Abstract
Prr/RegA response regulator is a global transcription regulator in purple bacteria Rhodobacter sphaeroides and Rhodobacter capsulatus, and is essential in controlling the metabolic changes between aerobic and anaerobic environments. We report here the structure determination by NMR of the C-terminal effector domain of PrrA, PrrAC. It forms a three-helix bundle containing a helix–turn–helix DNA binding motif. The fold is similar to FIS protein, but the domain architecture is different from previously characterised response regulator effector domains, as it is shorter than any characterised so far. Alignment of Prr/RegA DNA targets permitted a refinement of the consensus sequence, which contains two GCGNC inverted repeats with variable half-site spacings. NMR titrations of PrrAC with specific and non-specific DNA show which surfaces are involved in DNA binding and suggest residues important for binding specificity. A model of the PrrAC/DNA complex was constructed in which two PrrAC molecules are bound to DNA in a symmetrical manner.
INTRODUCTION
The purple, non-sulfur bacterium Rhodobacter sphaeroides has very versatile metabolic activities. Under low oxygen conditions, the global regulator Prr (homologous to Reg in Rhodobacter capsulatus) coordinatively controls a number of genes involved in the switch between aerobic and anaerobic lifestyles. It activates genes involved in photosynthesis, nitrogen and carbon fixation, and regulates other energy generating and consuming processes and its own expression (1).
The Prr/Reg system is a bacterial two-component system (2), and consists of the two proteins PrrB and PrrA. Proteins homologous to PrrB and PrrA have been found in other proteobacteria including non-photosynthetic bacteria, suggesting conserved mechanisms and/or properties, despite different in vivo functions (3,4). PrrB, a membrane- bound histidine kinase, is activated under low oxygen conditions, and autophosphorylates on a conserved histidine. It then transfers the phosphate to the response regulator (RR) PrrA, on a conserved aspartate, which increases its DNA-binding activity.
Sequence analysis suggests that PrrA consists of two domains. The N-terminal receiver domain (residues 1–130) is common to all bacterial TCS RRs, and there are a number of structures of these domains, e.g. FixJ (5) and NtrC (6). The C-terminal domain runs from approximately residue 141 to the C-terminus (184), with a short proline-rich linker between the two domains. Although sequence analysis of the C-terminal domain suggests a helix–turn–helix (HTH) DNA-binding motif, there are no structures of any close homologues despite important sequence identities with other RRs and transcription factors. The PrrA C-terminal sequence is highly conserved in a number of bacteria, defining a Prr/Reg family of RRs (3). The HTH motif (residues 159–179) is 100% conserved within the family, and ∼90% for the whole C-terminal domain (residues 139–184) (3). This degree of conservation is unusual for RRs, even for RRs having the same function in related organisms. Indeed, they are so highly conserved that PrrB and/or PrrA can be exchanged between organisms in the PrrA family, in vivo or in vitro, and still allow phosphate transfer and gene regulation (3,4). The DNA sequences recognized by this family have only weak sequence similarity, even within one organism, as observed with DNA sequences identified by DNase I protection with the constitutively active mutant RegA* (1) and SELEX selected DNA fragments recognised by the Bradyrhizobium japonicum RegR protein (7).
Although all RRs share a common receiver domain, and most are DNA-binding proteins, they function differently (2). Binding to DNA occurs in a symmetrical manner to inverted repeat sequences, as seen with NarL (8), but PhoB/OmpR family effector domains as well as Spo0A bind in a tandem arrangement to direct repeats (9,10). Many questions remain concerning RR function due to their high versatility.
The study of PrrA is of importance because of the lack of structural studies on the family despite considerable biochemical and genetic information. Furthermore, PrrAC is predicted to be one of the simplest folds so far for RR effector domains and does not belong to any of the three main effector domain families (OmpR/PhoB, NtrC/Dcdt, NarL/FixJ). Here we determine the structure of the C-terminal domain of PrrA (PrrAC); map how the domain binds to DNA; identify the residues involved in both specific DNA recognition and non-specific DNA binding; and present a model for PrrAC binding to its target DNA.
