SUMMARY
MLL1 fusion proteins activate HoxA9 gene expression and cause aggressive leukemias that respond poorly to treatment, but how they recognize and stably bind to HoxA9 is not clearly understood. In a systematic analysis of MLL1 domain recruitment activity, we identified an essential MLL1 recruitment domain that includes the CXXC domain and PHD fingers and is controlled by direct interactions with the PAF elongation complex and H3K4Me2/3. MLL1 fusion proteins lack the PHD fingers and require pre-binding of a wild type MLL1 complex and CXXC domain recognition of DNA for stable HoxA9 association. Together, these results suggest that specific recruitment of MLL1 requires multiple interactions and is a precondition for stable recruitment of MLL1 fusion proteins to HoxA9 in leukemogenesis. Since wild type MLL1 and oncogenic MLL1 fusion proteins have overlapping yet distinct recruitment mechanisms, this creates a “window of opportunity” that could be exploited for the development of targeted therapies.
INTRODUCTION
Rearrangements of the MLL1 (Mixed Lineage Leukemia 1) gene are associated with aggressive acute leukemias in both children and adults (Ayton and Cleary, 2001; Hess, 2004). The most common MLL1 rearrangements are cytologically visible chromosome translocations that fuse amino terminal sequences of MLL1 in frame with a wide variety of different partner genes creating novel fusion proteins (Ayton and Cleary, 2001 and see Figure 1A). The prognosis for leukemias containing MLL1 translocations can vary but is generally quite poor (Hess, 2004).
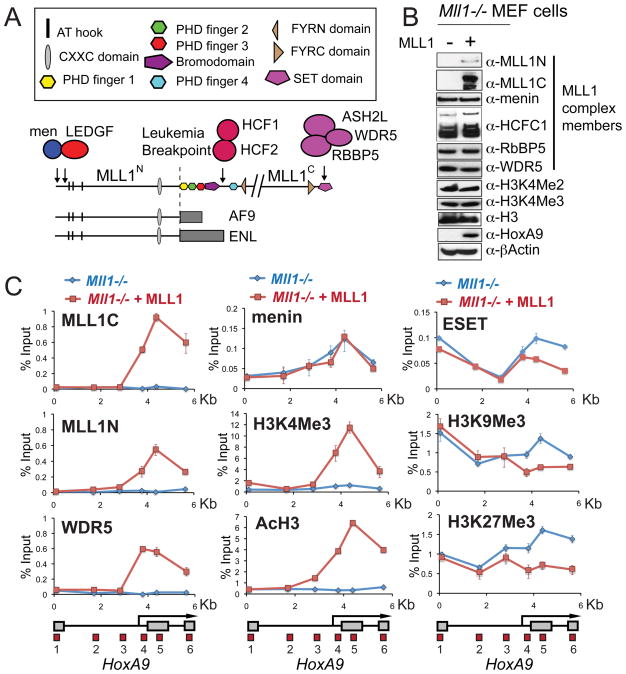
Figure 1. Recruitment of the MLL1 complex is required for HoxA9 activation.
(A) Important MLL1 domains and protein interactions. The MLL1 protein is proteolytically cleaved in the cell into N terminal (MLL1N) and C terminal (MLL1C) proteins. All MLL1 leukemogenic fusions break just before the first PHD finger (line, Leukemia Breakpoint). Two MLL1 fusion proteins MLL-AF9 and MLL-ENL are shown as examples. (B) HoxA9 protein expression depends on the expression of MLL1. Extracts were prepared from Mll1−/− mouse embryonic fibroblast (MEF) knockout cells (lane −) and Mll1−/− MEF cells that have been reconstituted with full length MLL1 (lane +). Western blots were probed with the antibodies indicated. (C) Chromatin immunoprecipitation (ChIP) experiments at HoxA9 in Mll1−/− (blue line) versus Mll1−/− reconstituted with MLL1 (red line) MEF cells using the antibodies indicated. A schematic of the mouse HoxA9 locus with red boxes indicating the positions of 6 primer/probe sets is shown at the bottom of the ChIP figures. ChIP results shown are typical for at least two independent experiments. Error bars represent standard deviation of three PCR reactions. See also Figure S1.
The normal function of the MLL1 protein is to maintain the activation of developmentally important genes such as the clustered Hox genes. Proper Hox gene expression is required for normal embryogenesis as well as for normal hematopoietic differentiation. Wild type MLL1 is a large protein that is proteolytically cleaved in vivo by the Taspase 1 protease into an N terminal 300 KDa fragment (MLL1N) and a C terminal 180 KDa fragment (MLL1C) (Hess, 2004). MLL1-mediated HOX gene activation is controlled in part by the enzymatic activity of a C-terminal SET domain that “writes” a methyl (Me) mark on lysine 4 (K4) of histone 3 (Milne et al., 2002; Nakamura et al., 2002). Histone lysines can be mono- (Me1), di- (Me2), or tri- (Me3) methylated, and it is the H3K4Me2 and 3 marks that are most highly correlated with Hox gene activation in vivo (Milne et al., 2005a; Ruthenburg et al., 2007a). H3K4Me2/3 promotes gene activation by acting as a docking site for PHD finger containing “reader” proteins that have a variety of different functions (Ruthenburg et al., 2007a). MLL1 fusion proteins lose the C terminal SET domain (see Figure 1A) and instead promote gene activation through recruitment of transcription elongation complexes (Yokoyama et al., 2010; Lin et al., 2010) and promotion of H3K79Me2 (Milne et al., 2005b; Krivtsov et al., 2008). H3K4Me3 and H3K79Me2 mediated activation of Hox genes is antagonized by marks such as H3K9Me3 and H3K27Me3, both of which have important roles in gene repression (Nottke, 2009).
HOXA9 is one of the more critical regulatory targets of MLL1 and MLL1 fusion proteins in human leukemia. During hematopoiesis, HOXA9 is normally expressed in more primitive cell types and is downregulated as differentiation proceeds. A hallmark of MLL1 mediated human leukemias is the presence of persistent HOXA9 expression (Krivtsov and Armstrong, 2007) and HoxA9 expression alone can cause leukemia in mice (Thorsteinsdottir et al., 2001). HoxA9 is directly bound and activated by MLL1 and MLL1 fusion proteins in leukemia cells (Milne et al., 2005a; Milne et al., 2005b; Yokoyama and Cleary, 2008) and continuous HOXA9 expression is essential for maintenance of the growth and leukemic potential of MLL1 rearranged human leukemias (Milne et al., 2005; Faber et al., 2009). Both wild type MLL1 and MLL1 fusion proteins are necessary for leukemic transformation (Thiel et al, 2010), but it is unknown if they specifically cooperate in maintaining HoxA9 expression.
