Abstract
Germ cells must safeguard, apportion, package, and deliver their genomes with exquisite precision to ensure proper reproduction and embryonic development. Classical genetic approaches have identified many genes controlling animal germ cell development, but only recently have some of these genes been linked to the RNA interference (RNAi) pathway, a gene silencing mechanism centered on small regulatory RNAs. Germ cells contain microRNAs (miRNAs), endogenous siRNAs (endo-siRNAs), and Piwi-interacting RNAs (piRNAs); these are bound by members of the Piwi/Argonaute protein family. piwi genes were known to specify germ cell development, but we now understand that mutations disrupting germline development can also affect small RNA accumulation. Small RNA studies in germ cells have revealed a surprising diversity of regulatory mechanisms and a unifying function for germline genes in controlling the spread of transposable elements. Future challenges will be to understand the production of germline small RNAs and to identify the full breadth of gene regulation by these RNAs. Progress in this area will likely impact biomedical goals of manipulating stem cells and preventing diseases caused by the transposition of mobile DNA elements.
Introduction
Small RNAs and the RNAi pathway are key components of gene regulation systems in eukaryotic cells. In animals, the proper development of somatic tissues is intimately dependent on a functioning RNAi pathway (Ambros, 2004). Although germ cells are uniquely distinct from somatic cells because they form gametes, germ cell development also appears to require RNAi. However, germ cells exhibit an unanticipated diversity of RNAi mechanisms and have become the richest environment for the discovery of small regulatory RNAs.
MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) are two prominent classes of small regulatory RNAs in eukaryotes (Figure 1). While miRNAs regulate many endogenous target messenger RNAs mainly through gene silencing (Bartel, 2009, Carthew and Sontheimer, 2009), siRNAs are thought to provide organisms with innate defenses against viruses and transposable elements (TEs or transposons). miRNAs were largely first characterized in somatic cells, but it was anticipated that germ cells would express certain miRNAs specific to the germline (Mishima et al., 2008, Murchison et al., 2007, Ro et al., 2007, Watanabe et al., 2006). However, the surprises lurking in germ cells were endogenous siRNAs (endo-siRNAs) not previously seen in somatic cells, and a germline-specific, abundant class of longer small RNAs called Piwi-interacting RNAs (piRNAs). Despite their abundance, piRNAs and endo-siRNAs have only recently been unveiled, and many aspects of their molecular functions are still undefined.
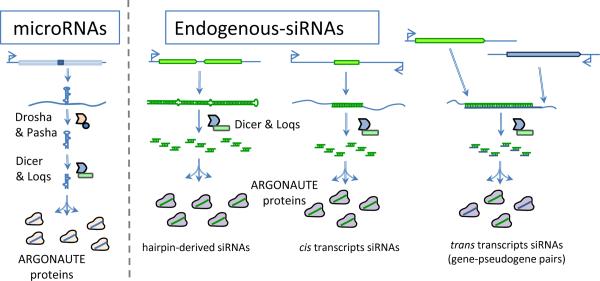
Figure 1.
Gonadal small RNAs that are generated by Dicer. MicroRNAs (miRNAs) and endogenous small interfering RNAs (endo-siRNAs) are processed into mature single-stranded RNAs that are incorporated into Argonaute (AGO) proteins.
To date, nearly all small regulatory RNAs function in a complex with an Argonaute (AGO) or PIWI protein. Most animal genomes contain multiple homologs of these proteins. Each possesses a PAZ domain, which binds the 3' end of small RNAs, and a PIWI domain, which folds into an RNase H-like fold and can cleave a target RNA strand that is matched to the guide RNA bound in the domain (“Slicer” activity, Tolia and Joshua-Tor, 2007). AGO proteins prefer to bind ~20–23 nucleotide (nt) miRNAs and siRNAs, while PIWI proteins prefer to bind ~24–31 nt long Piwi-interacting RNAs (piRNAs). miRNAs and siRNAs are processed by the Dicer endonuclease from precursors with double-stranded RNA (dsRNA) features; piRNAs are distinct because they are typically longer in length, their precursors, in general, lack dsRNA features, and their production does not require Dicer (Ghildiyal and Zamore, 2009).
Several recent reviews discuss the discovery of these small regulatory RNAs (Ghildiyal and Zamore, 2009, Malone and Hannon, 2009, Klattenhoff and Theurkauf, 2008). The miRNA and siRNA pathways have also been recently reviewed (Carthew and Sontheimer, 2009, Kim et al., 2009, Okamura and Lai, 2008). Therefore, this germlinecentric review of small regulatory RNAs will focus on the two newest classes of germ cell small RNAs: piRNAs and endo-siRNAs. I will discuss the history of how germline development studies have now intersected with the biology of small regulatory RNAs. Finally, I will address the current challenges in understanding small RNA biology within the context of germ cells and propose that germ-cell focused studies will have broad implications for understanding animal health and evolution.
Where it began: RNAi and the nematode germline
Our understanding of small RNA-mediated gene regulation in animals can be traced to pioneering discoveries in the nematode Caenorhabditis elegans (C. elegans), such as the revelation that dsRNA triggers RNAi (Fire et al., 1998) and that the developmental regulator genes lin-4 and let-7 encode miRNAs (Lee et al., 1993, Wightman et al., 1993, Reinhart et al., 2000). In C. elegans, it was shown that RNAi can be induced simply by feeding the nematodes bacteria expressing dsRNA, and that this effect persists into progeny long after the source of the dsRNA trigger is removed, suggesting transmission and amplification of siRNAs in the germline (Grishok et al., 2000, Sijen et al., 2001, Timmons and Fire, 1998).
The Plasterk laboratory isolated nematode mutants called mutators that displayed elevated mobilization of transposons; some of these mutants, such as mut-7, displayed a temperature-dependent sterility that was most evident when the homozygous mutation was carried by the hermaphrodite, suggesting that the MUT-7 protein was a maternally-deposited gene product (Ketting et al., 1999). Both the Mello and Plasterk laboratories found that some mutator strains were unable to mount an RNAi response when injected with dsRNA, while some RNAi-deficient (rde) mutant strains, such as rde-3, likewise displayed elevated transposon mobilization and were allelic with mutator genes (Ketting et al., 1999, Tabara et al., 1999).
In a different approach, the Maine laboratory isolated a C. elegans mutant that was severely defective in oogenesis and carried mutations in the gene ego-1, a homolog of plant RNA-dependent RNA polymerases (RdRP, Smardon et al., 2000). Plant and fungal RdRP genes are necessary for mounting an RNAi response to defend against viruses (Chen, 2009), while in nematodes, ego-1 was found to be necessary for proper meiosis and mitosis, as well as for responding to dsRNA to initiate RNAi. In addition to ego-1, three other RdRP homologs are encoded in the C. elegans genome: rrf-1, -2, and -3, and mutations in these genes all exhibit a dampened gene silencing response (Smardon et al., 2000). Converging on these RdRP genes as well as uncovering new genes, the Ruvkun and Plasterk laboratories discovered genes that enhance or suppress the RNAi response in somatic cells (Kennedy et al., 2004, Kim et al., 2005, Simmer et al., 2002). For example, mutations in rrf-3 and eri-1 enhance RNAi in neurons, and curiously cause a temperature-dependent sterility defect, analogous to the phenotype seen in mut-7.
Although the links between RNAi, transposon regulation, and germline development in the nematode were not clear at the time, an emerging hypothesis was that RNA-based gene silencing phenomena played a role in germ cell formation through putative endogenous siRNAs and protein factors. It was thought that while RNAi triggered by exogenous dsRNAs was robust in many somatic cells, it was also dampened when competing with endogenous RNAi pathways in the germline. Thus, mutations in endogenous RNAi pathways that lowered endo-siRNA levels were thought to liberate common limiting RNAi factors to be used for effective exogenous siRNA-mediated silencing.
Formal characterization of endo-siRNAs from C. elegans lagged behind the broader discovery of miRNAs, in part because the earliest small RNA cloning methods focused on the 5' monophosphate of miRNAs and exogenous siRNAs. However, the endo-siRNAs in C. elegans became apparent with cloning techniques that captured RNAs regardless of the status of the small RNAs' 5' end (Ambros et al., 2003, Lee et al., 2006, Pak and Fire, 2007) and in identification of new factors involved in small RNA biogenesis (Duchaine et al., 2006). An examination of secondary siRNA molecules generated by RdRPs in nematodes indicated that they contain 5' di- and triphosphates, and might be unprimed RdRP products from mRNAs (Pak and Fire, 2007, Sijen et al., 2007). Finally, a deep-sequencing study by the Bartel laboratory suggested that endo-siRNA populations are diverse in C. elegans, with 21-, 22-, and 26-nt species that preferentially start with a 5' guanosine (Ruby et al., 2006).
To further understand the repertoire of endo-siRNAs in the C. elegans germline, deep-sequencing efforts have recently been applied to embryos, gametes, mutants affecting germline development, and immunoprecipitates of nematode AGO proteins resident in the gonads (Claycomb et al., 2009, Gu et al., 2009, Han et al., 2009, van Wolfswinkel et al., 2009). These studies codified two classes of germline endo-siRNAs, 26G RNAs and 22G RNAs (Figure 2), which are distinct in their length, the factors that generate them, and the AGO proteins that bind them. Both 26G and 22G RNAs require an RdRP that likely synthesizes an unprimed antisense transcript to certain mRNAs expressed throughout the genome; however, the mechanism for selecting the specific mRNAs is currently unknown.
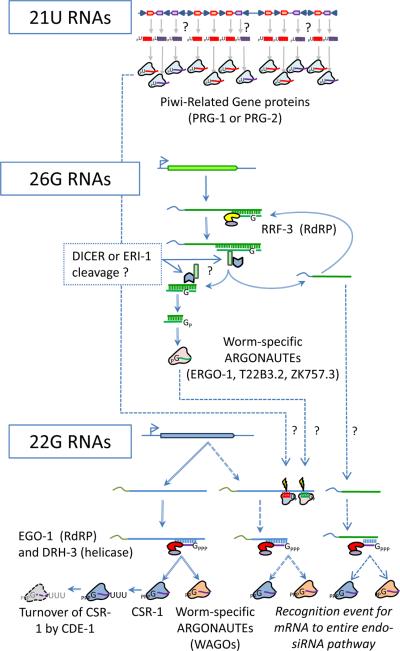
Figure 2.