MATERIALS AND METHODS
PrrA and PrrAC expression and purification
The full-length R.sphaeroides PrrA protein (184 residues), and the C-terminal domain PrrAC (125–184) were cloned into pET14b plasmids (Novagen) between NdeI and BamHI restriction sites and contain at the N-terminus an extra 21 residues with a His6-tag and a thrombin cleavage site (11). BL21 (DE3) pLysS and BL21 (DE3) Escherichia coli cells were transformed with pETprrA and pETprrAC, respectively. Transformed bacteria were grown at 37°C under agitation, induced with 0.4 mM IPTG at OD595nm = 0.8 and left at 30°C for 3 h. Cells were disrupted by sonication and the proteins were first purified with Ni–NTA chromatography (Qiagen) where proteins eluted between 60 and 100 mM imidazole. A final gel permeation chromatography step (G75, Pharmacia Biotech) produced PrrA and PrrAC to a purity >95% (SDS–PAGE Coomassie Blue stained). Uniformly 15N- and 15N-13C-labelled samples were produced using M9 minimal medium as a culture medium instead of LB, supplemented with 15N (NH4)2SO4, or 15N (NH4)2SO4 and 13C glucose. The proteins were studied in NaH2PO4 (50 mM) buffer pH 6.0, NaCl (50 mM), (NH4)2SO4 (200 mM) and DTT (10 mM). MgCl2 (10 mM) was added to ensure the presence of a magnesium ion in the active site of the full-length protein. Protein concentrations were 1 mM for PrrAC and 400–500 µM for PrrA. Ten per cent D2O was added to samples for NMR experiments and 20 µl of protease inhibitor cocktail complete (Roche) to PrrAC. PrrA was phosphorylated with BeF3–, a non-hydrolysable mimetic of the phosphate group (12), using BeCl2 (2 mM) and NaF (6 mM). Vivaspin (Sartorius) were used for buffer exchanges and concentration steps.
NMR experiments and structure calculation
The NMR experiments were recorded on Bruker DRX-500 and DRX-600 spectrometers at 275 K. The spectra acquired were 15N-HSQC, HNCA, HNCO, HN(CA)CO, CBCA(CO) NH, HNCACB, 15N-edited TOCSY-HSQC, 1H-TOCSY, CCH-TOCSY, HCCH-TOCSY, 13C-HSQC (aromatic and aliphatic versions), 15N and 13C-edited NOESY (100 ms mixing time), and 15N T1, T2 and NOE.
Backbone assignment was performed using the asstool package (Leicester University NMR group) by matching Cα, Cβ and CO chemical shifts. TALOS (13) dihedral angle restraints were introduced using ranges set at twice the TALOS standard deviations. NOE restraints were calibrated on known distances in the helices and divided into three categories: <2.8, <3.8 and <5 Å. For determination of hydrogen bonds involving amide protons, the PrrAC sample was diluted twice into D2O containing buffer, and hydrogen bonds were introduced for non-overlapping 15N-HSQC amide protons showing some protection after 60 min.
The final restraints list consisted of 942 unambiguous NOE distance restraints, 62 dihedral angle restraints obtained from TALOS (13), and 12 hydrogen bonds from H-D exchange experiments (see Table 1). Processing, viewing and analysis of the NMR spectra were performed with Felix2000 (Accelrys Inc., San Diego, CA). Molecule viewing and construction of the DNA/protein model was done with Pymol (Delaglio Scientific).
