) and by the Ministero dell’Istruzione dell’Università e della Ricerca (MIUR).
REFERENCES
- 1.Gradwohl G., Menissier de Murcia,J.M., Molinete,M., Simonin,F., Koken,M., Hoeijmakers,J.H. and de Murcia,G. (1990) The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc. Natl Acad. Sci. USA, 87, 2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikejima M., Noguchi,S., Yamashita,R., Ogura,T., Sugimura,T., Gill,D.M. and Miwa,M. (1990) The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J. Biol. Chem., 265, 21907–21913. [PubMed] [Google Scholar]
- 3.Caldecott K.W., Aoufouchi,S., Johnson,P. and Shall,S. (1996) XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res., 24, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackey Z.B., Niedergang,C., Murcia,J.M., Leppard,J., Au,K., Chen,J., de Murcia,G. and Tomkinson,A.E. (1999) DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J. Biol. Chem., 274, 21679–21687. [DOI] [PubMed] [Google Scholar]
- 5.D'Amours D., Desnoyers,S., D’Silva,I. and Poirier,G.G. (1999) Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J., 342, 249–268. [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor R.M., Whitehouse,C.J. and Caldecott,K.W. (2000) The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining. Nucleic Acids Res., 28, 3558–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrucco S., Volpi,G., Bolchi,A., Rivetti,C. and Ottonello,S. (2002) A nick-sensing DNA 3′-repair enzyme from Arabidopsis. J. Biol. Chem., 277, 23675–23683. [DOI] [PubMed] [Google Scholar]
- 8.de Murcia G. and Menissier de Murcia,J. (1994) Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci., 19, 172–176. [DOI] [PubMed] [Google Scholar]
- 9.Shall S. and de Murcia,G. (2000) Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res., 460, 1–15. [DOI] [PubMed] [Google Scholar]
- 10.Smith S. (2001) The world according to PARP. Trends Biochem. Sci., 26, 174–179. [DOI] [PubMed] [Google Scholar]
- 11.Whitehouse C.J., Taylor,R.M., Thistlethwaite,A., Zhang,H., Karimi-Busheri,F., Lasko,D.D., Weinfeld,M. and Caldecott,K.W. (2001) XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell, 104, 107–117. [DOI] [PubMed] [Google Scholar]
- 12.Betti M., Petrucco,S., Bolchi,A., Dieci,G. and Ottonello,S. (2001) A plant 3′-phosphoesterase involved in the repair of DNA strand breaks generated by oxidative damage. J. Biol. Chem., 276, 18038–18045. [DOI] [PubMed] [Google Scholar]
- 13.Ward J.E. (1998) DNA repair in higher eukaryotes. In Nickoloff,J.A. and Hoekstra,M.F. (eds), DNA Damage and Repair. Human Press, Inc., Totowa, NJ, Vol. 2, pp. 65–84. [Google Scholar]
- 14.Divine K.K., Gilliland,F.D., Crowell,R.E., Stidley,C.A., Bocklage,T.J., Cook,D.L. and Belinsky,S.A. (2001) The XRCC1 399 glutamine allele is a risk factor for adenocarcinoma of the lung. Mutat. Res., 461, 273–278. [DOI] [PubMed] [Google Scholar]
- 15.Sturgis E.M., Castillo,E.J., Li,L., Zheng,R., Eicher,S.A., Clayman,G.L., Strom,S.S., Spitz,M.R. and Wei,Q. (1999) Polymorphisms of DNA repair gene XRCC1 in squamous cell carcinoma of the head and neck. Carcinogenesis, 20, 2125–2129. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg E.C. (1995) Out of the shadows and into the light: the emergence of DNA repair. Trends Biochem. Sci., 20, 381. [DOI] [PubMed] [Google Scholar]
- 17.Sancar A. (1995) DNA repair in humans. Annu. Rev. Genet., 29, 69–105. [DOI] [PubMed] [Google Scholar]
- 18.Caldecott K.W. (2003) DNA single-strand break repair and spinocerebellar ataxia. Cell, 112, 7–10. [DOI] [PubMed] [Google Scholar]
- 19.Hoeijmakers J.H. (2001) Genome maintenance mechanisms for preventing cancer. Nature, 411, 366–374. [DOI] [PubMed] [Google Scholar]
- 20.Marintchev A., Mullen,M.A., Maciejewski,M.W., Pan,B., Gryk,M.R. and Mullen,G.P. (1999) Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nature Struct. Biol., 6, 884–893. [DOI] [PubMed] [Google Scholar]
- 21.Babiychuk E., Cottrill,P.B., Storozhenko,S., Fuangthong,M., Chen,Y., O’Farrell,M.K., Van Montagu,M., Inze,D. and Kushnir,S. (1998) Higher plants possess two structurally different poly(ADP-ribose) polymerases. Plant J., 15, 635–645. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholas K.B. and Nicholas,H.B.,JR (1997) GeneDoc: a tool for editing and annotating multiple sequence alignments. http://www.psc.edu/biomed/genedoc/index.html.