Gonadal small RNAs specific to the nematode Caenorhabditis elegans. In line with the expanded number of Argonaute-family proteins in the C. elegans genome, this nematode's gonad also contains an additional layer of small regulatory RNAs besides miRNAs and siRNAs. 21U RNAs may be orthologous to Piwi-interacting RNAs (piRNAs) based on the homology of Piwi-Related Gene-1 and -2 proteins to PIWI proteins of other animals. 26G and 22G RNAs are siRNAs most abundantly detected in gonads, and appear to require an RNA-dependent RNA Polymerase (RdRP) for initial steps of biogenesis. These RNAs are named after their signature length and the predominant 5' nucleotide in that class. While 21U and 26G RNAs likely often contain a 5' monophosphate, 22G RNAs predominantly contain a 5' di- or tri-phosphate. The model here speculates that 26G and 21U RNAs might serve as primary siRNAs/piRNAs that then promote generation of secondary siRNAs (22G RNAs).
Not just nematodes: insects and vertebrate gonadal endo-siRNAs
The first discovered miRNA, lin-4, was once thought to be unique to C. elegans because of lack of obvious animal homologs; this assumption may have also been extended to endo-siRNAs because higher animals lack an endogenous RdRP homologous to EGO-1 or RRF-1. Nevertheless, long dsRNAs that are substrates for endo-siRNA generation can be formed by single-stranded RNA precursors without an RdRP (Figure 1). However, most mammalian cells mount an aggressive innate immune response to intracellular dsRNA that is indicative of a viral infection (Interferon response, Sen and Sarkar, 2007), with the notable exception being germ cells, which do not activate the response when injected with dsRNA (Svoboda et al., 2000).
Using deep sequencing of small RNAs from many thousands of mouse oocytes, the Hannon and Watanabe laboratories identified not only many known miRNAs and some piRNAs, but also a substantial number of endo-siRNAs (Tam et al., 2008, Watanabe et al., 2008). The mouse oocyte endo-siRNAs are mainly 21–22 nt long, of which the majority map to transposons; in addition, a significant number can also be mapped to pseudogenes that either contain a large inverted repeat (e.g. the Ran-GAP pseudogene) or show extensive complementarity to an endogenous gene (e.g. Ppp4r1 and its pseudogene) (Figure 1). The groups postulated that these gene configurations produce dsRNA precursors that are processed by Dicer to generate endo-siRNAs that are loaded into the Argonaute-2 (Ago-2) complex (Tam et al., 2008, Watanabe et al., 2008).
To validate a function for oocyte endo-siRNAs, mRNAs were profiled in dicer-null and ago-2-null oocytes. Predicted target transcripts like Ran-GAP and Ppp4r1 were up-regulated 2–4 fold in mutant oocytes. Ran-GAP is a protein implicated in controlling the molecular gradient of Ran-GTP, which mediates spindle organization (Kalab and Heald, 2008). The substantial number of endo-siRNAs that target genes with microtubule-associated functions, like Ran-GAP, as well as the pronounced defects in meiotic spindle formation previously observed in dicer-null oocytes (Murchison et al., 2007, Tang et al., 2007), have led to the suggestion that RNAi can modulate the microtubule cytoskeleton, although this remains to be tested molecularly (Tam et al., 2008, Watanabe et al., 2008).
Shortly after mouse oocyte endo-siRNAs were described, multiple reports followed with deep sequencing of endo-siRNAs from Drosophila melanogaster (D. melanogaster) (Czech et al., 2008, Ghildiyal et al., 2008, Kawamura et al., 2008, Okamura et al., 2008a, Okamura et al., 2008b, Chung et al., 2008). Intriguingly, endo-siRNAs were detected not only in ovaries, but also in somatic cells like fly heads or Schneider-2 cultured cells, which are regarded as fly somatic cells of embryonic origin (Czech et al., 2008, Ghildiyal et al., 2008, Okamura et al., 2008a, Okamura et al., 2008b). Nuances on the origin and biogenesis of fly endo-siRNAs in somatic cells have been reviewed (Kim et al., 2009, Okamura and Lai, 2008, Ghildiyal and Zamore, 2009).
Interestingly, many endo-siRNAs from germline and somatic cells map to the same transposons that are targeted by piRNAs (e.g. mdg1, 297, blood, and others; Chung et al., 2008, Czech et al., 2008, Ghildiyal et al., 2008, Kawamura et al., 2008). When taking into account only uniquely mapping TE-associated siRNAs or normalizing TE-associated siRNA reads to the number of genomic loci, many endo-siRNA matches still accumulate on piRNA cluster loci (Chung et al., 2008, Czech et al., 2008). However, the number of endo-siRNAs mapping to piRNA clusters can be estimated to be an order of magnitude lower than piRNAs, and the distribution of piRNAs versus siRNAs mapping to consensus TE sequences differ significantly. For example, over 90% of the piRNAs corresponding to mdg1 and blood are antisense matches, while only 60% of the endo-siRNAs corresponding to these elements are antisense matches (Chung et al., 2008). It is unclear if endo-siRNAs are derived from the same transcriptional loci as piRNAs.
A small piece of piRNA background
Studies in fly and zebrafish embryos were the first to describe small RNAs complementary to repetitive transposable elements in animals; these were named repeat-associated siRNAs (rasiRNAs) (Aravin et al., 2001, Elbashir et al., 2001, Aravin et al., 2003, Chen et al., 2005). These small RNAs, as a class, have been reclassified as Piwi-interacting RNAs (piRNAs) because these rasiRNAs and other small RNAs co-purify with PIWI-group proteins from flies and mammals (Aravin et al., 2007a, Malone and Hannon, 2009, Ghildiyal and Zamore, 2009). The revolution in characterizing piRNAs has been next-generation deep sequencing technology (Morozova et al., 2009), which has revealed the extraordinary sequence diversity of individual piRNAs in flies and mammals. Recently, piRNAs have also been deeply sequenced in zebrafish, frog, monotremes, silkworm, sponges, and sea anemone (Grimson et al., 2008, Murchison et al., 2008, Armisen et al., 2009, Houwing et al., 2007, Kawaoka et al., 2009, Lau et al., 2009a), but studies in fly and mouse have yielded most of our understanding of piRNAs and their genomic origins.
Piwi-interacting RNAs can be classified into two main types based on where the piRNAs originate in the genome (Figure 3). Class I piRNAs predominantly begin with a 5' uridine (U), and generally map uniquely as clusters to genomic regions with a pronounced strand bias, such that single-stranded precursors ranging from 1–100 kilobases (kb) are thought to give rise to a huge variety of piRNAs. These piRNAs are typically abundant in adult rodent spermatocytes and spermatids when the male germ cells are progressing through the pachytene stage of meiosis (Aravin et al., 2006, Girard et al., 2006, Lau et al., 2006). In mammals, some of these class I piRNA clusters exhibit a bi-directional configuration that implies divergent transcription, with a 0.1–1kb region separating the two domains of piRNAs (Aravin et al., 2006, Girard et al., 2006, Lau et al., 2006). Mapping of class I piRNAs in D. melanogaster also revealed large strand-biased clusters of piRNAs described as “master control loci”, since mutations near these loci have broad effects on piRNA production and transposon control (Brennecke et al., 2007). However, these loci also contribute to the production of class II piRNAs because many transposon relics have accumulated in these regions.
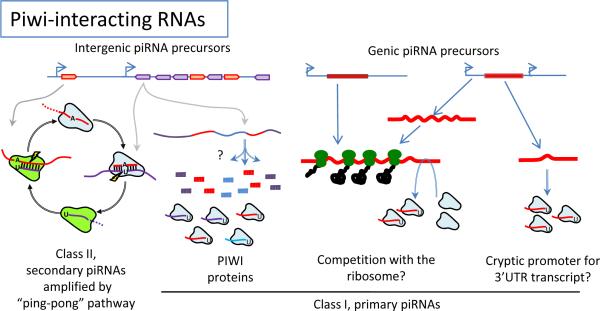
Figure 3.
Classification of Piwi-interacting RNAs. Multi-kilobase clusters of piRNAs lying in intergenic regions give rise to both class I (primary) piRNAs and class II (secondary) piRNAs. Class II piRNAs are thought to be generated by a “ping-pong” mechanism, whereas biogenesis of class I piRNAs is unclear. However, a group of class I piRNAs derived from mRNAs (genic piRNA precursors) appear to preferentially arise from the 3' untranslated region (3' UTRs) of the mRNAs, suggesting PIWI proteins might compete with the ribosome for access to mRNAs or cryptic promoters may reside upstream of certain 3' UTRs that promote transcription of an independent transcript.
Class II piRNAs generally map to multiple genomic loci because they have a higher propensity to match repetitive elements like transposons. These piRNAs are composed of one form that often begins with a 5' U and is antisense to TE messages, while a second form contains adenine (A) at position 10 and is the same sense strand as TE messages (Figure 3). The class II piRNA forms can base-pair to each other in the first 10 nt, such that the U of the antisense piRNA matches the A of the sense piRNA. In D. melanogaster, one PIWI homolog, Aubergine (Aub), mainly binds piRNAs that are antisense to TEs, while piRNAs that are sense to TEs are predominantly bound by another piRNA-binding protein, Argonaute-3 (AGO3) (Brennecke et al., 2007, Gunawardane et al., 2007).
When Aub-piRNA complexes seek transcripts from active TEs, the Slicer activity in Aub that is guided by the piRNA can cleave the TE transcript to define the 5' end of a new piRNA for incorporation into AGO3. The AGO3-piRNA complex reciprocates on a transcript made from master control loci to define the 5' ends of piRNAs incorporated into Aub. Additional processing events not yet well understood help define the 3' end of the piRNA. This circular model has been called the “ping-pong” amplification mechanism (Figure 3), because two PIWI proteins “ping-pong” off of each other to amplify piRNA biogenesis (Kim et al., 2009, Malone and Hannon, 2009). Although Aub and AGO3 clearly bind class II piRNAs, it is not clear whether Piwi, the founding member of PIWI proteins, prefers class I over class II piRNAs. However, its piRNAs are also antisense to TEs and can accumulate abundantly even when AGO3-associated piRNAs are poorly expressed or absent (Gunawardane et al., 2007, Lau et al., 2009b).