Table 1. Structural statistics for PrrAC structure determination.
| Unrefineda | H2O refineda | |
|---|---|---|
| Restraint violations | ||
| NOE violations >0.5 Å | 0.6 ± 0.7 | 1.7 ± 1.0 |
| NOE violations >0.3 Å | 2 ± 1 | 4 ± 1 |
| Dihedral violations >5° | 0 | 0 |
| RMSD from experimental restraints | ||
| Distance restraints (Å)b | 0.06 ± 0.02 | 0.06 ± 0.02 |
| Dihedral restraints (°)c | 0.5 ± 0.1 | 0.5 ± 0.1 |
| Coordinate precision (residues 143–179) (Å)d | ||
| Backbone | 0.4 ± 0.3 | 0.4 ± 0.3 |
| All heavy atoms | 1.0 ± 0.8 | 1.0 ± 0.7 |
| Ramachandran analysise | ||
| Most favoured region (%) | 81.9 | 83.1 |
| Additionally allowed region (%) | 14.3 | 13.9 |
| Generously allowed region (%) | 2.3 | 1.4 |
| Disallowed region (%) | 1.6 | 1.6 |
| Energyf kcal/mol | ||
| Overall | 521 ± 15 | –1502 ± 63 |
| Bond | 6.1 ± 0.7 | 20 ± 3 |
| Angle | 46 ± 4 | 96 ± 10 |
| Dihedral | 329 ± 5 | 323 ± 7 |
| VdW | 77 ± 5 | –105 ± 18 |
| Electrostatics | NA | –1995 ± 70 |
| NOE | 55 ± 13 | 115 ± 22 |
| Dihedral (TALOS) | 1.0 ± 0.3 | 1.0 ± 0.6 |
| Difference from ideal values | ||
| Bond (Å) | 0.0024 ± 1 × 10–4 | 0.0044 ± 3 × 10–4 |
| Angle (°) | 0.40 ± 0.02 | 0.58 ± 0.03 |
| Improper (°) | 0.28 ± 0.02 | 1.4 ± 0.2 |
aValues are calculated for 20 best structures energetically.
b438 intra-residue, 211 sequential, 188 medium-range 2 ≤ d ≤ 4, 105 long range d > 4.
c62 phi and psi restraints (TALOS).
dAfter alignment of backbone atoms of residues 143–179.
eCalculated with Procheck-NMR (37).
fFrom Aria 1.2.
CNS 1.1 (14) was used for structure calculation with the anneal standard protocols using the protein-allhdg parameters. ARIA 1.2 (15) was used for its water refinement protocol re_h2o described in (16). NOE restraints were used as unambiguous data to calculate 100 structures with the refine protocol. The 25 best energy structures have been water refined and the 20 best energy structures retained as final structures. Five of these structures have been deposited in the Protein Data Bank under the accession code 1umq.
DNA titration experiments with PrrAC
DNA sequences were chosen from sequences identified by DNase I protection experiments using the RegA* (A95S) mutant (17). Two sequences were chosen, one from the R.sphaeroides cycAP2 promoter sequence 5′-tcgttgtgcggcaatccgtcatata-3′ (18) and one from the R.capsulatus puc promoter sequence 5′-actgcggcaaattcggccacccccg-3′ (17). Both sequences were chosen to have ∼5 bp on each side of the prrA consensus sequence and a total length of 25 bp. The two puc half-sites were designed as follows with the underlined regions corresponding with the wild-type oligonucleotide: pucR (5′-actgcggcaaatttttttttttttt-3′) and pucL (5′-aaaaaaaaaaattcggccacccccg-3′). A non-specific 25 bp sequence (ACTG repeat) was also chosen (5′-actgactgactgactgactgactga-3′).
Specific sequences (puc and cycAP2) were purchased from Genosphere (Paris, France) after RP-HPLC purification. The ACTG repeat and the pucR and pucL oligonucleotides were synthesised by Dr A. Moir (Sheffield University, UK). The DNA duplexes were formed by heating at 95°C for 5 min in TE buffer (10 mM Tris, pH 8.0, 50 mM NaCl, 1 mM EDTA) followed by a slow cooling for 3 h. The presence of double-stranded DNA was checked with 15% PAGE. The DNA duplexes were then exchanged against the PrrAC buffer using Vivaspin (Sartorius) and concentrated for NMR titration experiments. PrrAC (∼200 µM) and DNA stock solution (∼1 mM) concentration were similar in each experiment to minimize differences due to sample dilution. 15N-HSQC titrations were recorded by adding DNA to 60 nmol of PrrAC in steps of 0.25 molar equivalent up to 1.5 equivalents DNA/protein for specific DNA and up to two equivalents in steps of 1 molar equivalent for the ACTG repeat. Chemical shift changes were classified by a [(δH)2 + (δN/10)2]1/2 weighted difference.