In mammals, MLL1 belongs to a family of H3K4 methyltransferases that also includes MLL2 (aka MLL4), MLL3 (aka hALR), MLL4 (aka ALR), SET1A and SET1B (Ruthenburg et al., 2007a). These methyltransferases all reside in distinct protein complexes that contain the common subunits ASH2L, RBBP5 and WDR5 (Dou et al., 2005 and Figure 1A). Despite having similar enzymatic activities, these H3K4Me writer proteins regulate different gene targets (Wang et al., 2009b; Issaeva et al., 2007; Lee et al., 2006; Glaser et al., 2005). MLL1 is specifically required for HoxA9 expression (Milne et al., 2002; Milne et al., 2005a) and is the only H3K4Me writer that has been directly implicated in human leukemia. In fact, despite a very similar domain architecture and overall homology, fusion proteins made using MLL2 in place of MLL1 are unable to transform mouse bone marrow (Bach et al., 2009). It remains an open question why MLL1 is specifically required for HoxA9 expression, but one possibility is that there are distinct, HoxA9 specific recruitment mechanisms for MLL1 and MLL1 fusion proteins.
Menin and LEDGF/p75 have been implicated in MLL1 recruitment to HoxA9 (Yokoyama and Cleary, 2008; Thiel et al, 2010). However, since menin (and by extension LEDGF/p75) are components of both MLL1 and MLL2 complexes (Hughes et al., 2004; Yokoyama and Cleary, 2008), these interactions alone can’t explain specific recruitment of MLL1 to HoxA9. In Drosophila, recruitment of the MLL1 homolog Trithorax is partially disrupted by mutations in the FACT elongation complex (Petruk et al., 2006), while in yeast, the MLL1 homolog SET1 is recruited by gene activation and the PAF elongation complex (Krogan et al., 2003; Ng et al., 2003). Although it has been recognized that a mammalian PAF elongation complex (PAF1C) associates with methyltransferase activity (Rozenblatt-Rosen et al., 2005), neither of these recruitment mechanisms have been fully characterized or shown to function in mammals (Lee and Skalnik, 2008) or to differentiate between mammalian MLL family members.
Here, we set out to systematically determine which protein domain interactions contribute specifically to both wild type and MLL1 fusion protein recruitment to HoxA9. We have identified a minimal recruitment domain that requires the CXXC/RD1 region and the PHD fingers of MLL1. A CXXC/RD1 mediated direct interaction with PAF1C that is not conserved with either hSET1 or with MLL2 is required for MLL1 recruitment to HoxA9. In addition, MLL1 PHD3 binds directly to H3K4Me2/3 and this is also necessary for wild type MLL1 recruitment. MLL1 fusion proteins lack the PHD fingers and are recruited to HoxA9 through direct interactions with PAF1 and CXXC domain recognition of DNA, but this is only possible in the presence of an actively bound wild type MLL1 complex. Since MLL1 and MLL1 fusion proteins display overlapping yet distinct requirements for stable target gene binding, details about these interactions could provide potential therapeutic “windows of opportunity” between wild type and oncogenic versions of MLL1.
RESULTS
Wild type MLL1 is specifically required for activation of HoxA9 and recruitment of MLL1 complex members
To study MLL1 recruitment and HoxA9 regulation in vivo, we used Mll1− − mouse embryonic fibroblast (MEF) cells and an Mll1−/− cell line that had full length MLL1 added back (Mll1−/− + MLL1) for ChIP analysis across 6 Kb of the HoxA9 locus (Figure S1A). MLL1 complex members are expressed in Mll1−/− cells (Figure 1B), but with the exception of the menin protein, none of the complex members were detected bound to the HoxA9 locus (Figures 1C and S1C, blue line). Expression of wild type MLL1 (Figure 1B) recruits the MLL1 complex to the downstream region of HoxA9 (Figures 1C and S1C, blue line, primer sets 4,5 and 6), increases canonical HoxA9 transcript expression (Figure S1B), and drastically increases HoxA9 protein levels (Figure 1B). Global levels of H3K4Me2/3 are unaffected by MLL1 expression (Figure 1B), but levels of H3K4Me2/3 and H3 acetylation (AcH3) are significantly increased at HoxA9 in the region of the bound MLL1 complex (Figures 1C and S1C, red line, primer sets 4,5 and 6).
Binding of the MLL1 complex and activation of HoxA9 also causes a decrease in the repression marks H3K9Me3 and H3K27Me3 (Figure 1C, red versus blue line, primer sets 4, 5 and 6). Consistent with this, there is a decrease in binding of the ESET H3K9 methyltransferase in the same region (Figure 1C, red versus blue line, primer sets 5 and 6). Continued repression of the upstream transcript of HoxA9 (Figure S1B) is strongly correlated with a lack of MLL1 complex binding, a lack of H3K4Me2/3 or AcH3 and maintenance of the H3K9Me3 and H3K27Me3 repression marks (Figures 1C and S1C red line, primer sets 1, 2 and 3). These results show that wild type MLL1 recruitment is necessary for robust activation of HoxA9 and that factors required for MLL1 recruitment function only in the downstream (covered by primer sets 4, 5 and 6) but not the upstream region of HoxA9 in these cells.
Multiple MLL1 domains are necessary for recruitment to HoxA9 in vivo
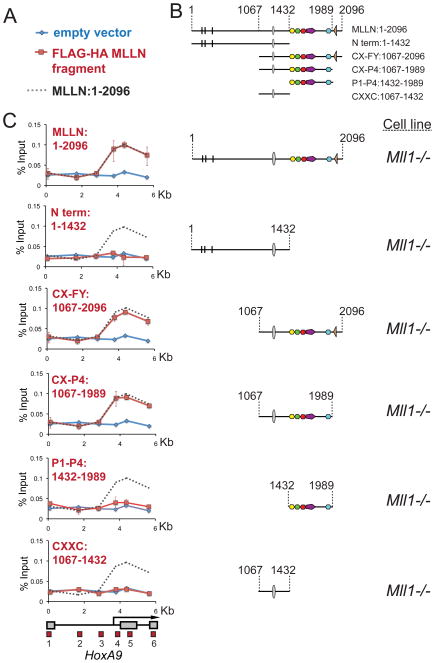
From the results in Figure 1, we determined that Mll1−/− cells provide a useful system for analyzing MLL1 recruitment to HoxA9. To identify a minimal HoxA9 recruitment domain, we made a series of constructs that express FLAG and HA double-tagged MLL1N fragments in Mll1−/− MEF cells (Figure 2B and Figure S2A) and used an anti-HA antibody for ChIP across the HoxA9 locus (Figure 2C red line versus blue line empty vector control). Unexpectedly, we find that a fragment (CX-P4:1067–1989) that lacks the menin and LEDGF/p75 major interaction sites but contains the CXXC domain and PHD fingers (Figure 1A and Figure S2D), is sufficient for targeted recruitment to HoxA9 in vivo (Figure 2C, CX-P4:1067–1989 panel, red line). Additionally, a CXXC/PHD fragment that lacks the HCFC1 interaction site, is also sufficient for recruitment to HoxA9 (Figure Figure S2C and D). Fragments containing the CXXC region alone or PHD fingers 1–4 alone fail to associate with HoxA9 (CXXC:1067–1432 and P1-P4:1432–1989, Figure 2C, red line). This suggests that MLL1 recruitment to HoxA9 may require multiple interactions with different MLL1 domains.