Mammals also contain three PIWI homologs, and in mice the Miwi, Mili, and Miwi2 proteins are all expressed in the male germline, whereas oocytes appear to express only Mili (Tam et al., 2008, Watanabe et al., 2008). In early stages of spermatogenesis, Mili, Miwi2, and class II piRNAs are expressed in the primordial germ cells that develop and divide by mitosis into primary spermatogonia (Aravin et al., 2003, Aravin et al., 2008, Carmell et al., 2007, Aravin et al., 2007b). When spermatocytes enter meiosis to become spermatids, they express Miwi as well as a burst of class I piRNAs. Class II piRNAs are generally more difficult to detect in mouse, perhaps due to the small number of early germ cells, whereas class I piRNAs in adult mouse testes are so abundant that they can be detected by ethidium bromide staining (Aravin et al., 2006, Girard et al., 2006). In contrast to fly master control loci, which yield both class I and class II piRNAs, the clusters of mouse class II piRNAs are often distinct, more numerous, and smaller in genomic coverage than the class I piRNA clusters (Aravin et al., 2008, Aravin et al., 2007b, Kuramochi-Miyagawa et al., 2008)
Recently, new distinctions in class I piRNAs have come into focus in flies and mice. In contrast to adult mouse testes, D. melanogaster ovaries express abundant levels of class II piRNAs, of which many are maternally deposited into the embryo. Three groups using different approaches to partition class II piRNAs have determined that a cohort of class I piRNAs are present in the somatic follicle cells of the ovary, and many derive from a master control locus called flamenco (Lau et al., 2009b, Li et al., 2009, Malone et al., 2009). The flamenco piRNAs and bulk of class I piRNA clusters in mouse adult testes can be considered intergenic piRNA clusters. The Lau and Lai laboratories have now observed that significant numbers of class I piRNAs across diverse animals can also derive from many specific mRNAs, such that the piRNAs preferentially map to the 3' UTRs of the mRNAs (Robine et al., 2009). These recent findings underlie additional complexities in this germline gene regulatory pathway that remain to be uncovered.
Nematode piRNAs are different: 21U RNAs
Most animal piRNAs conform to the categories of class I and class II types; however, an atypical class of piRNAs were revealed in C. elegans and named 21U RNAs because of their uniform 21 nt length, their initial 5'U, and their genomic origin as a cohort from large domains of chromosome IV (Ruby et al., 2006). A consistent motif upstream of the sequence encoding the 21U RNAs was detected for each individual 21U specimen, but whether this motif serves as a transcription promoter or a processing signal is currently unknown.
At first glance, 21U RNAs might seem like endo-siRNAs in their length and the apparent derivation from either genomic strand, a hallmark for a dsRNA precursor. However, three groups recently demonstrated that 21U RNAs are actually Piwi-interacting RNAs: mutants lacking functional Piwi-related gene-1 (prg-1) lack 21U RNAs (Batista et al., 2008, Das et al., 2008, Wang and Reinke, 2008), the immunoprecipitates (IPs) of PRG-1 contain these small RNAs, and the biogenesis of 21U RNAs is Dicer-independent (Batista et al., 2008, Das et al., 2008). Although the length and genomic characteristics of 21U RNAs are quite divergent from other animal piRNAs, PRG-1 exhibits many similarities with PIWI proteins besides amino acid sequence similarity. PRG-1 expression is germline-restricted and concentrated in germ cell foci called P-granules, and mutants exhibit temperature-sensitive sterility due to germ cell loss (Batista et al., 2008, Das et al., 2008, Wang and Reinke, 2008). Curiously, loss of 21U RNA expression only up-regulates the Tc3 class of transposons in the worm germline (see below, Batista et al., 2008, Das et al., 2008). Thus, 21U RNAs and their loci from chromosome IV represent an extreme configuration of a piRNA class.
Are we near the saturation point for discovery of small RNAs? Perhaps not, since abundant RNAs masked the discovery of the less abundant species in past deep-sequencing efforts. In mice, the class I piRNAs obscured initial detection of class II piRNAs, but class II piRNAs became apparent in neonatal and young testes libraries, when class I piRNAs were absent (Aravin et al., 2007b, Aravin et al., 2008, Kuramochi-Miyagawa et al., 2008). The Zamore laboratory has suggested a class of “piRNA-like” RNAs detectable in D. melanogaster heads when ago-2 is genetically ablated and the “background” of endo-siRNAs has been cleared away (Ghildiyal et al., 2008). Finally, a new class of transcription start site small RNAs have been uncovered in mouse embryonic stem (ES) cells, but these species represent a miniscule proportion of the sequenced libraries (Seila et al., 2008, Fejes-Toth K, 2009). Increasing the depth of library sequencing may be a straightforward approach to future small RNA discovery. However, focused library construction from specific stages or cell types, organelles, or biochemical fractions of ribonucleoproteins will also yield insight into the diversity of small RNAs.
The riddles of germline small RNA biogenesis
Although our picture of miRNA/siRNA biogenesis is detailed (Figures 1 and 2; Kim et al., 2009), we only have a few insights into piRNA biogenesis (Figure 3). Studies in flies, zebrafish, and nematodes demonstrated that piRNA production does not require Dicer (Vagin et al., 2006, Houwing et al., 2007, Batista et al., 2008, Das et al., 2008); this suggests that piRNA biogenesis is uncoupled from miRNA and siRNA biogenesis in animals. The ping-pong model for amplification of piRNA biogenesis explains why piRNA biogenesis does not require Dicer and how the prevalent 5' U of piRNAs can be defined (Brennecke et al., 2007, Gunawardane et al., 2007). Nevertheless, several questions remain: How does efficient primary piRNA production occur without the ping-pong mechanism? How exactly are piRNA 3' ends defined? How are piRNA precursor transcripts selected? It has been hypothesized that the nuclease domain-containing proteins Zuc and Squ might play a role in defining the 3' end of piRNAs; however, this remains to be molecularly demonstrated (Pane et al., 2007, Ghildiyal and Zamore, 2009, Klattenhoff and Theurkauf, 2008, Li et al., 2009).
Interestingly, the 3' ends of piRNAs (and endo-siRNAs) in all animals examined are processed to contain a 2' hydroxyl (2'OMe) modification (Houwing et al., 2007, Kirino and Mourelatos, 2007, Ohara et al., 2007, Vagin et al., 2006, Ruby et al., 2006, Grimson et al., 2008). In D. melanogaster, this is mediated by a ribose methylase called DmHen1/Pimet (Horwich et al., 2007, Saito et al., 2007). Although a 2'OMe modification of plant siRNAs and miRNAs is necessary to stabilize these small RNAs (Yu et al., 2005), the role of DmHen1/Pimet and the 2'OMe modification is unclear because the phenotypes of the null mutant are modest (Horwich et al., 2007, Saito et al., 2007).
Many PIWI-associated factors and genes genetically linked with piRNA biogenesis have been described, but their direct impact on piRNA function still needs to be evaluated. For example, among the proteins encoded by the fly genes implicated in piRNA biogenesis (the RNA helicases vasa, spn-E, and armi; the putative nucleases zuc and squ; and nuage components krimp and mael; Klattenhoff et al., 2007, Lim and Kai, 2007, Pane et al., 2007, Vagin et al., 2006), only Vasa, Zuc and Squ have been shown to interact with Aub (Megosh et al., 2006, Thomson et al., 2008, Pane et al., 2007). The mouse homologs of vasa and mael have also been genetically implicated in piRNA biogenesis (Kuramochi-Miyagawa et al., 2004, Soper et al., 2008), while several candidate PIWI-associated factors in mice (RecQ1, Kif17b, eIF3a) await further physiological characterization (Kotaja et al., 2006, Lau et al., 2006, Unhavaithaya et al., 2008).
The interest in PIWI proteins has set off a race to identify additional interacting factors. Recently, several laboratories performing proteomics of PIWI protein complexes from fly, frog, and mouse germ cells have all simultaneously identified a protein factor with single or multiple Tudor domain(s) (Kirino et al., 2009a, Kirino et al., 2009b, Lau et al., 2009a, Reuter et al., 2009, Vagin et al., 2009, Chen et al., 2009, Nishida et al., 2009, Vasileva et al., 2009, Wang et al., 2009). The founding member of the Tudor gene family is D. melanogaster tudor, some mutants of which produce sterile progeny and display abnormal germ cell development due to improper localization of maternal components (Thomson and Lasko, 2005). The structure and biochemical function of the Tudor protein domain itself has been shown to bind symmetrically dimethylated arginines (sDMAs; Thomson and Lasko, 2005), and the presence of multiple sDMAs has been determined in the N-terminal regions of Mili, Miwi, Aub, AGO3, and Piwi (Chen et al., 2009, Kirino et al., 2009a, Nishida et al., 2009, Reuter et al., 2009, Vagin et al., 2009). The association of Tudor proteins and PIWI proteins may be important for the localization of the PIWI/piRNA complex to germ plasm in oocytes or the chromatoid body in mammalian sperm (Kirino et al., 2009a, Kirino et al., 2009b, Lau et al., 2009a, Nishida et al., 2009, Reuter et al., 2009, Vagin et al., 2009, Vasileva et al., 2009, Wang et al., 2009). However, ablation of individual Tudor genes in flies and mice causes varied reduction of piRNA biogenesis and PIWI protein stability (Kirino et al., 2009a, Kirino et al., 2009b, Nishida et al., 2009, Reuter et al., 2009, Vagin et al., 2009). Thus the molecular function of D. melanogaster Tudor and vertebrate Tudor homologs remains to be fully elucidated.
Given the diversity of small RNA types in animal gonads, several conundrums vex the field regarding how germ cells and associated gonadal cells regulate small RNA expression and production. First, how is the transcription of germ cell-specific miRNA, siRNA, and piRNA precursors regulated? The promoter elements for these non-coding RNA genes are not defined, and motif prediction can be imprecise if the transcription start site (TSS) is unknown. The region that separates the piRNA domains in bi-directional adult mammalian piRNA clusters may harbor promoter elements (Aravin et al., 2006, Girard et al., 2006, Lau et al., 2006). In contrast to the motif detected upstream of 21U RNAs, however, no obvious sequence motif shared by mammalian bi-directional clusters has been detected. A recent analysis of histone modifications and transcription factors in mouse ES cells has implicated CpG-rich genomic regions as the promoters of specific miRNA genes (Marson et al., 2008). Perhaps this methodology can also ascertain the promoters of germ cell-specific small RNA genes.
Other questions regarding transcriptional regulation of piRNA clusters include: Which promoting factors and which RNA polymerases mediate piRNA precursor transcription? How are pre-pachytene piRNA clusters regulated differently from adult pachytene piRNA clusters during germ cell development? Does a given germ cell express all piRNA clusters simultaneously, or does it pick one or several piRNA clusters to express, analogous to how lymphocytes or olfactory neurons pick one immunoglobulin gene or odorant receptor gene, respectively, for expression? This last question has recently been explored in a single-cell analysis of small RNAs from Xenopus tropicalis oocytes that compared the overall profile of piRNA cluster expression between different eggs from different individual females (Lau et al., 2009a). Interestingly, each egg expressed multiple piRNA clusters simultaneously, and the cluster expression profiles were similar in eggs derived from one mother but were different from eggs from a different mother. This suggests that piRNA cluster expression profiles can vary significantly between different individual animals.