RESULTS
NMR assignment of PrrAC
The PrrAC protein corresponding to residues 125–184 of R.sphaeroides PrrA was expressed and purified uniformly labelled for NMR experiments (see Materials and Methods). NMR experiments on PrrAC were acquired at 2°C in the presence of 200 mM (NH4)2SO4 to obtain optimum quality NMR spectra combined with maximum sample solubility and stability. Backbone assignment is complete. Several extra peaks in the 15N-HSQC and the backbone triple-resonance experiments were rejected because they represented minor conformations of the protein, mainly in the His6-tag region and the disordered N-terminus of PrrAC. Side chain assignment is almost complete. His170 aromatic protons could not be identified, even via through-space connectivities, probably because of overlaps with the His6-tag signal. Arg145 and 184 Hε could not be assigned because of overlaps. The hydroxyl proton Thr163 Hγ1 had slow enough solvent exchange to be identifiable in NOESY spectra, probably because the residue is completely inaccessible to solvent. To date it has not been possible to identify a likely hydrogen bonding partner of Thr163 Hγ1. NMR assignments have been deposited with BioMagResBank (BMRB accession code 5920).
Structure calculation
The protein studied here, PrrAC, consists of an N-terminal 21-residue His-tag, the last few residues of the N-terminal domain (residues 125–130), the inter-domain linker (residues 131–140) and the C-terminal domain of PrrA (residues 141–184). The initial structure calculation was carried out using CNS 1.1 (14) with only 644 unambiguous NOE restraints (391 intra-residue, 136 sequential, 62 medium range and 51 long range) and 76 TALOS restraints (13) based on 13C chemical shifts (see Materials and Methods). More NOE restraints could be assigned, based on distances in the first structures obtained, and were added progressively. The final set of restraints is indicated in Table 1. TALOS restraints at the edge of the α-helices were systematically causing NOE violations and forbidden phi and psi angles and were removed. Hydrogen bond restraints were only introduced in the later stages of the structure determination. Among the hydrogen bonds introduced as restraints, two involve loop residues: Tyr153 O-R158 HN, forming an (i, i + 5) connectivity, and His170 O-Leu174 HN. These are particularly important in helping the loop residues to adopt torsion angles in the accepted regions of the Ramachandran plot. The final structures using CNS did not present any significant nor systematic NOE or dihedral violations. In order to refine the structure in explicit solvent, the final set of restraints was used to calculate 100 structures using the ARIA 1.2 standard annealing protocol (16), and the 25 lowest energy structures of these were refined using the ARIA water refinement protocol (15). The 20 structures of best total energy are shown overlaid in Figure 1 with the structural statistics of the refined and unrefined structures in Table 1. The water refinement process increases the number of NOE violations but improves the Ramachandran plot, as expected (15).
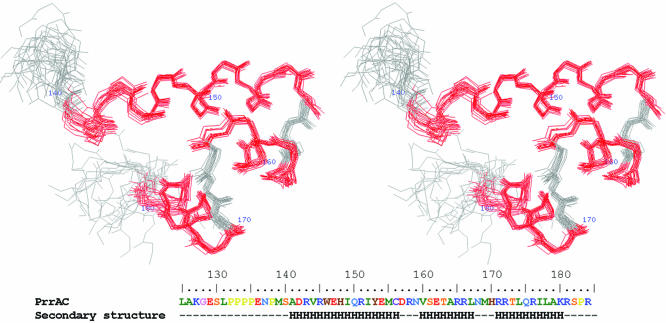
Figure 1.
Stereo view of 20 overlaid PrrAC structures. The 20 best energy, water refined, structures of PrrAC calculated from NMR restraints, with the residues in helices coloured in red. The residues N-terminal to the area represented (137–184) are disordered. PrrAC amino acid sequence and secondary structure are indicated.