- 24.Karimi-Busheri F., Daly,G., Robins,P., Canas,B., Pappin,D.J., Sgouros,J., Miller,G.G., Fakhrai,H., Davis,E.M., Le Beau,M.M. et al. (1999) Molecular characterization of a human DNA kinase. J. Biol. Chem., 274, 24187–24194. [DOI] [PubMed] [Google Scholar]
- 25.LeCam E., Fack,F., Menissier-de Murcia,J., Cognet,J.A., Barbin,A., Sarantoglou,V., Revet,B., Delain,E. and de Murcia,G. (1994) Conformational analysis of a 139 base-pair DNA fragment containing a single-stranded break and its interaction with human poly(ADP-ribose) polymerase. J. Mol. Biol., 235, 1062–1071. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y.H., Barker,P. and Griffith,J. (1992) Visualization of diagnostic heteroduplex DNAs from cystic fibrosis deletion heterozygotes provides an estimate of the kinking of DNA by bulged bases. J. Biol. Chem., 267, 4911–4915. [PubMed] [Google Scholar]
- 27.Soldatenkov V.A., Trofimova,I.N., Rouzaut,A., McDermott,F., Dritschilo,A. and Notario,V. (2002) Differential regulation of the response to DNA damage in Ewing’s sarcoma cells by ETS1 and EWS/FLI-1. Oncogene, 21, 2890–2895. [DOI] [PubMed] [Google Scholar]
- 28.Sastry S.S., Buki,K.G. and Kun,E. (1989) Binding of adenosine diphosphoribosyltransferase to the termini and internal regions of linear DNAs. Biochemistry, 28, 5670–5680. [DOI] [PubMed] [Google Scholar]
- 29.Dantzer F., Schreiber,V., Niedergang,C., Trucco,C., Flatter,E., De La Rubia,G., Oliver,J., Rolli,V., Menissier-de Murcia,J. and de Murcia,G. (1999) Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie, 81, 69–75. [DOI] [PubMed] [Google Scholar]
- 30.Molinete M., Vermeulen,W., Burkle,A., Menissier-de Murcia,J., Kupper,J.H., Hoeijmakers,J.H. and de Murcia,G. (1993) Overproduction of the poly(ADP-ribose) polymerase DNA-binding domain blocks alkylation-induced DNA repair synthesis in mammalian cells. EMBO J., 12, 2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber V., Hunting,D., Trucco,C., Gowans,B., Grunwald,D., De Murcia,G. and De Murcia,J.M. (1995) A dominant-negative mutant of human poly(ADP-ribose) polymerase affects cell recovery, apoptosis and sister chromatid exchange following DNA damage. Proc. Natl Acad. Sci. USA, 92, 4753–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masson M., Niedergang,C., Schreiber,V., Muller,S., Menissier-de Murcia,J. and de Murcia,G. (1998) XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol., 18, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okano S., Lan,L., Caldecott,K.W., Mori,T. and Yasui,A. (2003) Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol., 23, 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volker M., Mone,M.J., Karmakar,P., van Hoffen,A., Schul,W., Vermeulen,W., Hoeijmakers,J.H., van Driel,R., van Zeeland,A.A. and Mullenders,L.H. (2001) Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell, 8, 213–224. [DOI] [PubMed] [Google Scholar]