Overall, our grasp of piRNA biogenesis still lags behind our understanding of miRNA and siRNA biogenesis, partly because in vitro cultured cells expressing miRNAs and siRNAs have facilitated biochemical analysis, while culture systems containing piRNAs had been lacking. Since piRNA expression is largely restricted to gonads, in particular to meiotic germ cells in mammals, finding a mitotic cell line that retained true gonadal characteristics seemed daunting. However, two insect cell lines derived from ovaries have recently been found to endogenously express abundant levels of piRNAs and PIWI proteins. An Ovary Somatic Sheet (OSS) cell line that likely derives from D. melanogaster ovary follicle cells expresses abundant levels of class I piRNAs because it only expresses Piwi (Lau et al., 2009b). Complementing the OSS cells is the BmN4 cell line derived from the ovaries of the silkworm, Bombyx mori, which expresses both Siwi (i.e., silkworm Piwi) and BmAGO3; thus this line appears to express class II piRNAs predominantly (Kawaoka et al., 2009). Since both these cell lines also express miRNAs and siRNAs, these systems will be invaluable for functional dissection of piRNA biogenesis and determining how cells partition miRNAs, siRNAs, and piRNAs into different AGO/PIWI proteins.
The impact of small RNAs in germline development
Although animal germline development is complex (Cinalli et al., 2008, Lin, 1997, Seydoux and Braun, 2006), some general principles have emerged. In the earliest stages of embryonic development, primordial germ cells (PGCs) are sequestered and proliferate in a niche that includes somatic support cells. During prenatal and juvenile development, PGCs transition into germline stem cells (GSCs), which proliferate by self-renewing and/or differentiating into secondary germ progenitor cells. Somatic support cells provide signals and molecules to germ cells that commit germ cells to enter meiotic phases like leptotene, zygotene, and, most critically, pachytene, when synapsis of sister chromatids occurs. Afterwards, germ cells further differentiate into haploid gametes: the sperm in males and the oocytes in females. During terminal differentiation, sperm shed their cytoplasm and streamline into DNA delivery vectors. In contrast, oocytes bulk up on maternal components such as energy-rich molecules and materials that are utilized for building the embryo and for establishing the next generation of PGCs. Most animal oocytes become polarized during growth, such that maternal components concentrate in special cytoplasm called germ plasm and serve as determinants in the embryo to specify which blastomeres become PGCs.
The influence of miRNAs and endo-siRNAs
Although it is known that individual small regulatory RNAs can broadly impact animal development (Ambros, 2004), the impact of small RNAs as a class on germ cell development is only now coming into focus. Studies where dicer or ago genes are mutated in the germline have demonstrated that miRNAs and siRNAs are crucial for germ cell development. For example, dicer (dcr-1) mutations in nematodes result in deformed oocytes (Knight and Bass, 2001), whereas dcr-1 mutations specifically in the D. melanogaster female germline produce GSCs that fail to self-renew but are able to differentiate into gametes (Hatfield et al., 2005, Jin and Xie, 2007). Mutations in loqs, ago-1, and mei-p26, a regulator of ago-1, all likely disrupt miRNA function and exhibit similar defects in GSC maintenance through the loss of GSC self-renewal (Forstemann et al., 2005, Park et al., 2007, Neumuller et al., 2008, Yang et al., 2007). Although the role of the miRNA pathway was deemed to be intrinsic to germ cells because somatic support cells expressing functional Ago-1 or Loqs failed to rescue the maintenance of mutant GSCs (Park et al., 2007, Yang et al., 2007), proliferation of somatic support cells also seems dependent on miRNAs (Jin and Xie, 2007). Together, these female germline studies suggest that miRNAs down-regulate differentiation genes in order to promote germ cell self-renewal and proliferation. However, the validation of genes predicted to be down-regulated by miRNAs in the germline remains to be explored.
Mouse male and female germ cells equally depend on miRNAs and siRNAs for proper development, but for reasons that may be different from fly germ cells. Although conditional oocyte-specific Dicer knockout mice are infertile, ovary morphology and oocyte numbers are similar to wild-type mice, suggesting that Dicer is not required for mammalian oocyte growth (Murchison et al., 2007, Tang et al., 2007). However, the mutant oocytes are incapable of meiotic progression because the spindles are malformed, while some transposons are activated (Murchison et al., 2007, Tang et al., 2007). In contrast to females, male mice with dicer mutations in the germline exhibit reduced PGC proliferation and a dramatic loss of spermatocytes developing into round spermatids (Maatouk et al., 2008, Hayashi et al., 2008). Nevertheless, these mutant males are not completely sterile and produce a few motile sperm, but these may result from germ cells that escape the conditional gene ablation (Hayashi et al., 2008). As in flies, the somatic support cells of both the male and female mouse germline require a functioning miRNA pathway to ensure that germ cells properly develop (Gonzalez and Behringer, 2009, Hong et al., 2008, Nagaraja et al., 2008).
Although the importance of miRNAs in fly and mice germ cells is apparent, zebrafish buck this trend because fish mutants lacking dicer in both the zygotic genome and in maternal contribution are able to produce viable germ cells that can fertilize and form an initial zygote (Giraldez et al., 2005). The maternal-zygotic dicer null (MZdicer) embryos eventually fail to gastrulate properly, but an injection of a double-stranded miR-403b duplex into the one-cell MZdicer mutant embryo rescues embryonic development (Giraldez et al., 2005). A possible explanation for this may be that piRNAs and PIWI proteins can compensate for miRNAs and siRNAs in zebrafish germ cell development, since they are likely maternally deposited in the egg and do not depend on Dicer for production (Houwing et al., 2007). Alternatively, maternal transcripts in zebrafish germ cells may have evolved mechanisms that obviate a need for regulation by miRNAs (Mishima et al., 2006).
In mammals, it is more difficult to differentiate the impact of miRNAs versus endo-siRNAs on germ cell development because there is a single Dicer that produces both miRNAs and endo-siRNAs, while all mammalian AGOs bind both miRNAs and siRNAs. However, a Drosha germline mutant, which should only lack miRNAs, might indicate if endo-siRNAs are sufficient for germ cell development. In D. melanogaster, the miRNA and siRNA pathways are genetically separable because Dcr-2 and Ago-2 produce endo-siRNAs independently of the Dcr-1 and Ago-1 production of miRNAs (Forstemann et al., 2007, Lee et al., 2004, Okamura et al., 2004, Tomari et al., 2007). However, the role that endo-siRNAs play in fly development remains mysterious - endogenous mRNA levels can be altered when endo-siRNAs are depleted, yet dcr-2 and ago-2 mutant flies that likely lack most endo-siRNAs do not appear overtly different from wild-type flies (Lee et al., 2004, Okamura et al., 2004). TE-associated piRNAs might compensate for TE-associated endo-siRNAs in the germline, and perhaps the endosiRNA pathway impacts behaviors and responses to natural selective pressures that are typically absent in laboratory conditions.
The impact of endo-siRNAs on the C. elegans germline is much more evident, as many mutations that abrogate endo-siRNA production or stability cause varying degrees of sterility. Mutants with decreased 26G endo-siRNAs (eri-1, rrf-3, and a double mutant of the Argonautes T22B3.2 and ZK757.3) exhibit temperature-dependent sterility, presumably due to overexpression of target genes (Han et al., 2009). In addition, mutations that affect 22G endo-siRNA accumulation (cde-1, csr-1, drh-3, ego-1, and ekl-1) result in broad gametogenesis defects because mitotic and meiotic chromosomes fail to segregate properly (Claycomb et al., 2009, She et al., 2009, van Wolfswinkel et al., 2009, Gu et al., 2009). The protein products of these genes appear to coalesce around chromosomes in gametes and blastomeres, while CSR-1, the AGO protein that binds a subset of 22G endo-siRNAs, directly interacts with chromatin targets. Bearing similarity to heterochromatic siRNAs in plants and fungi, which guide AGOs to modulate chromatin dynamics (Grewal and Moazed, 2003), components of the CSR-1 endo-siRNA pathway might also direct methylation on histone H3. However, in contrast to heterochromatin targets in fungi and plants, CSR-1 is primarily enriched on coding genes (Claycomb et al., 2009, She et al., 2009). Interestingly, the dosage and turnover of CSR-1 may be highly fine-tuned, because mutants in cde-1, which encodes a nucleotidyltransferase that uridylates 22G endo-siRNAs for possible degradation, have increased levels of 22G endo-siRNAs and exhibit misaligned mitotic chromosomes, perhaps due to ectopic histone methylation (She et al., 2009, van Wolfswinkel et al., 2009).
A conserved role of keeping transposons at bay
RNAi had been recognized as a mechanism to explain the phenomenon of cosuppression, where the silencing of multi-copy transgenes causes the silencing of endogenous genes, presumably from aberrant dsRNAs arising from the repetitive array of transgenes (Henikoff, 1998, Birchler et al., 2000). This hypothesis led to the idea that RNAi could also silence endogenous genomic repetitive elements like transposons, which can number in the thousands of copies per element in animal genomes. Given the mutagenic capabilities of transposon mobilization, transposon control is essential for the long term fitness of almost all organisms, and RNAi could have evolved as an elegant means to recognize these genomic invaders with small RNAs (Goodier and Kazazian, 2008). This model was confirmed by the discovery of transposon-directed siRNAs in fungi and plants (Birchler et al., 2000, Zaratiegui et al., 2007) and rasiRNAs from flies and fish, all of which appear to arise from both strands of transposons (Aravin et al., 2003, Aravin et al., 2001, Chen et al., 2005). These studies suggested that dsRNA is the common trigger for programming RNAi to target transposons, and identification of RdRP genes important for transposon control in plants and nematodes and the endo-siRNA pathway in fly further supported the model (Ghildiyal and Zamore, 2009, Malone and Hannon, 2009, Okamura and Lai, 2008).
Transposon control may be most critical in animal germ cells since, ultimately, germ cells are the essential vehicles for transposon replication. However, instead of relying on the conventional RNAi pathway, animal germs cells have evolved the PIWI/piRNA pathway to regulate transposons. The function and primacy of the PIWI pathway in controlling transposons in the germline was first demonstrated by fly mutants that lack piRNAs, which show highly elevated TE transcript levels in ovaries and testes (Siomi and Kuramochi-Miyagawa, 2009). In contrast, mutations affecting endo-siRNA production in flies do not reveal observable defects in gametogenesis (Ghildiyal and Zamore, 2009, Malone and Hannon, 2009, Okamura and Lai, 2008). The exact mechanism for how PIWI proteins and piRNAs silence transposon transcripts is not fully elucidated, but it is presumed that PIWI proteins use piRNAs that are antisense to transposons to recognize TE transcripts for cleavage and degradation. This hypothesis is supported by the conservation of catalytic residues between PIWI and AGO proteins necessary for Slicer activity (Tolia and Joshua-Tor, 2007), and the observation of Slicer activity in vitro by PIWI/piRNA complexes (Lau et al., 2006, Saito et al., 2006).