Figure 2. The CXXC domain and PHD fingers are sufficient for targeting MLL1 to the HoxA9 locus in vivo.
(A) Legend for the ChIP experiments in C. Blue line= ChIP in cells transfected with an empty vector, Red line= ChIP in cells transfected with the constructs indicated and Grey dots = the binding pattern of full length MLLN for comparison. (B) Schematics showing the series of deletion constructs. (C) The regions containing the MLL1 CXXC domain and the PHD fingers are both necessary for targeting MLL1 to the HoxA9 locus. Different fragments of the MLL1 N terminus were FLAG and HA (fh) double tagged and expressed in Mll1−/− MEFs and α-HA ChIP experiments were performed. ChIP results shown are typical for at least three independent experiments. Error bars represent standard deviation of three separate PCR reactions. See also Figure S2.
The CXXC/RD1 domain of MLL1 interacts with the PAF complex
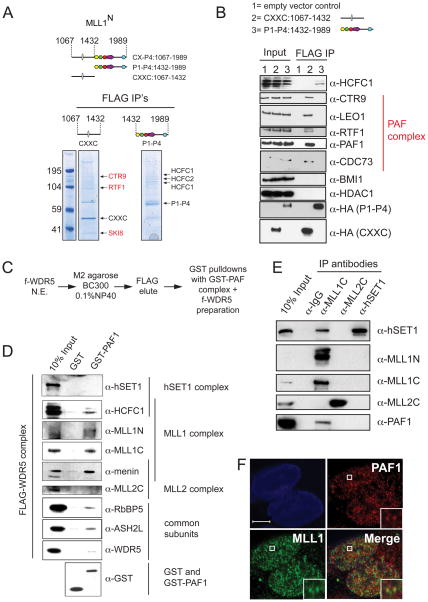
From the results in Figure 2, we determined that there are potentially one or more essential recruitment activities in the PHD fingers as well as in the region containing the CXXC domain of MLL1. To identify proteins that could potentially be involved in recruitment, FLAG-HA (fh) tagged CXXC:1067–1432 and fh-P1-P4:1432–1989 MLLN fragments were stably expressed in 293 cells and subjected to a one-step FLAG purification under mild wash conditions. Consistent with previous more rigorous purifications (Dou et al., 2005), mass spectroscopy analysis identified the core complex members HCFC1 and HCFC2 (Figure 3A). Interestingly, several CXXC interacting proteins were also identified (CTR9, RTF1, SKI8, red, Figure 3A), all of which are components of the PAF elongation complex (PAF1C, Kim et al, 2009; 2010). Western blots confirmed the specificity of the PAF1C and HCFC1 interactions (Figure 3B). Sequences surrounding the MLL1 CXXC domain have previously been shown to interact directly with the repressor proteins HDAC1 and BMI1 (Xia et al., 2003), but under our IP conditions, we failed to detect this interaction (Figure 3B).
Figure 3. The MLL1 complex interacts specifically with the PAF elongation complex.
(A) 293 cells stably expressing either FLAG-HA tagged (fh) CXXC:1067–1432 or fh-P1-P4:1432–1989 were subjected to a one step FLAG purification. Mass spectroscopy analysis identified components of the MLL1 core complex (HCFC1 and HCFC2) and components of PAF1C (shown in red) (B) FLAG IP’s were done from 293 cell nuclear extracts expressing either an empty vector (1), fh-CXXC:1067–1432 (2) or fh- P1-P4:1432–1989 (3). Western blots were probed with the antibodies indicated. (C) Scheme for purification of multiple MLL family complexes from HeLa extracts expressing a FLAG (f-) tagged WDR5 protein. (D) GST-PAF1C specifically interacts with components of the MLL1 complex. The f-WDR5 preparation was subjected to GST pulldowns in the presence of either GST or a GST-hPAF1C and the results were blotted and probed with the antibodies indicated. (E) PAF1 interacts with the MLL1 complex in vivo. 293 nuclear extracts were used for IP’s with the antibodies indicated and western blots were probed with the antibodies indicated. (F) Immunofluorescence of PAF1 (red) and MLL1 (green) in 293 cells. PAF1 and MLL1 overlap in only discrete regions, a blowup of a region of overlap is shown as an example (white box). The white bar represents 5μm. See also Figure S3.
The PAF1C interaction is specific for MLL1
If PAF1C is required for recruitment of MLL1 to HoxA9, we might expect that the interaction would be specific for MLL1 and not involve other MLL family members. To test this possibility, we used a FLAG tagged common subunit (f-WDR5) to purify multiple MLL family complexes (Figure S3A) which were then incubated with either GST alone or a recombinant, purified GST-PAF1 complex (Figure S3B). GST-PAF1C specifically interacts with MLL1 complex components (Figure 3D), but not with either SET1 or MLL2 (Figure 3D). Immuno-precipitation (IP) experiments in 293 cell nuclear extracts confirm this result and show a specific interaction between MLL1 and PAF1 (Figure 3E). Interestingly, under these conditions, hSET1 also IP’s with MLL1C (Figure 3E, lane 3), suggesting that there may be an interaction between these two methyltransferase complexes.
Only a small amount of PAF1 and MLL1 IP together (Figure 3E), suggesting that either this interaction is weak or that it occurs at only a few loci. To differentiate between these two possibilities, we used MLL1 and PAF1 specific antibodies for immunofluorescence experiments in 293 cells (Figure 3F) and in Mll1−/− + MLL cells (Figure S3C). Interestingly, overlap between PAF1 and MLL1 is relatively rare (See Figure 3F, inset for an example), suggesting that the interaction between MLL1 and the PAF complex occurs only at a small number of specific sites in vivo.
The CXXC containing RD1 region of MLL1 interacts directly with PAF1
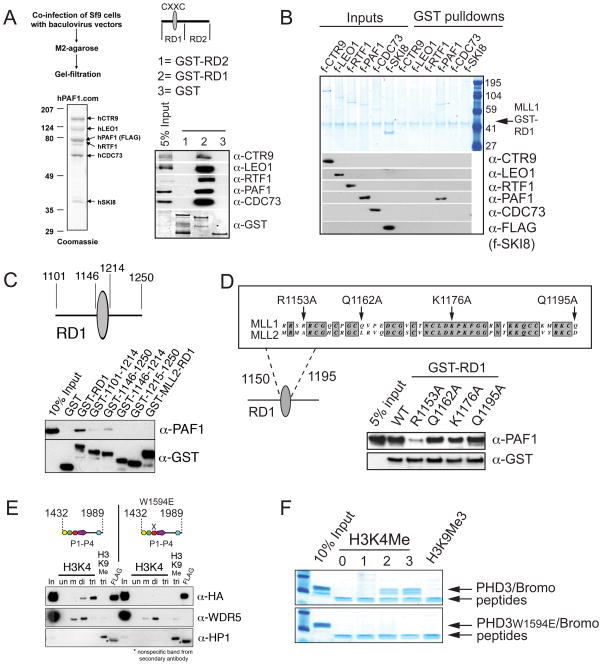
For direct interaction studies, the MLL1 CXXC region was divided into GST-RD1 and GST-RD2 subfragments as in Xia et al., 2003 (Figure 4A). GST pull-down experiments with a recombinant, purified PAF complex (Figure 4A) shows that PAF1C interacts specifically with RD1, the region that contains the CXXC domain (Figure 4A and Figure S4A). Pull-down experiments with GST-RD1 and individually expressed FLAG (f-) tagged recombinant components of PAF1C reveal that it is the PAF1 component of the complex that interacts specifically and directly with RD1 (Figure 4B). We next wanted to characterize the PAF1 interaction more carefully in order to design point mutations for in vivo studies.