The N-terminus of the expressed protein (which contains parts of the N-terminal domain and the inter-domain linker) is disordered, and the folded domain extends between M139 and K180. Rapid timescale motion of PrrAC was studied using 15N relaxation experiments (T1, T2 and 1H-15N NOE), which show disordered N- and C-termini and no particularly mobile residues in the structured part, even in the loops. Residues E136 to M139 are poorly defined in the structures due to the absence of restraints, but the relaxation data show that the backbone here is not as mobile as the rest of the N-terminus. This suggests that these residues retain interactions with the folded domain and are not fully disordered in solution.
PrrAC is a three-helix bundle HTH
PrrAC forms a three-helix bundle (residues 141–156, 160–167 and 171–180), each helix forming about a 30° angle with the next one. Helices α7 and α8 form the predicted HTH motif, of which the recognition helix, α8, is expected to insert into the major groove of DNA. The domain is structured via the formation of an extensive hydrophobic core involving I149, I152, C156, T163, L167, M169, L174 and L178, in which side chains are completely buried but also W146, Y153, V160, R166 and I177, which are only partially buried.
PrrAC belongs to the abundant family of three-helix bundle HTH DNA-binding domains, and the program Dali shows that it presents structural homologies with prokaryotic and eukaryotic DNA-binding domains such as DNA polymerase and many transcription factors (19), but with <20% identity on average. Despite low sequence similarities with known structures, 3DPSSM (20) predicts that PrrAC belongs to the E.coli Fis family (Factor for Inversion Stimulation) (21). Using Dali (19), PrrAC and Fis have 19% identity and an RMSD of 2.2 Å using Cα atoms between residues 140 and 182 from PrrA and residues 54–96 from Fis. The most structurally similar RR effector domain is from NtrC (22), a domain highly similar to Fis. PrrAC is less homologous to Salmonella typhimurium NtrCC than to Fis, having only 14% identity and 3 Å RMSD on the Cα of residues 140–184 of PrrA.
Phosphorylated PrrA and PrrAC bind DNA
PrrA is regulated by phosphorylation, and it has been suggested that it is able to bind DNA even when unphosphorylated (23). Phosphorylation of PrrA by a phosphate analogue, BeF3– (12), results in PrrA dimerisation (Laguri, Phillips-Jones and Williamson, manuscript in preparation), similarly to many RRs. Binding of PrrA, phosphorylated PrrA and PrrAC proteins to a 25 bp section of the cycAP2 promoter known to be bound by RegA (18) was tested by 1D 1H-NMR. Line broadening on addition of DNA shows that PrrA + BeF3– and PrrAC significantly bind DNA, PrrA + BeF3– apparently with the higher affinity (data not shown). The lack of line broadening for PrrA could indicate weak or no binding. This confirms previous suggestions that the N-terminal domain has an inhibitory activity on DNA binding of the C-terminal domain in unphosphorylated PrrA. PrrA + BeF3– does not produce an NMR spectrum of sufficient quality to investigate DNA binding at a molecular level and therefore PrrAC was used to investigate DNA binding and specific DNA recognition at a molecular level.