In addition to the slicing mechanism postulated for silencing transposons, work on mouse PIWI genes has suggested an additional transcriptional silencing pathway for maintaining transposon silencing. In the testes of mili and miwi2 mutant mice, there is elevated expression of retrotransposons like the LINE element L1 and the LTR element IAP (Aravin et al., 2008, Aravin et al., 2007b, Carmell et al., 2007, Kuramochi-Miyagawa et al., 2008). Interestingly, miwi2 and mili mutant mice also display loss of DNA methylation at L1 and IAP loci, whereas the profiles of IAP-targeting piRNAs are altered in Dnmt3L knockout mice, which lack a putative regulator of DNA methyltransferase activity during de novo establishment of DNA methylation patterns in the fetal male germline (Aravin and Bourc'his, 2008). Although Mili and Miwi are predominantly in the cytoplasm, Miwi2 is localized in the nucleus in specific fetal germ cell stages, and this localization is dependent on functional Mili (Aravin et al., 2008).
These results have suggested that Mili and Miwi2 control transposons at both the transcriptional and post-transcriptional levels. Although a biochemical interaction among Mili, Miwi2, Dnmt3L, and other DNA methyltransferases has not been shown, the genetic links between class II piRNAs and DNA methylation events are compelling, and might explain how silencing of TEs can be established and maintained in somatic cells when piRNA expression is restricted to the germline. In addition to these findings in mouse, zebrafish oocytes deficient in Ziwi and Zili also display elevated TE transcripts, and it is hypothesized that mobilized TEs cause genomic damage that triggers apoptosis (Houwing et al., 2008, Houwing et al., 2007). However, it is unknown if DNA methylation is affected in ziwi and zili mutants, and DNA methylation mechanisms are not thought to exist in invertebrates like D. melanogaster and C. elegans.
Nevertheless, plant small RNAs clearly guide DNA methyltransferases to TE loci, and both entities are critical for heterochromatin formation (Zaratiegui et al., 2007, Chen, 2009). RdRPs in plants also help to enforce TE silencing by converting TE transcripts into dsRNAs that are processed into secondary siRNAs. In this respect, the C. elegans piRNA pathway shares similarity with plants. Very few 21U RNAs appear to target the major Tc1 and Tc3 transposable elements, but secondary siRNAs targeting Tc3 are lost in the prg-1 mutant (Batista et al., 2008, Das et al., 2008). Thus, it has been proposed that 21U RNAs might help signal RdRPs in C. elegans to prime dsRNA production from aberrant TE transcripts (Figure 2), and this dsRNA becomes the source of secondary siRNAs that ultimately silence TEs, perhaps at the chromatin level (Batista et al., 2008, Das et al., 2008). Whereas TE silencing is a conserved function for the piRNA pathway in animals, the mechanistic details of piRNA and PIWI function may be quite varied among animal species.
Beyond simply controlling TE mobility, fly genetics has also revealed that TE control can extend to telomere regulation and possibly to speciation of organisms. In the first example, D. melanogaster telomeres contain specific retrotransposons (HeT-A, TAHRE, TART) and TAS repeats that likely recombine with each other to maintain telomere length (George et al., 2006). To stabilize telomere structure, the transposons and TAS repeats appear to generate piRNAs which direct Piwi and Aub to genetically impose heterochromatic marks onto the telomeres (Savitsky et al., 2006, Yin and Lin, 2007, Klenov et al., 2007). The heterochromatic marks that are induced by piRNAs then repress telomeric TEs from mobilizing and can silence transgenes inserted into nearby telomeres. piRNA pathway mutants (piwi, aub, spn-E) appear to lose the ability to package telomeres into stable heterochromatin, allowing expression and mobilization of the telomeric TEs and loss of silencing of transgenes integrated near telomeres (Josse et al., 2007, Savitsky et al., 2006, Yin and Lin, 2007, Klenov et al., 2007).
A second interesting manifestation of TE control by the piRNA pathway is hybrid dysgenesis, which describes a genetic syndrome of gonadal atrophy and sterility in hybrid progeny derived from a cross of wild-type D. melanogaster males and females from isolated laboratory strains (Kidwell et al., 1977). This syndrome has been determined to result from an inability of laboratory females to repress certain transposons. Some have postulated that a homology-based mechanism could be the system for mediating this genetic phenotype (Chaboissier et al., 1998, Jensen et al., 1999). Only recently have many groups begun to converge on the notion that siRNAs and piRNAs may be the critical maternal factors that endow daughters with the ability to resist the dysgenic syndrome by quenching TEs (Blumenstiel and Hartl, 2005, Brennecke et al., 2008, Chambeyron et al., 2008, Pelisson et al., 2007, Sarot et al., 2004). Published reviews further detail the link between piRNAs and hybrid dysgenesis (Malone and Hannon, 2009, Shpiz and Kalmykova, 2009).
In addition to the facet of transposon control, hybrid dysgenesis mediated by piRNAs and the PIWI pathway can also be viewed as an evolutionary force that drives speciation through modulating fertility. The Hannon laboratory demonstrated that fly strains can be genetically identical but epigenetically dissimilar on the basis of piRNA content (Brennecke et al., 2008). Since the piRNA pathway is intact and the bulk of piRNAs are present in sterile dysgenic hybrids, the study may imply that subtle interplays between TEs and piRNAs can have a profound and rapid impact in generating a reproductive barrier that could ultimately drive animal speciation. This study also provokes the question of whether piRNA profiles might impact progeny health, given that individual differences in piRNA cluster expression may be quite prevalent even between wild-type animals (Lau et al., 2009a). Finally, parameters like reduced growth temperature and increased parent age can suppress the dysgenic syndrome (Kidwell et al., 1977); thus, it will be interesting to see whether piRNA profiles or TE activity become altered by environmental conditions.
Classic mutants but new twists: fly genes affecting the piRNA pathway
Many genes affecting germline development in fly were previously discovered before they were known to affect piRNA function. Screens for female sterility and/or disrupted oocyte polarity identified aub, cuff, vasa, zuc, squ (Schupbach and Wieschaus, 1991); spn-E (Gillespie and Berg, 1995); mael (Clegg et al., 1997); and more recently armi (Cook et al., 2004) and krimp (Lim and Kai, 2007). These genes all affect piRNA accumulation and can be grouped as a class of related mutations where oocytes still form, but most of these oocytes cannot be fertilized and develop abnormally (Gillespie and Berg, 1995, Klattenhoff et al., 2007, Clegg et al., 1997, Lim and Kai, 2007, Chen et al., 2007). These mutations also affect RNA localization mechanisms that help establish polarity in the oocyte. For example, the maternal mRNA for oskar, which nucleates germ plasm, is not properly localized to the posterior end of the oocytes in these mutants, possibly because the normally polarized microtubule network between nurse cells and oocytes is disrupted (Chen et al., 2007, Clegg et al., 1997, Cook et al., 2004, Lim and Kai, 2007, Pane et al., 2007, Wilson et al., 1996). Interestingly, the proteins from these germline genes all co-localize at the nuage, a perinuclear zone in nurse cells believed to be an active region of RNA organization (Lim and Kai, 2007, Klattenhoff and Theurkauf, 2008, Kloc and Etkin, 2005, Lim et al., 2009).
How do piRNAs fit into the observed phenotypes of germline development mutations? The Schupbach and Therkauf laboratories have shown that mutants that are unable to accumulate piRNAs fail to prevent TEs from mobilizing, which results in elevated numbers of DNA breaks in the female germline (Chen et al., 2007, Klattenhoff et al., 2007, Pane et al., 2007). The aberrant DNA breaks then trigger damage signaling checkpoint pathways, which in turn affect microtubule network organization. Genetic support for this hypothesis comes from the suppression of microtubule disorganization observed when checkpoint factors are also mutated in the background of a piRNA pathway mutant (Chen et al., 2007, Klattenhoff et al., 2007, Pane et al., 2007). However, these double mutants in checkpoint pathways do not suppress the loss of Oskar localization (Klattenhoff and Theurkauf, 2008, Navarro et al., 2009), suggesting that RNA transport, perhaps utilizing microtubule machinery, may be a direct role for PIWI proteins and piRNAs. Indeed, a sea urchin and a frog PIWI homolog both interact with microtubules (Rodriguez et al., 2005, Lau et al., 2009a), while small RNA pathways have been shown to rely on microtubule regulation (Brodersen et al., 2008, Parry et al., 2007).
The piwi gene was discovered in separate screens for mutants with few dividing germline stem cells (Lin and Spradling, 1997). Although ovaries initially develop normally in piwi mutants, the adult GSCs fail to self-renew and only differentiate into oocytes that can be fertilized but are incompetent for embryogenesis. This function of piwi draws similarity to the GSC maintenance functions of ago-1 and dcr-1, but the mechanism for this function is still obscure. In contrast to the cytoplasmic and perinuclear localization of Aub in nurse cells, Piwi is predominantly nuclear localized in both germ cells and somatic support cells (Cox et al., 1998, Cox et al., 2000, Brennecke et al., 2007, Saito et al., 2006, Lau et al., 2009b). Additionally, piwi has been implicated in regulation of multi-copy transgene silencing through genetic or direct interactions with chromatin-associated proteins like Polycomb group (PcG) proteins and heterochromatin protein 1 (HP1; Brower-Toland et al., 2007, Grimaud et al., 2006, Pal-Bhadra et al., 2004, Yin and Lin, 2007). Since PcG proteins are integral to the self-renewing capacity of mouse ES cells due to their role in silencing differentiation genes at the chromatin level (Boyer et al., 2006), it is tempting to speculate that piwi might also serve an analogous function in the nucleus of GSCs or other stem cells of the fly germline.
Spermatogenesis in flies also depends on piwi and aub function, but the regulatory mechanisms are somewhat distinct from those in oocytes. piwi mutants lack sperm because GSCs fail to self-renew in the germinal niche (Lin and Spradling, 1997), but aub mutant sperm divide yet fail to mature because protein crystals encoded by Stellate (Ste) gene repeats accumulate and probably poison the spermatocytes (Aravin et al., 2001, Schmidt et al., 1999, Stapleton et al., 2001, Aravin et al., 2004). In wild-type fly testes, the Suppressor of Stellate [Su(Ste)] gene, a locus with homology to Ste repeats, produces piRNAs primarily antisense to Su(Ste), which then presumably direct Aub to destroy aberrant Ste transcripts (Nishida et al., 2007, Vagin et al., 2006, Palumbo et al., 1994). The oocyte nuage component genes armi, spn-E, zuc, and squ also influence Ste silencing (Aravin et al., 2004, Aravin et al., 2001, Pane et al., 2007, Schmidt et al., 1999, Stapleton et al., 2001, Tomari et al., 2004), suggesting that the core Aub-piRNA mechanism operates similarly in males and females even though Ste silencing is male-specific (Palumbo et al., 1994). Interestingly, loqs mutations also de-repress Ste silencing in addition to affecting miRNA and endo-siRNA production (Czech et al., 2008, Forstemann et al., 2005, Okamura et al., 2008b), suggesting that either endo-siRNAs contribute to Ste silencing, or loqs might participate in the function of all three major small RNA types in fly germ cells.