Figure 4. The MLL1 CXXC/RD1 domain interacts directly with PAF1 and PHD finger 3 interacts directly with H3K4Me2/3.
(A) PAF1C interacts directly with the CXXC containing RD1 fragment of MLL1. The MLL1 CXXC region was divided into two fragments (RD1 and RD2) and expressed as recombinant GST proteins. Purified GST (1), GST-RD1 (2) and GST-RD2 (3) were used in GST pulldown experiments with a recombinant purified PAF1C. Westerns were probed with the antibodies indicated. (B) The PAF1 component of PAF1C interacts directly with MLL RD1. GST-RD1 was used in a GST pulldown experiment with individual FLAG (f-) tagged components of human PAF1C as shown. The upper panel is a gel stained with colloidal blue and the lower panels are western blots of the same pulldowns using the antibodies indicated. (C) Only full length MLL1-RD1 interacts efficiently with PAF1, while MLL2-RD1 does not. A series of deletions of the MLL1 RD1 region were made and subjected to GST pulldowns with purified f-PAF1. Westerns were probed with the antibodies indicated. (D) Top: conservation with MLL2 and the positions of point mutations made in the MLL1 CXXC domain are shown. Bottom: The R1153A point mutation disrupts the direct interaction between purified GST-RD1 and purified f-PAF1 protein. GST pulldowns were done as in (A). (E) PHD fingers 1 to 4 with and without a point mutation in PHD3 (W1594E) were FLAG and HA double tagged and expressed in 293 cells. Nuclear extracts were subjected to peptide pulldowns as indicated and probed with the antibodies shown. (F) Recombinantly expressed and purified dual PHD3/Bromodomain of MLL1 binds specifically to H3 lysine 4 di and trimethyl peptides (top panel). A W1594E point mutation in PHD3 abolishes this interaction (bottom panel). See also Figure S4.
An R1153A point mutation in MLL1 disrupts the interaction between PAF1 and MLL1
To narrow down the PAF1/MLL1 interaction domain, we constructed a series of deletions across the RD1 region (Figure 4C), but efficient binding to PAF1 was only seen with the entire intact RD1 region (Figure 4C). The equivalent of the CXXC containing RD1 regions of MLL2 and CFP1 (the CXXC containing component of the hSET1 complex) do not interact with PAF1 (Figure 4C and Figure S4B), providing further evidence that PAF1 interacts specifically with MLL1 and does not directly recruit the MLL2 or hSET1 complexes.
The CXXC domain has previously been shown to bind to unmethylated CpG rich DNA and structural analyses have identified the specific residues that are directly involved in DNA binding (Allen et al., 2006; Cierpicki et al., 2009). Residues on the opposite face of the DNA binding pocket are potentially available for interactions with other proteins (Allen et al., 2006; Cierpicki et al., 2009). Using these structural analyses, we made three point mutations (R1153A, Q1162A and Q1195A) in residues that specifically have no effect on DNA binding (Allen et al., 2006), do not disrupt the overall fold of the CXXC domain (Allen et al., 2006), are not conserved between MLL1 and MLL2 (Figure 4D), and map to the opposite face of the DNA binding fold (Allen et al., 2006). The R1153A point mutation was especially interesting to us because it converts an R in MLL1 to a matching A residue in MLL2. Since the in vitro DNA binding properties of the MLL1 and MLL2 CXXC domains are similar (Bach et al., 2009), this is further evidence that the R1153A point mutation has no effect on DNA binding. As a control, we also used a point mutation (K1176A, Figure 4D) that does not affect the three-dimensional folding of the CXXC domain, but is essential for DNA binding (Allen et al., 2006).
In GST pull-down experiments, the R1153A point mutation in the CXXC domain disrupts the RD1/PAF1 interaction and reduces binding significantly (Figure 4D, bottom) while point mutations in either the DNA binding fold (K1176A) or the Q1162A or Q1195A residues have no effect on the RD1/PAF1 interaction (Figure 4D, bottom). This result is especially significant considering that the R1153A mutation makes MLL1 more closely mimic MLL2, which does not interact with PAF1.
The third PHD finger of MLL1 binds specifically and directly to H3K4Me2/3
In Figure 2 we showed that the PHD fingers are also necessary for recruitment of MLL1 to HoxA9. Close examination of the amino acid sequences of the four PHD fingers of MLL1 suggested that PHD 3, but not PHD 1, 2 or 4 has H3K4Me binding activity (Ruthenburg et al., 2007a and Figure S4C). An MLL1 fragment containing all 4 PHD fingers bound specifically to H3K4Me2/3 (Figure 4E). A PHD3 W1594E point mutation abolished the H3K4Me2/3 interaction (Figure 4E), indicating that neither PHD1, 2 nor 4 can interact with H3K4Me. Specific PHD3 binding to H3K4Me2/3 was confirmed with recombinant, purified PHD3/Bromodomain protein (Figure 4F) and is further supported by a recently determined PHD3/H3K4Me structure (Wang et al., 2010).