A refined consensus PrrA DNA-binding sequence
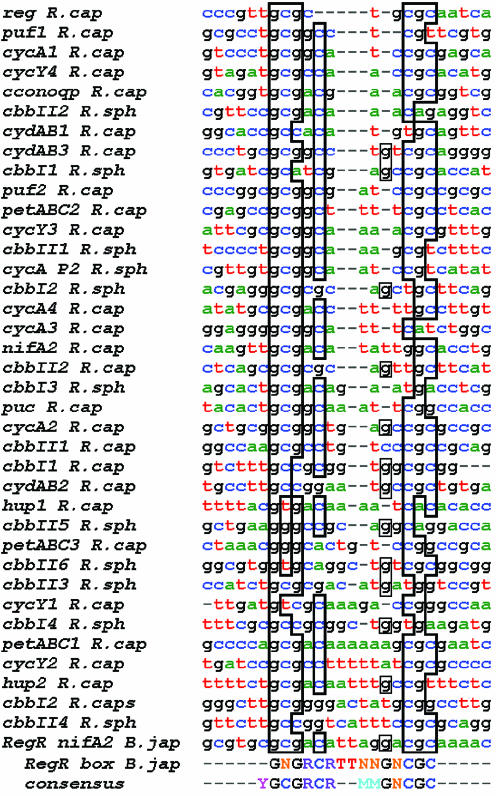
The PrrB/PrrA system is a global regulator in R.sphaeroides and R.capsulatus. PrrA-binding sites in gene promoters known to be regulated by the PrrB/PrrA system have been identified by DNase I protection experiments using a hyperactive PrrA mutant from R.capsulatus, RegA* (1,17,18,24–27), and by in vitro selection experiments involving gel retardation assays with wild-type B.japonicum RegR (7). The study on the RegR NifA2-binding site defined a 17 bp minimum binding sequence containing an inverted GCG repeat with an A/T-rich section between the two repeats and 11 positions critical for DNA binding (the RegR box shown at the bottom of Fig. 2). The alignment of PrrA-binding sequences is difficult due to the low conservation of the recognition elements but also because of the variable distances between the half-sites. Alignments were performed with Tcoffee (28) and additional manual adjustments using the previously determined consensus sequences (1,7), and maintaining the alignments within the DNase protection regions. The new alignment is shown in Figure 2.
Figure 2.
Alignment of DNA sequences recognised by PrrA/RegA. DNA sequences are identified by DNase I protection with R.capsulatus RegA* (1,17,18,24–27) and by gel retardation assays with B.japonicum RegR (7). The consensus sequence found for RegR, the RegR box, is indicated. A consensus sequence determined from this alignment is also indicated and the identities with this sequence outlined. N stands for any nucleotide, Y for pyrimidine, R for purine and M for A or T. The sequences have been sorted by increasing distance (in bp) between the GCG and CGC repeats, ranging from 3 (top) to 9 (bottom).
The consensus sequence found is YGCGRCRxTATAx GNCGC (Y = pyrimidine, R = purine, N = any nucleotide and x a variable number of bases) and is in agreement with the ‘RegR box’ alignment (5′-GNGRCRTTNNGNCGC-3′) (7). It differs from the alignment performed by Swem and colleagues where the different spacing between the recognition elements was not taken into account and where some aligned sequences were outside the DNase protection data (1). Because of the poor conservation of the consensus sequence the alignment should be considered to be tentative, not least because sometimes several PrrA-binding sites are possible within the same DNA sequence. The main features of the PrrA-binding sites presented in Figure 2 are: the palindromic GCGNC… GNCGC consensus that we may safely assume forms the specific recognition elements for the binding of two PrrA monomers, probably as a symmetric dimer; the central AT-rich section; and the variable distance between the left and right sites. It can also be noticed on the alignment that the left and right sites are not usually symmetrical. Although the consensus sequence is an imperfect repeat, the PrrB/A gene cluster site itself forms a perfect palindrome.
The number of bases between GCG and CGC motifs, which are found more often in the sequences than the entire palindrome and are thought to be the main recognition elements, range from 3 (for the PrrA cluster sequence) to 9 nt. The sequences have been represented in Figure 2 with an increasing distance between the GCG inverted repeats. This variable distance puts the recognition elements at different relative positions on the B-DNA helix and suggests that the PrrA dimer and/or the DNA itself would have to adopt different conformations to adapt for different spacings, probably with different affinities.
A careful study of DNA/protein complexes (29) showed that in most such complexes the DNA is bent. Furthermore, the bending is not usually continuous but shows kinks at discrete sites. The kink sites are generally formed by pyrimidine–purine (YR) steps [CA (= TG), TA or CG], a particularly flexible combination (30), which for proteins that bind DNA in adjacent major grooves, as many dimers do, are usually found about one helix turn apart (8–10 bp) (31). In the PrrA-binding sequences, whatever the distance between the right and left repeats there are always two YR steps 8–11 bases apart. We therefore suggest that the binding of PrrA to DNA fits this model and is accompanied by kinking. In support of the kink hypothesis, we note that GC-rich regions and AT-rich regions favour compression of the major and the minor groove, respectively, therefore are likely to encourage the bending process (32).