Two newly characterized D. melanogaster mutants with deficits in the piRNA pathway have recently been shown to display gametogenic defects similar to those seen in aub and piwi mutants. Isolation of the ago3 null mutant by the Zamore laboratory required the use of reverse genetics, possibly due to the location of the gene in proximity to heterochromatin (Li et al., 2009). Consistent with the hypothesis of AGO3 acting in the ping-pong piRNA amplification pathway, the bulk of class II piRNAs are lost. A second mutant called rhino was also found to be deficient in class II piRNAs, primarily those deriving from master control loci producing piRNAs from both genomic strands, like the 42AB cluster (Klattenhoff et al., 2009). Rhino was determined to be a rapidly evolving variant of heterochromatin protein 1 (HP1), and chromatin immunoprecipitation of Rhino indicated its presence at the 42AB piRNA cluster (Klattenhoff et al., 2009, Vermaak et al., 2005). In contrast to a chromatin silencing role attributed to HP1, Rhino instead appears necessary to stimulate expression of piRNAs from clusters.
Although much interesting biology remains to be garnered from the ago3 and rhino mutants, an important insight revealed by these mutants is the partitioning of class I and class II piRNA biogenesis mechanisms (Klattenhoff et al., 2009, Li et al., 2009). Since class II piRNAs are depleted and class I piRNA biogenesis remains intact in these mutants, the class I piRNAs surface in small RNA libraries, allowing confident classification of the flamenco locus as a class I piRNA cluster. Independent studies looking at maternal contribution of piRNAs (Malone et al., 2009) or examining a follicle cell line (Lau et al., 2009b) have also confirmed flamenco as a primary generator of class I piRNAs; however, the functional partition of the two classes of piRNAs in vertebrates remains unclear, since there are fewer vertebrate piRNA mutants and the vertebrate orthologs of AGO3 and Rhino are not obvious.
PIWI proteins and vertebrate gametogenesis
Mouse male germ cells initially develop synchronously before the first wave of meiosis begins at 10 days postpartum (dpp), during which the class II pre-pachytene piRNAs are made. After the onset of meiosis, class I piRNAs become the dominant species in late spermatocytes and spermatids. Constitutive gene targeting of the three PIWI homologs in mice, Miwi, Mili, and Miwi2, has demonstrated that PIWI proteins and piRNAs are essential for male fertility, since the testes are diminished in each knock-out mutant due specifically to the apoptosis of germ cells and not the somatic support cells (Carmell et al., 2007, Deng and Lin, 2002, Kuramochi-Miyagawa et al., 2004). Mili is expressed as early as PGC and GSC formation and persists throughout spermatogenesis, while Miwi2 is only detected in germ cells in the 7 days between 15 days post coitus (dpc) and 3 dpp (Aravin et al., 2008, Carmell et al., 2007, Kuramochi-Miyagawa et al., 2004, Kuramochi-Miyagawa et al., 2008). This stage-specific regulation of Miwi2, Mili, and Miwi expression correlates with the order of the developmental stage defects in mutants: early spermatocytes arrest in early prophase of meiosis I in miwi2 and mili homozygous knock-outs, while miwi mutant germ cells arrest in the round spermatid stage. As the mutant mice age, arrested germ cells undergo apoptosis, and, through unknown causes, earlier spermatocytes and spermatogonia also progressively become depleted (Carmell et al., 2007, Deng and Lin, 2002, Kuramochi-Miyagawa et al., 2004).
Mouse PIWI mutant neonates contain wild-type numbers of spermatogonia, indicating that GSC proliferation is initially unaffected. Thus, at the physiological level, mouse piwi genes do not exactly share the same stem cell maintenance role of D. melanogaster piwi. Also, in contrast to fly piwi, mouse piwi genes are dispensable for female fertility (Carmell et al., 2007, Deng and Lin, 2002, Kuramochi-Miyagawa et al., 2004), although long-term fertility studies of homozygous female mutants have not been described. Since female germ cells only appear to express Mili, AGO proteins and endo-siRNAs may play a redundant role within mouse oocytes (Tam et al., 2008, Watanabe et al., 2008). However, mammalian oogenesis is also fundamentally different from oogenesis in other vertebrates and spermatogenesis in general, because mammalian oogonia proliferate only during gestation and are committed to meiosis as primary oocytes at birth.
How do piwi genes impact female germline development in non-mammalian vertebrates, where oogenesis, like in D. melanogaster, depends upon mitotic proliferation and self-renewal of GSCs? The zebrafish piwi homologs, ziwi and zili, are expressed in both male and female gonads, but mutant analyses of these genes has not provided a clear answer. Null mutations in ziwi and zili cause sterility and germ cell loss during juvenile development, but this event also masculinizes zebrafish (Houwing et al., 2008, Houwing et al., 2007). Zebrafish sex determination is not driven by sex chromosomes but by environmental and internal cues; however, a hypomorph of zili allows female germ cells to develop into sterile oocytes. Compared to zili, the ziwi mutant phenotypes are generally more severe: by 3 weeks post fertilization (wpf) ziwi mutant germ cells universally undergo apoptosis, while zili mutant gonads at 6 wpf still retain a few germ cells expressing Ziwi (Houwing et al., 2008, Houwing et al., 2007). Additionally, the zili hypomorph causing female sterility is male fertile. Perhaps future analysis with the genetically-tractable medaka fish may address how PIWI proteins directly affect oogenesis in vertebrates, since medaka sex determination is genetically specified (Wittbrodt et al., 2002).
More to piRNAs than transposon silencing?
Although transposon control is the primary biological function for PIWI proteins and piRNAs, evidence for additional functions for PIWI proteins and piRNAs are also emerging, such as mRNA localization or broad gene expression control. For example, the specific posterior localization of oskar mRNA and other maternal factors in the D. melanogaster oocyte might be regulated by the PIWI pathway beyond simply the activation of DNA damage checkpoint mechanisms (Klattenhoff and Theurkauf, 2008, Navarro et al., 2009). Interestingly, overexpression of piwi in D. melanogaster females results in embryos with greater Oskar expression, which increases the number of pole cells, the fly equivalent of PGCs (Megosh et al., 2006). This suggests that Piwi may directly interact and recruit more oskar mRNA to the posterior end of the egg. It would be interesting to see if overexpression of aub or ago3 recapitulates this result.
In contrast to the >60% of D. melanogaster piRNAs corresponding to transposons (Brennecke et al 2006, Yin & Lin, 2007), only 18–34% of adult vertebrate piRNAs map to repetitive elements (Aravin et al., 2006, Girard et al., 2006, Houwing et al., 2007, Lau et al., 2006). So, what might be the role for the bulk of class I piRNAs? Translational control has been discussed as a significant mechanism for germ cell gene regulation, of which part of the cellular mechanism depends upon RNA organization and localization events (Figure 4; Mendez and Richter, 2001, Kloc and Etkin, 2005, Kotaja and Sassone-Corsi, 2007). aub and piwi mutations affecting oskar mRNA localization is one example of the piRNA pathway intersecting with a translational regulation mechanism (Cook et al., 2004, Harris and Macdonald, 2001). In addition, the localization of PIWI/piRNA complexes in the fly germline may be highly dynamic, involving processing bodies and dynein transport components (Lim et al., 2009, Navarro et al., 2009).
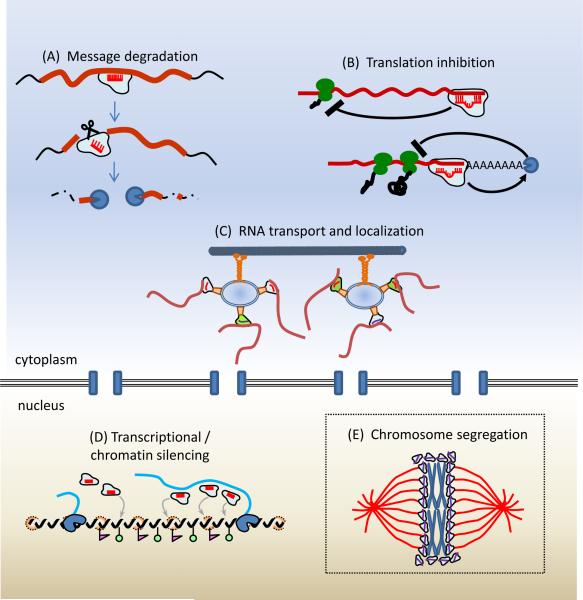
Figure 4.
Mechanistic models of gene regulation by animal germline small RNAs. (A) Transposable element silencing by piRNAs and siRNAs and silencing of certain miRNA targets may operate at the level of mRNA degradation. (B) Many miRNA targets and potential piRNA and siRNA targets are also regulated at the level of translation. (C) miRNA/AGO complexes or piRNA/PIWI complexes have the potential to direct target RNAs to sequestration in particular cellular compartments, possibly through active RNA transport along cytoskeletal tracks. (D) siRNAs and piRNAs may have the capacity to inhibit and possibly activate transcription at the chromatin level, either at specific loci, or more broadly at the genomic level through regulation of chromosome segregation (E).
The Lin laboratory has proposed that Miwi and Mili may promote translation of male germline transcripts. Their support for this hypothesis includes co-sedimentation of Miwi and Mili with polysomes in density gradients, the association of Miwi and Mili with mRNAs and proteins that bind mRNA caps, and reduced rates of translation initiation in mili knockout testes (Deng and Lin, 2002, Grivna et al., 2006a, Grivna et al., 2006b, Unhavaithaya et al., 2008). In spermatids, Miwi and Mili concentrate in the chromatoid body, a perinuclear organelle similar to nuage and germ plasm and thought to be an RNA processing center (Kotaja et al., 2006, Kotaja and Sassone-Corsi, 2007). Although more biochemical studies are needed to assess the effect of Miwi and Mili on translation, the Xenopus tropicalis PIWI homolog, Xiwi, localizes to germ plasm and also associates with components of the translation machinery, including mRNAs (Lau et al., 2009a). Additionally, in zebrafish, a zili hypomorph is capable of transposon silencing, yet the oocytes are unable to complete meiosis normally, resulting in sterility (Houwing et al., 2008). Perhaps the meiotic defect in this zili hypomorph could also be attributed to a loss of translation regulation of protein factors that drive oocyte maturation, since translation regulation is integral to oocyte maturation in Xenopus laevis (Mendez and Richter, 2001). The prevalence of translation regulation mechanisms in animal germ cells and the commonalities between nuage, germ plasm, and the chromatoid body suggest there may be a conserved RNA processing and translation regulation role for PIWI proteins.