The MLL1 recruitment domain requires an interaction with PAF1 and H3K4Me2/3 to bind to HOXA9 in vivo
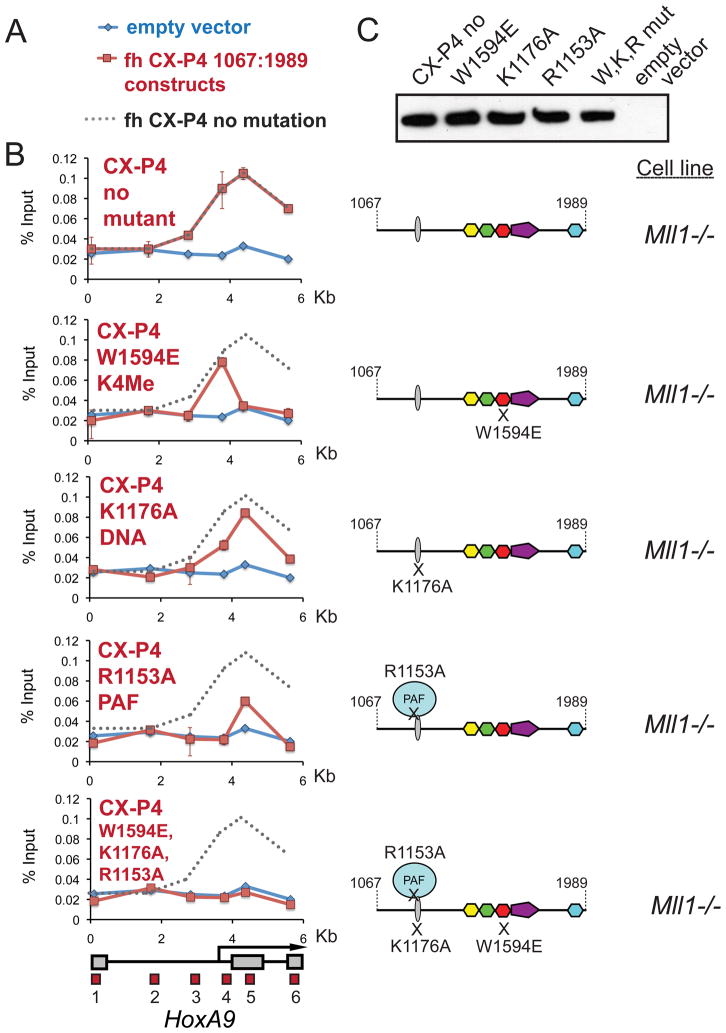
Having characterized several interactions, we used α-HA ChIP to determine the relative importance of each interaction for binding of the MLL1 recruitment domain to HoxA9. Disruption of the DNA binding activity of the CXXC domain has only a mild effect on binding to HoxA9 (Figure 5B, K1176A panel), while the PAF1 mutation almost completely disrupts recruitment to HoxA9 (Figure 5B, R1153A panel). Surprisingly, even though H3K4Me2/3 levels are quite low at HoxA9 in Mll1−/− cells (see Figure 1C and Figure S1D), disruption of H3K4Me binding with the W1594E mutation abolishes CX-P4 1067:1989 binding to the coding region, although not the promoter, of HoxA9 (Figure 5B, W1594E panel). Binding of CX-P4 1067:1989 has no effect on H3K4Me3 or AcH3 levels at HoxA9 (Figure S5A), suggesting that the small peak of H3K4Me2/3 in the coding region at HoxA9 (Figures S1D and S5A, compare probes 4, 5 and 6 to 1, 2 and 3) is sufficient for promoting the recruitment of MLL1. Point mutations that disrupt PHD fingers 1,2 and 4 have no effect on binding of CX-P4 1067:1989 to HoxA9 (Figure S5B and C and see also Figure S2C), which suggests that the other PHD fingers of MLL1 are not required for recruitment. Significantly, all three point mutations combined completely abolish binding of the MLL1 recruitment domain to HoxA9 (Figure 5B,W1594E-K1176A-R1153A bottom panel). Taken together, these results suggest that DNA binding, H3K4Me2/3 binding and the PAF1 interaction are all necessary to recapitulate the wild type MLL1 binding pattern at HoxA9 in Mll1−/− cells.
Figure 5. Binding to H3K4Me and PAF1 are both necessary for recruitment of MLL1 across HoxA9 in vivo.
(A) Legend for the ChIP experiments in C. Blue line= HA ChIP in cells transfected with an empty vector, Red line= HA ChIP in cells transfected with the CX-P4 constructs indicated and Grey dots = the binding pattern of CX-P4 with no mutation. (B) PAF1 and H3K4Me interactions are important for recruiting MLL1 to HoxA9. Using α-HA ChIP, recruitment of the MLL1 fh-CX-P4 protein to HoxA9 in Mll1−/− MEF cells (no mutant) was compared to individual PHD3 W1594E, K1176A and R1153A point mutations and a triple mutant. ChIP results shown are typical for at least two independent experiments. Error bars represent standard deviation of three separate PCR reactions. (C) An α-HA western blot showing expression of the wild type and mutant constructs from the experiments in B. W,K,R mut = the W1594E, K1176A, R1153A triple mutant. See also Figure S5.
The MLL-AF9 leukemogenic fusion protein requires pre-binding of the MLL1 wild type complex for recruitment to an active HOXA9 locus
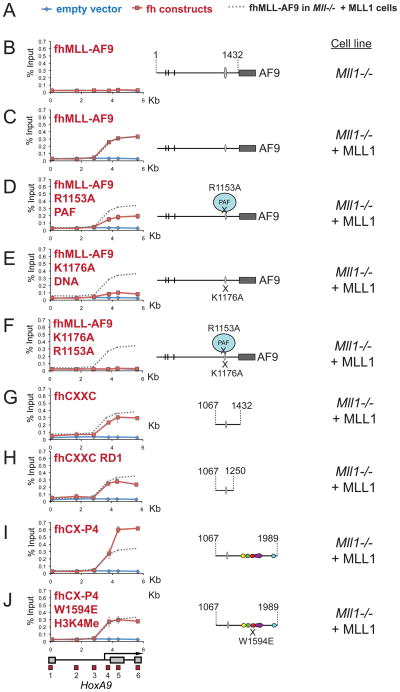
Our data shows that both the PHD fingers and CXXC domain are necessary for recruitment of MLL1 to HoxA9 in Mll1−/− cells. MLL1 fusion proteins, however, retain the CXXC domain but not the PHD fingers of MLL1 (see Figure 1A). Consistent with these results, a FLAG-HA (fh) tagged MLL-AF9 fusion protein cannot bind to or activate the HoxA9 locus in Mll1−/− cells (Figures 6B and S6A and B). Conversely, when fh-MLL-AF9 is expressed in Mll1−/− + MLL1 cells (see Figure S6C), fh-MLL-AF9 is able to bind to the downstream region of HoxA9 (Figure 6C) where wild type MLL1 is bound (see Figure 1). These results suggest that MLL1 fusion protein recruitment to HoxA9 requires pre-binding of the wild type MLL1 complex to create an “open” chromatin state and provides a mechanistic explanation for the observation that wild type MLL1 is necessary for fusion protein mediated leukemogenesis (Thiel et al., 2010).
Figure 6. The MLL-AF9 fusion protein needs wild type MLL1, PAF1 and DNA binding for recruitment to HoxA9.
(A) Legend for the ChIP experiments in B-J. Blue line= HA ChIP in cells transfected with an empty vector, Red line= HA ChIP in cells transfected with the constructs indicated and Grey dots = the binding pattern of fh-MLL-AF9 in Mll1−/− + MLL cells for comparison. (B) – (J) α-HA ChIP across HoxA9 for the constructs and in the specific cell lines as indicated. ChIP results shown are typical for at least three independent experiments. Error bars represent standard deviation of three separate PCR reactions. See also Figure S6.