Specific and non-specific DNA binding of PrrAC
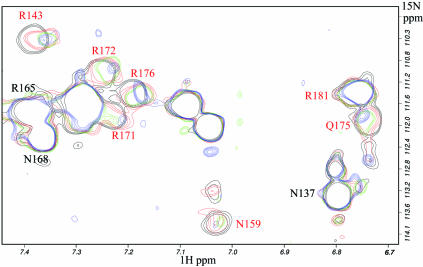
15N-HSQC titrations of PrrAC with 25 bp DNA fragments were recorded. Two specific PrrA targets were chosen for this study, the R.sphaeroides cycAP2 (18) and R.capsulatus puc (17) binding sites (Fig. 2), as well as a non-specific fragment, which does not contain the consensus sequence, as a control (see Materials and Methods). The 15N, 1H backbone (HN) and side chain (R Hε, N Hδ2 and Q Hε2; Fig. 3) chemical shift changes upon DNA binding are very similar for cycAP2 and puc (Fig. 4). The largest variations involve the HTH motif and especially the first half of the recognition helix, but also the α6–α7 loop and to a lesser extent the beginning of α6. Binding to the puc sequence appears stronger than to cycAP2; the chemical shift changes are larger, and some peaks also experience exchange broadening, suggesting a more intermediate exchange regime (Fig. 3). Some peaks disappear before a 0.5 DNA/protein molar ratio (N159 Hδ2 and V160, R171 and R172 HNs).
Figure 3.
15N-HSQC titration of PrrAC with puc DNA fragment: side chain region of the spectrum. Increasing quantities of the 25 bp DNA fragment were added to PrrAC; in black before addition, and in red, green and blue for 0.5, 1 and 1.5 DNA/protein molar ratios, respectively. The residue numbers of the side chains involved in puc binding are represented in red. Residues involved in DNA binding have peaks exhibiting chemical shift variations and/or exchange broadening, which alters the peak shapes. The intensity of the peaks are corrected for dilution.
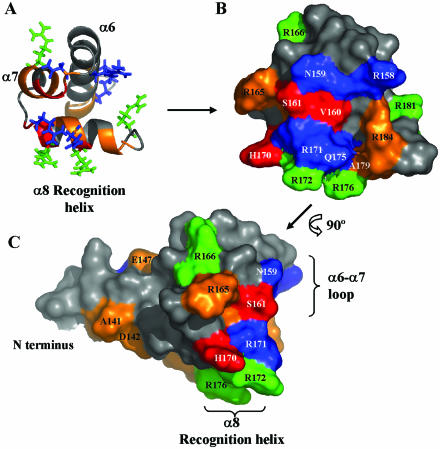
Figure 4.
Representation of chemical shift variations of PrrAC (residues 137–184) upon binding to R.capsulatus puc DNA sequence. The variations were comparable for puc and cycAP2 promoter sequences. Large and medium backbone HN chemical shift variations are, respectively, in red and orange. Side chains experiencing chemical shift variations are shown, in green for the ones affected only by puc binding and in blue the ones affected by both specific and non-specific DNA binding. The protein is shown with the same orientation in (A) and surface representation (B). A continuous surface is affected by DNA binding: the recognition helix α8 and the α6–α7 loop, plus the beginning of α6. A perpendicular view (C) close to the orientation that PrrAC would have when bound in the major groove of DNA (see Fig. 6) shows that most of the contact made by PrrAC would mainly involve the floor and one side of the major groove. The main difference between puc and cycAP2 titrations is the Q175 Hε side chain, which is affected only by binding to puc.
PrrAC is also able to bind to a non-specific sequence, but more weakly than to puc or cycAP2. The backbone residues perturbed are similar to the ones affected by puc and cycAP2, but the pattern of chemical shift changes for the side chains is quite different. In particular, the side chain resonances of R166, R172, R176 and R181 seem to be only affected in the complexes with specific DNA, whereas R143, R158, R171 and N159 change in both specific and non-specific complexes. This difference most likely reflects the different interactions involved in sequence-specific contacts, and contacts with the DNA phosphate backbone, respectively. Specific and non-specific contacts are discussed in the model presented below.