If PIWI proteins do regulate germ cell mRNAs, then how might piRNAs from intergenic clusters or TEs mechanistically modulate this function? The substrate recognition rules between piRNAs and mRNAs is unknown – do piRNAs recognize targets with imperfect bulges like miRNAs, or do they only act upon perfectly matched substrates like siRNAs (Figure 4)? Two recent papers suggest another possible mechanism of gene regulation by PIWI proteins processing primarily the 3' UTRs of select mRNAs into piRNAs (Robine et al., 2009, Saito et al., 2009). Both studies uncovered the D. melanogaster traffic jam mRNA as a precursor of primary piRNAs, which one study suggests may regulate downstream genes that enforce follicle cell development (Saito et al., 2009). The other study demonstrated that this piRNA biogenesis pathway from the 3' UTRs of mRNAs is broadly conserved from flies to vertebrates and may have evolved to select many specific mRNAs for piRNA processing as a possible mode of gene regulation (Robine et al., 2009). The implications of these two new pathways on germ cell development remain to be further investigated.
Evolutionary and concluding perspectives
RNAi is an ancient mode of gene regulation, conserved from animals and plants to archaea and protists. Almost all eukaryotes possess AGO and Dicer proteins and a population of small RNAs, including miRNAs and siRNAs (Ghildiyal and Zamore, 2009). Interestingly, fungi and plants lack piRNAs since their AGO proteins, based on their amino acid sequence, do not fit in the same subclade as animal PIWI proteins (Seto et al., 2007). However, the nebulous kingdom of protozoans possess homologs to the PIWI subclade, and many protozoan small RNAs also serve to target repetitive sequences like transposons, although the genesis of these small RNAs requires a Dicer enzyme (Couvillion et al., 2009, Mochizuki and Gorovsky, 2004, Shi et al., 2004, Shi et al., 2006, Ullu et al., 2005, Zhang et al., 2008). Although plant germ cells are quite different from animal germ cells, plants also utilize their own repertoire of specific small RNAs to regulate germline tissue development (Slotkin et al., 2009). Despite this common theme, the miRNA and siRNA pathways in animals are distinct from the piRNA pathway in terms of the restricted tissue expression, independence from Dicer for biogenesis, and possibly in final regulatory mechanisms. Perhaps this distinction arose with the PIWI pathway evolving separately and more recently in animal lineages, because siRNAs are found in nearly all eukaryotes, while PIWI genes are only known in animals and some protozoans (Seto et al., 2007, Grimson et al., 2008).
In bilaterian animals, miRNAs can be highly conserved. For example, the miRNA let-7 is perfectly conserved at the primary sequence level between nematodes and humans (Pasquinelli et al., 2000). This may be because miRNAs regulate hundreds of targets in essential gene regulatory networks via imperfect base pairing interactions between the miRNA and target 3' UTRs (Bartel, 2009). Neither individual endo-siRNAs nor individual piRNAs likely exhibit the deep primary sequence conservation seen in miRNAs. This may be attributed to the scattered biogenesis of multitudes of endo-siRNAs and piRNAs from a precursor versus a single pair of miRNA/miRNA* from a miRNA hairpin precursor, and possibly to a much more limited regulatory role for piRNAs and siRNAs compared to miRNAs.
However, conservation of entire piRNA clusters is maintained at the level of synteny within mammals. Syntenic alignments of chromosomes between mouse, rat, and human illustrate the conserved presence and configuration of pachytene piRNA clusters. Nevertheless, very little primary sequence conservation is detected between rodent and human piRNAs (Girard et al., 2006, Lau et al., 2006). This suggests that piRNA production is essential, but the sequence identity of the piRNAs can evolve rapidly, possibly to combat transposons that also evolve quickly. Although PIWI protein components and germline functions of piRNAs may be conserved among animals, the piRNA clusters can vary significantly (i.e. D. melanogaster master control loci versus C. elegans 21U RNA clusters).
A recent analysis of two non-bilaterian animals that are at a basal point of the evolutionary tree has indicated that the major proportion of small RNAs in basal animals exhibit qualities of piRNAs (Grimson et al., 2008). The genomes of the anemone Nematostella vectensis and the sponge Amphimedon queenslandica each encode three PIWI homologs, and class II and class I piRNAs could be identified based upon reads that form ping-pong pairs and clusters of reads that map in a strand-biased fashion, respectively (Grimson et al., 2008). Whether these piRNAs are restricted to the germ cells of these non-bilaterians remains to be established. However, sponges and anemones contain not typical germ cells, but rather derivatives of somatic pluripotent stem cells, which lend to these organisms' extraordinary regenerative capabilities (Extavour et al., 2005, Muller, 2006). Interestingly, one class of bilaterian animals, planarians, also exhibit extensive regenerative capacities and possess somatic pluripotent stem cells called neoblasts, which can give rise to planarian germ cells. Small RNA studies of planaria might relate to the biology of non-bilaterians.
The planarian Schmidtea mediterranea also contains three piwi homologs, smedwi-1, -2,and -3, which are mainly expressed in the neoblasts, and both piRNAs and miRNAs have been cloned from this animal (Palakodeti et al., 2006, Palakodeti et al., 2008, Friedlander et al., 2009). Although full genomic characterization of planarian piRNAs is still pending, these piRNAs are a bit longer than other animal piRNAs (mainly 30–32 nt), and several piRNAs also target transposons (Palakodeti et al., 2008). RNAi knockdown studies of smedwi-2 and smedwi-3 indicate that these genes affect the regenerative capacity of planaria after amputation, such that neoblast proliferation is not affected, but differentiation and tissue repair by neoblasts are blocked (Palakodeti et al., 2008, Reddien et al., 2005). Knockdown of smedwi-2 and smedwi-3 also reduces piRNA but not miRNA levels, suggesting that planarian neoblasts rely upon piRNAs for differentiation (Palakodeti et al., 2008), in contrasts to mouse ES cells, which rely on miRNAs for differentiation (Bernstein et al., 2003). The fact that planaria and basal animals such as sponges and sea anemones contain pluripotent neoblasts/neoblast-like cells together with abundant populations of piRNAs suggests that an ancestral function of the piRNA pathway might have been to modulate tissue regeneration in simple animals.
Research into the biology of pluripotent stem cells, like ES cells, may enable a revolution in treating human diseases (Yu and Thomson, 2008). Given the ethical and practical limitations of accessing embryos, other tissue types have been explored for pluripotent stem cells, including male gonads. Since germ cells give rise to the embryo and GSCs are totipotent, investigations on mouse and human male gonads have also yielded pluripotent stem cells (Conrad et al., 2008, Kanatsu-Shinohara et al., 2004). The impact of the piRNA pathway on stem cell function in vertebrates and mammals is only now being explored, and while piRNAs have not been detected in mouse ES cells, class II piRNAs have been detected in certain mouse spermatogonial stem cell lines (Lau, unpublished data). Since the piRNA pathway plays an integral role in GSC and neoblast development, it is encouraging to speculate that other regenerative processes involving stem cells might be impacted by piRNAs or other germline small RNAs.
Beyond stem cells, further investigation of transposon control by piRNAs may also impact our understanding of human aging. For example, maintaining the stability of genomes is a critical component of cancer prevention and avoiding developmental abnormalities. Comparing TE control in germ cells versus TE regulation in somatic cells might reveal the differences in pluripotency between GSCs and somatic stem cells. Perhaps epigenetic programming events during gametogenesis via small RNAs help to establish chromatin conformations necessary for proper embryogenesis.
In multicellular organisms, germ cells may not be essential to the health of the individual, but they are essential to the propagation and survival of the species. In the recent past, germ cell biology has become evidently indispensable for investigations into small RNA function. As future experiments reveal the molecular underpinnings of how PIWI proteins and piRNAs work, the insights gained may lead to applications useful for human therapies.