The MLL-AF9 leukemogenic fusion protein minimal recruitment domain requires DNA binding and the PAF1 interaction for recruitment to an active HoxA9 locus
Since MLL1 fusion proteins contain only the CXXC “half” of the MLL1 recruitment domain, we wanted to determine if the CXXC domain interactions are important for MLL1 fusion protein recruitment. We compared binding of full length fh-MLL-AF9 to fh-MLL-AF9 with CXXC mutations in either K1176A (DNA binding) or R1153A (PAF1 interaction) in Mll1−/− + MLL1 cells (Figures 6C-F). Using an α-HA antibody, ChIP experiments reveal that the R1153A/PAF1 disruption reduces fh-MLL-AF9 binding by about two-fold (Figure 6D), while a K1176A/DNA binding mutation drastically reduces fh-MLL-AF9 binding across HoxA9 (Figure 6E). This result contrasts that with the wild type MLL1 protein, which has only a minimal requirement for the DNA binding activity of the CXXC domain (Figure 5B). Significantly, an R1153A/K1176A double mutant completely abolishes binding across HoxA9 (Figure 6F), suggesting that these two interactions are both important to recapitulate MLL-AF9 binding across HoxA9. Amazingly, expression of the minimal fragment necessary to interact with PAF1 and bind to DNA (fh-CXXC RD1:1067–1250), is sufficient to recapitulate full length MLL-AF9 binding across HoxA9 in Mll1−/− + MLL cells (Figure 6H).
We have found that without the PHD fingers, MLL1 cannot be recruited to HoxA9 in Mll1−/− cells, but in Mll1−/− + MLL1 cells the CXXC RD1:1067–1250 domain alone is sufficient for recruitment to HoxA9. In order to see if PHD3 binding to H3K4Me2/3 contributes to HoxA9 binding in Mll1−/− + MLL1 cells, we compared binding of the fh-CX-P4 1067:1989 recruitment domain (which contains both the CXXC domain and PHD fingers) with fh-MLL-AF9 in Mll1−/− + MLL1 cells. The fh-CX-P4 fragment shows about a two fold increase in binding compared to fh-MLL-AF9 (Figure 6I, probes 4 and 5) in the peak region of H3K4Me (compare Figure 6I, probes 4 and 5 with H3K4Me2/3 in Figure 1C and Figure S1C). The W1594E mutation in PHD3 disrupts this PHD mediated increase in binding (Figure 6J, probes 4 and 5).
Recruitment of the MLL-AF9 fusion protein to the HoxA9 locus is essential for HoxA9 gene activation in leukemogenesis
In leukemia, except in a few rare instances, MLL1 fusion protein translocations involve only one allele of MLL1 and leave one wild type copy of MLL1 untouched (Ayton and Cleary, 2001). We have previously used an in vitro assay to show that leukemogenic oncogenes have the ability to transform mouse bone marrow cells (Wang et al., 2009a and summarized in Figure 7A top).
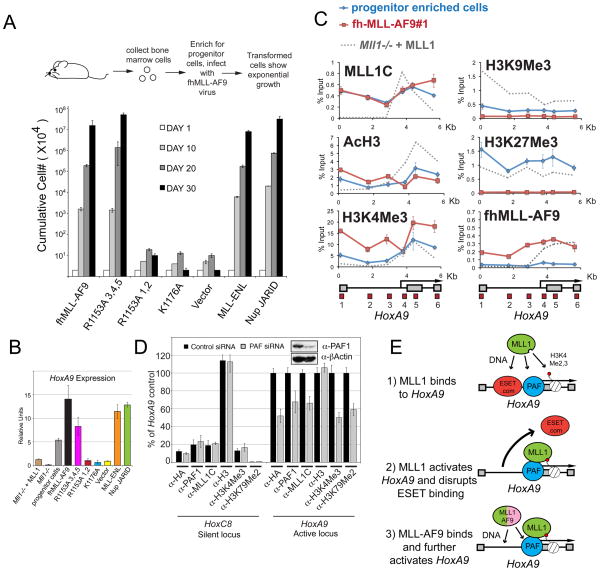
Figure 7. Recruitment of fh-MLL-AF9 to HoxA9 in bone marrow cells is essential for leukemic transformation.
(A) Leukemogenic transformation of MLL-AF9 constructs. Retrovirally expressed FLAG and HA tagged (fh-) MLL-AF9 fusion protein constructs that were either normal for MLL-AF9 or had R1153A (PAF1 disruption), or K1176A (DNA binding disruption) point mutations were transduced into murine bone marrow cells, along with empty vector, untagged MLL-ENL, and triple FLAG tagged NUP98-Jarid controls. Error bars represent the standard deviation of averaged, cumulative cell numbers across five independent lines for each construct. (B) HoxA9 expression in transformed and control cell lines. Cells from (A) were tested for HoxA9 expression and the average values across all five independent cell lines per construct are shown. Error bars represent standard deviation. HoxA9 expression in Mll1−/− and Mll1−/− + MLL cell lines are also shown for comparison. (C) ChIP in progenitor enriched (Blue line) and fh-MLLAF9 (line #1, Red line) cells with the antibodies indicated. ChIP experiments in Mll1−/− + MLL cells are shown for comparison (Grey dots). (D) PAF1 is required for MLL1 and MLL-AF9 binding to HoxA9. The fh-MLLAF9#1 cell line was treated with control vs PAF1 siRNA’s and the cells were subjected to western blot (inset) and ChIP analysis using the antibodies indicated. α-HA antibody recognizes the HA tagged MLL-AF9 fusion protein. ChIP results were quantified using a primer/probe set for the silent Hoxc8 locus or the mouse HoxA9 coding region (primer/probe set 5 from D). ChIP signal was quantified to inputs and then relative to the HoxA9 control signal which was arbitrarily set to 100. (E) 1= wild type MLL1 is recruited to HoxA9 through interactions with H3K4Me, PAF1 and DNA. 2= MLL1-mediated activation of the HoxA9 locus disrupts binding of repressors such as ESET and creates a more “open” chromatin conformation. 3= An MLL1 fusion protein can now bind through interactions with PAF1 and with CpG rich DNA. ChIP results shown are typical for at least two independent experiments. Error bars represent standard deviation of three separate PCR reactions. See also Figure S7.
To test whether MLL-AF9 recruitment to HoxA9 was important for leukemic transformation, we transduced mouse bone marrow cells with normal fh-MLL-AF9, fh-MLL-AF9 R1153A/PAF1 (R1153A) point mutants, or fh-MLL-AF9 K1176A/DNA (K1176A) point mutants. An empty vector was used as a negative control and untagged MLL-ENL and a FLAG tagged NUP98-Jarid fusion protein were used as positive controls. The NUP98-Jarid fusion protein is unrelated to MLL1 fusion proteins, but it causes leukemia in a HoxA9 dependent manner (Wang et al., 2009a). For all of the fh-MLL-AF9 constructs, five independent retroviral transductions were tested and cell counts and gene expression assays were averaged across lines where appropriate.