Acknowledgments
I am grateful to Michael Blower, Julie Claycomb, Andrew Grimson, Eric Lai, Sivanne Pearl, Christina Post, and Dianne Schwarz for discussions and critical reading of this manuscript. I apologize for citations omitted due to space and publication constraints. This work is supported by a K99/R00 award from the NICHD and the NIH (HD057298). I dedicate this manuscript to the memory of my father, Wing-yan Lau, whose passion for science lives on in my studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Bourc'his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22:970–975. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007a;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Armisen J, Gilchrist MJ, Wilczynska A, Standart N, Miska EA. Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome Res. 2009;19:1766–1775. doi: 10.1101/gr.093054.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte D, Jr., Luo S, Schroth GP, Carrington JC, Bartel DP, Mello CC. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Bhadra MP, Bhadra U. Making noise about silence: repression of repeated genes in animals. Curr Opin Genet Dev. 2000;10:211–216. doi: 10.1016/s0959-437x(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Blumenstiel JP, Hartl DL. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci U S A. 2005;102:15965–15970. doi: 10.1073/pnas.0508192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 Is Essential for Spermatogenesis and Repression of Transposons in the Mouse Male Germline. Dev Cell. 2007 doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboissier MC, Bucheton A, Finnegan DJ. Copy number control of a transposable element, the I factor, a LINE-like element in Drosophila. Proc Natl Acad Sci U S A. 1998;95:11781–11785. doi: 10.1073/pnas.95.20.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci U S A. 2008;105:14964–14969. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, Tenaglia E, Xu C, Gish G, Min J, Pawson T. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci U S A. 2009;106:20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, Sander C, Zavolan M, Tuschl T. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinalli RM, Rangan P, Lehmann R. Germ cells are forever. Cell. 2008;132:559–562. doi: 10.1016/j.cell.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, Conte D, Jr., Mello CC. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–134. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg NJ, Frost DM, Larkin MK, Subrahmanyan L, Bryant Z, Ruohola-Baker H. maelstrom is required for an early step in the establishment of Drosophila oocyte polarity: posterior localization of grk mRNA. Development. 1997;124:4661–4671. doi: 10.1242/dev.124.22.4661. [DOI] [PubMed] [Google Scholar]
- Conrad S, Renninger M, Hennenlotter J, Wiesner T, Just L, Bonin M, Aicher W, Buhring HJ, Mattheus U, Mack A, Wagner HJ, Minger S, Matzkies M, Reppel M, Hescheler J, Sievert KD, Stenzl A, Skutella T. Generation of pluripotent stem cells from adult human testis. Nature. 2008;456:344–349. doi: 10.1038/nature07404. [DOI] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Couvillion MT, Lee SR, Hogstad B, Malone CD, Tonkin LA, Sachidanandam R, Hannon GJ, Collins K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev. 2009;23:2016–2032. doi: 10.1101/gad.1821209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, Hannon GJ, Brennecke J. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, Matthews N, Berezikov E, Ketting RF, Tavare S, Miska EA. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr., Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, Yates JR, 3rd, Mello CC. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour CG, Pang K, Matus DQ, Martindale MQ. vasa and nanos expression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol Dev. 2005;7:201–215. doi: 10.1111/j.1525-142X.2005.05023.x. [DOI] [PubMed] [Google Scholar]
- Fejes-Toth K SV, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, Gingeras TR. Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedländer MR, Adamidi C, Han T, Lebedeva S, Isenbarger TA, Hirst M, Marra M, Nusbaum C, Lee WL, Sánchez Alvarado A, Kim JK, Rajewsky N. High-resolution discovery and profiling of planarian small RNAs. Proc. Natl. Acad. Sci. USA. 2009;106:11546–51. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George JA, DeBaryshe PG, Traverse KL, Celniker SE, Pardue ML. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 2006;16:1231–1240. doi: 10.1101/gr.5348806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie DE, Berg CA. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Gonzalez G, Behringer RR. Dicer is required for female reproductive tract development and fertility in the mouse. Mol Reprod Dev. 2009 doi: 10.1002/mrd.21010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–2497. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006a;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A. 2006b;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D, Jr., Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, Chen CC, Chaves DA, Duan S, Kasschau KD, Fahlgren N, Yates JR, 3rd, Mitani S, Carrington JC, Mello CC. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, O'Carroll D, Das PP, Tarakhovsky A, Miska EA, Surani MA. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays. 1998;20:532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Hong X, Luense LJ, McGinnis LK, Nothnick WB, Christenson LK. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology. 2008;149:6207–6212. doi: 10.1210/en.2008-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27:2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF. A Role for Piwi and piRNAs in Germ Cell Maintenance and Transposon Silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Jensen S, Gassama MP, Heidmann T. Taming of transposable elements by homology-dependent gene silencing. Nat Genet. 1999;21:209–212. doi: 10.1038/5997. [DOI] [PubMed] [Google Scholar]
- Jin Z, Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr Biol. 2007;17:539–544. doi: 10.1016/j.cub.2007.01.050. [DOI] [PubMed] [Google Scholar]
- Josse T, Teysset L, Todeschini AL, Sidor CM, Anxolabehere D, Ronsseray S. Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet. 2007;3:1633–1643. doi: 10.1371/journal.pgen.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, Toyoshima M, Niwa O, Oshimura M, Heike T, Nakahata T, Ishino F, Ogura A, Shinohara T. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 2004;119:1001–1012. doi: 10.1016/j.cell.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, Tomari Y, Shimada T, Katsuma S. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA. 2009;15:1258–1264. doi: 10.1261/rna.1452209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, Plasterk RH. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Kidwell JF, Sved JA. Hybrid Dysgenesis in DROSOPHILA MELANOGASTER: A Syndrome of Aberrant Traits Including Mutation, Sterility and Male Recombination. Genetics. 1977;86:813–833. doi: 10.1093/genetics/86.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, Vidal M, Ruvkun G. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol. 2009a;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2'-O-methylated at their 3' termini. Nat Struct Mol Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA. 2009b doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, Seitz H, Zamore PD, Weng Z, Theurkauf WE. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. RNA localization mechanisms in oocytes. J Cell Sci. 2005;118:269–282. doi: 10.1242/jcs.01637. [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Lin H, Parvinen M, Sassone-Corsi P. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J Cell Sci. 2006;119:2819–2825. doi: 10.1242/jcs.03022. [DOI] [PubMed] [Google Scholar]
- Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC. (unpublished data)
- Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J. 2009a;28:2945–2958. doi: 10.1038/emboj.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Robine N, Martin R, Chung WJ, Niki Y, Berezikov E, Lai EC. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009b;19:1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol. 2009;186:333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. The tao of stem cells in the germline. Annu Rev Genet. 1997;31:455–491. doi: 10.1146/annurev.genet.31.1.455. [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD. Dicer1 is required for differentiation of the mouse male germline. Biol Reprod. 2008;79:696–703. doi: 10.1095/biolreprod.108.067827. [DOI] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Mishima T, Takizawa T, Luo SS, Ishibashi O, Kawahigashi Y, Mizuguchi Y, Ishikawa T, Mori M, Kanda T, Goto T. MicroRNA (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction. 2008;136:811–822. doi: 10.1530/REP-08-0349. [DOI] [PubMed] [Google Scholar]
- Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev. 2004;14:181–187. doi: 10.1016/j.gde.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Morozova O, Hirst M, Marra MA. Applications of new sequencing technologies for transcriptome analysis. Annu Rev Genomics Hum Genet. 2009;10:135–151. doi: 10.1146/annurev-genom-082908-145957. [DOI] [PubMed] [Google Scholar]
- Muller WE. The stem cell concept in sponges (Porifera): Metazoan traits. Semin Cell Dev Biol. 2006;17:481–491. doi: 10.1016/j.semcdb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Kheradpour P, Sachidanandam R, Smith C, Hodges E, Xuan Z, Kellis M, Grutzner F, Stark A, Hannon GJ. Conservation of small RNA pathways in platypus. Genome Res. 2008;18:995–1004. doi: 10.1101/gr.073056.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja AK, Andreu-Vieyra C, Franco HL, Ma L, Chen R, Han DY, Zhu H, Agno JE, Gunaratne PH, DeMayo FJ, Matzuk MM. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol. 2008;22:2336–2352. doi: 10.1210/me.2008-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C, Bullock S, Lehmann R. Altered dynein-dependent transport in piRNA pathway mutants. Proc Natl Acad Sci U S A. 2009;106:9691–9696. doi: 10.1073/pnas.0903837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–245. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, Siomi MC. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009 doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA. 2007;13:1911–1922. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K. The 3' termini of mouse Piwi-interacting RNAs are 2'-O-methylated. Nat Struct Mol Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 2008a;15:998. doi: 10.1038/nsmb0908-998c. [DOI] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008b;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Graveley BR. MicroRNAs from the Planarian Schmidtea mediterranea: a model system for stem cell biology. RNA. 2006;12:1640–1649. doi: 10.1261/rna.117206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo G, Bonaccorsi S, Robbins LG, Pimpinelli S. Genetic analysis of Stellate elements of Drosophila melanogaster. Genetics. 1994;138:1181–1197. doi: 10.1093/genetics/138.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr Biol. 2007;17:533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- Parry DH, Xu J, Ruvkun G. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr Biol. 2007;17:2013–2022. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sanchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- Ro S, Song R, Park C, Zheng H, Sanders KM, Yan W. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA. 2007;13:2366–2380. doi: 10.1261/rna.754207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC. A broadly conserved pathway generates 3' UTR-directed primary piRNAs. Current Biology. 2009;19 doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Seipel SA, Hamill DR, Romancino DP, M DIC, Suprenant KA, Bonder EM. Seawi--a sea urchin piwi/argonaute family member is a component of MT-RNP complexes. RNA. 2005;11:646–656. doi: 10.1261/rna.7198205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2'-O-methylation of Piwi- interacting RNAs at their 3' ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20:345–354. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Palumbo G, Bozzetti MP, Tritto P, Pimpinelli S, Schafer U. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics. 1999;151:749–760. doi: 10.1093/genetics/151.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GC, Sarkar SN. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr Top Microbiol Immunol. 2007;316:233–250. doi: 10.1007/978-3-540-71329-6_12. [DOI] [PubMed] [Google Scholar]
- Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell. 2007;26:603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- She X, Xu X, Fedotov A, Kelly WG, Maine EM. Regulation of heterochromatin assembly on unpaired chromosomes during caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 2009;5:e1000624. doi: 10.1371/journal.pgen.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Djikeng A, Tschudi C, Ullu E. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol Cell Biol. 2004;24:420–427. doi: 10.1128/MCB.24.1.420-427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Tschudi C, Ullu E. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA. 2006;12:2063–2072. doi: 10.1261/rna.246906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Kalmykova A. Epigenetic transmission of piRNAs through the female germline. Genome Biol. 2009;10:208. doi: 10.1186/gb-2009-10-2-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Kuramochi-Miyagawa S. RNA silencing in germlines--exquisite collaboration of Argonaute proteins with small RNAs for germline survival. Curr Opin Cell Biol. 2009;21:426–434. doi: 10.1016/j.ceb.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton W, Das S, McKee BD. A role of the Drosophila homeless gene in repression of Stellate in male meiosis. Chromosoma. 2001;110:228–240. doi: 10.1007/s004120100136. [DOI] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Lasko P. Tudor and its domains: germ cell formation from a Tudor perspective. Cell Res. 2005;15:281–291. doi: 10.1038/sj.cr.7290297. [DOI] [PubMed] [Google Scholar]
- Thomson T, Liu N, Arkov A, Lehmann R, Lasko P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev. 2008;125:865–873. doi: 10.1016/j.mod.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Lujan HD, Tschudi C. Small sense and antisense RNAs derived from a telomeric retroposon family in Giardia intestinalis. Eukaryot Cell. 2005;4:1155–1157. doi: 10.1128/EC.4.6.1155-1157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H. MILI, a piRNA binding protein, is required for germline stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2008 doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol. 2009;19:630–639. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Henikoff S, Malik HS. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Saxe JP, Tanaka T, Chuma S, Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol. 2009;19:640–644. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- Wittbrodt J, Shima A, Schartl M. Medaka--a model organism from the far East. Nat Rev Genet. 2002;3:53–64. doi: 10.1038/nrg704. [DOI] [PubMed] [Google Scholar]
- Yang L, Chen D, Duan R, Xia L, Wang J, Qurashi A, Jin P. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development. 2007;134:4265–4272. doi: 10.1242/dev.009159. [DOI] [PubMed] [Google Scholar]
- Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Thomson JA. Pluripotent stem cell lines. Genes Dev. 2008;22:1987–1997. doi: 10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Zhang H, Ehrenkaufer GM, Pompey JM, Hackney JA, Singh U. Small RNAs with 5'-polyphosphate termini associate with a Piwi-related protein and regulate gene expression in the single-celled eukaryote Entamoeba histolytica. PLoS Pathog. 2008;4:e1000219. doi: 10.1371/journal.ppat.1000219. [DOI] [PMC free article] [PubMed] [Google Scholar]