The fh-MLL-AF9 lines and the MLL-ENL and Nup JARID positive controls all showed exponential growth (Figure 7A) and increased HoxA9 expression (Figure 7B). fh-MLL-AF9 lines with the K1176A/DNA mutation, R1153A lines #1 and 2 and cells transformed with an empty vector failed to grow (Figure 7A) and showed an inability to maintain HoxA9 expression (Figure 7B). R1153A/PAF mutant lines #3,4 and 5 all showed exponential growth (Figure 7A), although they only maintained an intermediate level of HoxA9 expression (Figure 7B) and have increased expression of the Gr1 cell surface marker (fh-MLL-AF9 line#1 vs. R1153A#1, Figure S7B), suggesting that they are slightly more differentiated than WT controls. Most of the fh-MLL-AF9 transduced cells expressed MLL-AF9 at roughly equal levels (Figure S7A), but R1153A cell lines 3,4 and 5 had MLL-AF9 expressed at 4–5 fold higher levels relative to normal fh-MLL-AF9 (Figure S7A). These results suggest two things: 1) since the R1153A mutation only disrupts MLL-AF9 binding by about half (see Figure 6D), this defect can be partially overcome by higher level expression levels of the MLL-AF9 construct, and 2) the K1176A mutation that severely disrupts fhMLL-AF9 recruitment (see Figure 6E), also completely abolishes the ability of this protein to cause leukemic transformation in bone marrow cells (Figure 7A).
Our results above would predict that for efficient leukemic transformation, the HoxA9 locus in progenitor enriched cells should have MLL1 already bound to it. Using ChIP, we found that wild type MLL1 was bound across the entire HoxA9 locus in progenitor enriched cells (Figure 7C, blue line). AcH3, H3K4Me3 and H3K27Me3 are spread across the entire HoxA9 locus, while there is almost no detectable H3K9Me3 in progenitor cells at HoxA9 (Figure 7C, blue line). Levels of H3K27Me3 may be due to small amounts of differentiated cells present. Using line#1 for ChIP analysis, we found that fh-MLL-AF9 binds across the entire HoxA9 locus in a pattern that mirrors wild type MLL1 binding (Figure 7C) and causes an increase in H3K4Me3 and H3K79Me2 levels (Figure 7C and Figure S7C, red line versus blue line). These results indicate that, similar to Mll1−/− + MLL cells, in progenitor enriched cells HoxA9 has wild type MLL1 bound to it and has low levels of H3K9Me3.
To further test whether the PAF1 protein is required for MLL-AF9 and/or MLL1 binding to HoxA9, we used siRNA’s to partially knockdown PAF1 in fh-MLL-AF9 line#1 (Figure 7D). By ChIP analysis, PAF1 binding to the HoxA9 locus was reduced by about 30% (Figure 7D), and both MLL1 and fh-MLL-AF9 binding was reduced about 40–50% (Figure 7D). Overall H3 levels were not affected by the PAF1 siRNA, but there was a reduction in H3K4Me3 and H3K79Me2 that mirrored the loss of MLL1 and fh-MLL-AF9 binding (Figure 7D). Thus in transformed myeloblast cells, PAF1 is required for stable binding of both MLL1 and MLL1 fusion proteins and for H3K4 and H3K79 methylation at HoxA9. Taken together, these results show that both DNA binding and the PAF1 interaction contribute to recruitment of the MLL-AF9 fusion protein in leukemia cells.
DISCUSSION
Wild type MLL1 protein is recruited to HoxA9 in vivo through interactions with PAF1 and H3K4Me2/3
MLL family members regulate a wide range of different gene targets, but little is known about the mechanisms that control preferential recruitment of MLL1 or MLL1 fusion proteins to the HoxA9 locus in leukemia. In this study, we have identified a minimal MLL1 recruitment domain that is controlled by a CXXC/RD1 domain interaction with PAF1 and an MLL1 PHD3 direct interaction with H3K4Me2/3.
Mammalian PAF1C is composed of 6 subunits that includes CTR9, LEO1, RTF1, PAF1, CDC73 and SKI8 (Kim et al, 2009; 2010). Detailed in vitro studies have indicated that PAF1C has a distinct role in directly facilitating transcription elongation, as well as promoting H2BK120 ubiquitylation (uH2BK120) by recruiting a BRE1 complex (Kim et al., 2009; 2010). uH2BK120 is required for global H3K4Me3 and H3K79Me2 levels (Kim et al., 2009), which suggests that PAF1C has an important role in controlling histone methylation through uH2BK120. In yeast, there is evidence that PAF1C can also recruit H3K4Me activity through recruitment of SET1, but a direct interaction between the proteins/complexes has not been established (Krogan et al., 2003). Although it still remains formally possible that other MLL family members interact with PAF1, our studies failed to observe an MLL2 or human SET1 interaction with PAF1C (also see Lee and Skalnik, 2008). Instead, we provide evidence of a direct link between PAF1C and H3K4Me through recruitment of MLL1.
The specificity of this interaction suggests that PAF1 could be part of the mechanism for specifically recruiting MLL1 to HoxA9. Interestingly, since overlap between MLL1 and PAF1 in vivo is not complete (Figure 3F), this suggests that the MLL1/PAF1 interaction alone is not sufficient for MLL1 recruitment to all target genes. Instead, Ruthenburg et al (2007b) have suggested that chromatin-associated proteins and the complexes that they reside in could be recruited to gene targets through multiple weak or transient interactions that together produce specificity and stable binding. In the case of MLL1, binding to either H3K4Me or PAF1 alone may not be sufficient for recruitment, but the combined interactions could produce stable and specific recruitment of MLL1 to HoxA9. Our data do not rule out the possibility that other interactions also contribute to this effect or that a single MLL1 protein interacts sequentially with PAF1 and H3K4Me, creating transient subcomplexes.
MLL1 fusion proteins are dependent on wild type MLL1 binding for recruitment to HoxA9
We have shown that the MLL-AF9 fusion protein can only bind to HoxA9 in regions where MLL1 is present (Figure 6B and C and Figure 7C). We have no evidence for a direct protein-protein interaction between MLL1 and MLL-AF9, so we instead favour the idea that H3K9Me mediated repression prevents binding of the MLL1 fusion protein while higher levels of gene activation promote recruitment of the fusion protein. More specifically, the order of events that we envision is that wild type MLL1 is recruited to HoxA9 through interactions with H3K4Me, PAF1 and DNA (Figure 7E 1) and are together able to stabilize MLL1 binding even though repressors such as ESET are present (Figure 7E 1). MLL1-mediated activation of the HoxA9 locus and binding to CpG rich DNA disrupts binding of repressors such as ESET (Figure 7E 2). This allows an MLL1 fusion protein to bind through interactions with PAF1 and with CpG rich DNA (Figure 7E 3) and also perhaps through other uncharacterized interactions. MLL1 fusion proteins may be more dependent on the DNA binding activity of the CXXC domain because they lack PHD fingers and do not have the additional stabilizing effect of binding to H3K4Me. It is not clear exactly how the presence of H3K9Me blocks MLL1 fusion protein binding, but the CXXC domain containing MBD1 protein represses HoxA cluster genes and correlates with sites of H3K9Me (Sakamoto et al., 2007). Thus, MBD1 could block MLL-AF9 fusion protein binding by competing for CpG rich DNA binding sites.
Our findings call attention to differences in recruitment and stable binding between the wild type MLL1 protein and leukemogenic MLL1 fusion proteins, which could, in turn, be exploited for the development of targeted therapies. For instance, we predict that inhibitors designed to specifically target the DNA binding pocket of the CXXC domain 19 would be more disruptive on MLL1 fusion proteins than the wild type MLL1 protein itself. We look forward to further experiments aimed at testing this and related hypotheses as the “rules of recruitment” become better understood for MLL1 and other such chromatin remodeling activities that lie at the heart of normal and abnormal pathways of development and differentiation
Experimental Procedures
Transfection of Cell lines
Mll1−/− MEF cells were transfected with various constructs using an Amaxa Nucleofector machine according to the manufacturer’s instructions. 293 cells were transfected using Fugene 6 (Roche).
Chromatin Immunoprecipitation (ChIP) Experiments
The ChIP protocol was followed as outlined in Milne et al., 2009 using a formaldehyde fixation protocol for histone ChIP and a DMP/formaldehyde double fixation protocol for MLL1 complex members. Results were quantified relative to inputs as explained in detail in Milne et al., 2009 and using a 7300 ABI Real Time PCR machine. Taqman primer/probe sets are listed in supplemental procedures.
Gene expression
cDNA was made and gene expression was quantified for Mll1−/− MEF cells and mouse bone marrow cells as in Milne et al., 2005b. Taqman primer/probe sets are listed in supplemental procedures.
FLAG one step purifications and immunoprecipitations
Nuclear extracts expressing either FLAG-HA tagged CXXC or P1-P4 were incubated with M2 beads in 50mM Tris, pH 7.5, 300mM KCl and 20% Glycerol. Beads were washed 4X in 50mM Tris, pH 7.5, 150mM KCl, 0.05% NP-40, and 20% glycerol. IP’s of native MLL family complexes were done with the antibodies listed in supplemental procedures and with 5X washes of 50mM Tris, pH 7.5, 200mM KCl, 0.1% NP-40, and 20% glycerol. Nuclear extracts expressing FLAG tagged WDR5 were incubated with M2 beads in 50mM Tris, pH 7.5, 300mM KCl and 20% Glycerol and washed 3X in binding buffer along with 0.1% NP40. Bound proteins were eluted with FLAG peptide.
Immunofluorescence staining
Immunofluorescence staining was performed using a standard protocol and the following antibody dilutions: rabbit PAF1 1:500 (Bethyl), mouse MLL1 1:100 (Millipore). Details are provided in supplemental procedures.
Reconstitution of the hPAF Complex
As outlined in Kim et al, 2010.
GST pulldown Assays
For GST-pull down assays, 500ng of GST-fused proteins and 200 ng of purified factors were mixed with glutathione-Sepharose 4B beads in binding buffer (20 mM Tris-Cl [pH 7.9], 300 mM KCl, no EDTA, 20% glycerol, 0.1% NP-40) and washed 3X in the same buffer. Bound proteins were analyzed by western blotting and/or colloidal blue staining.
Peptide pull-down assay
Pull-downs using biotinylated histone peptide and recombinant protein was performed as described (Wang et al., 2009a). Results were run on a gel and stained with coomassie blue or blotted and probed with an α-HA antibody.
RNA interference
The WT#1 myeloblast cell line was treated with control or mouse PAF siRNA duplex (Dharamacon) using an Amaxa Nucleofector machine according to the manufacturer’s instructions.
Haematopoietic cell transformation assays
Protocols for the culture of primary murine haematopoietic stem/progenitor cells were previously described (Wang et al, 2009a). In brief, 100,000 lineage-negative bone marrow stem/progenitor cells were subjected to retroviral infection, followed by kinetics analyses of proliferation versus differentiation
FACS analysis
Approximately 5×10^5–1×10^6 cells were washed and stained with antibodies from BD Pharmingen before FACS analysis and were compared to unstained control cells. Anti-cKit-FITC was used at 1:1000, anti-Gr1-PE at 1:500, and anti-Mac1-FITC at 1:5,000.
Antibodies used, primer/probe sequences and mass spec techniques are in Supplemental procedures
Supplementary Material
Acknowledgments
This work was supported by a Leukemia and Lymphoma Society SCOR grant and by National Institutes of Health (NIH) grants to C.D.A. and R.G.R. T.A.M was supported by a Canadian Institutes of Health Research (CIHR) fellowship and a Leukemia and Lymphoma Society SCOR grant. J.K. was supported by a Senior Fellowship in Biomedical Sciences from the Charles H. Revson Foundation. We thank Christina Hughes for helpful comments on the manuscript and Dr. Jay Hess for sharing of results before publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, Freund SM, Bycroft M, Warren AJ. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. Embo J. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- Bach C, Mueller D, Buhl S, Garcia-Cuellar MP, Slany RK. Alterations of the CXXC domain preclude oncogenic activation of mixed-lineage leukemia 2. Oncogene. 2009;28:815–823. doi: 10.1038/onc.2008.443. [DOI] [PubMed] [Google Scholar]
- Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol. 2010;17:62–8. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, Zwaan CM, Kung AL, Armstrong SA. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- Hess JL. Mechanisms of transformation by MLL. Crit Rev Eukaryot Gene Expr. 2004;14:235–254. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.10. [DOI] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee DK, Dou Y, Lee J, Lee B, Kwak E, Kong YY, Lee SK, Roeder RG, Lee JW. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci U S A. 2006;103:15392–15397. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28:609–618. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci U S A. 2005a;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005b;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Milne TA, Zhao K, Hess JL. Chromatin immunoprecipitation (ChIP) for analysis of histone modifications and chromatin-associated proteins. Methods Mol Biol. 2009;538:409–423. doi: 10.1007/978-1-59745-418-6_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007a;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007b;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto Y, Watanabe S, Ichimura T, Kawasuji M, Koseki H, Baba H, Nakao M. Overlapping roles of the methylated DNA-binding protein MBD1 and polycomb group proteins in transcriptional repression of HOXA genes and heterochromatin foci formation. J Biol Chem. 2007;282:16391–16400. doi: 10.1074/jbc.M700011200. [DOI] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21:224–234. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009a;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009b doi: 10.1128/MCB.00924-09. advanced online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ. Molecular events underlying MLL1 regulation by H3K4me3 readout and cyclophilin-mediated proline isomerization. Cell. 2009c in press. [Google Scholar]
- Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci U S A. 2003;100:8342–8347. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.