PrrA forms a dimer when activated and is likely to bind to the two inverted GCG motifs cooperatively, as in many DNA-binding proteins. In the titration assays with PrrAC described above, it is difficult to assess if the domain is able to bind one or two sites, and also if the relative affinities vary depending on the similarities with the consensus sequence. We investigated this by repeating the NMR titrations, but using the two half-sites of the puc sequence separately. For each DNA sequence, the other half-site was replaced by a string of T or A to make an efficient binding site possible only on one of the half-sites for each DNA fragment. Furthermore, the size of the DNA fragment was maintained to 25 bp to produce a molecular complex of the same size, hence similar broadening effects (see Material and Methods). PrrAC was found to bind to both half-sites with comparable affinity, since the chemical shift variations were of similar amplitude. However, the decrease in 15N HSQC peak intensity during the titrations, reflecting the formation of a protein/DNA complex through line broadening, goes in the order pucL < pucR < puc. The main contribution to peak broadening is coming from pucR (due to slower chemical exchange) but is more important when both sites are present, probably due to the formation of a larger complex when two PrrAC molecules are bound to DNA at the same time. The chemical shift variations between the two half-sites are significantly different, mainly in the areas involved in DNA binding, which exhibit more perturbation for pucR than for pucL (R143, T163, R166, T173, R176, A179 and R184 HN, R171, R172, R176 Hε). In a similar way as between puc and the ACTG repeat binding, more side chains are involved in binding pucR than pucL. This is expected from the sequences of the two sites, since pucR contains the consensus sequence (YGCGRCR) whereas pucL exhibits less conservation, and the putative PrrAC-binding site is hard to predict within the sequence (see Materials and Methods).
The residues involved in sequence-specific DNA recognition are located for HTH domains in the recognition helix, and are in contact with the bases on the major groove floor. In the PrrA recognition helix, R172, R176 and R181 have different behaviours between specific and non-specific DNA binding, while T173, R176, A179 HNs and R171, R172 and R176 Hεs differ between pucR and pucL. R171, R172 and R176 positions on the recognition helix (Fig. 4B and C) associated with their involvement in specific binding are proposed to be the main residues important for DNA-binding specificity. Other side chains (R143, R158 and N159) are implicated in non-specific binding, presumably to the phosphate backbone.
Model of PrrAC bound to DNA and discussion on the mode of PrrA DNA binding
A model of the PrrAC/DNA complex was constructed, using the NMR-derived structure of PrrAC and DNA taken from the structure of 434CRO complexed with DNA (33). 434CRO-bound DNA represents a good model for PrrA binding because it is bent by kinking upon protein binding at two YR steps 10 bases apart (32), a characteristic suggested here to be shared by PrrA-bound DNA. Standard linear B-DNA models, as well as several other kinked DNA structures, could be fitted with PrrAC (e.g. 434REP, NarL). Some other kinked structures, including a model of DNA bound to Fis (34), fitted less well. In the successful fittings, the most important characteristic is a sufficient width of the major groove of the DNA to be able to accommodate PrrAC.
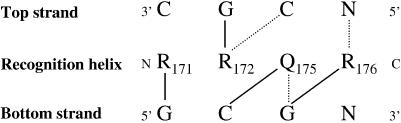
In order to define a model in which the residues important for binding specificity would be positioned in close contact to the DNA bases, a code for residue-base specific contacts in HTH proteins has been used (35). It defines a binding score combining a chemical merit, from observed contacts in DNA complexes between residue types and base types, and a stereochemical merit defined from observed contacts in HTH/DNA complexes between residues of the recognition helix and bases from the top and bottom strands of DNA, depending on the size of the amino acid side chain (small, medium or large). The HTH proteins usually recognise four bases and the stereochemical code involves four residues from the recognition helix (residues 1 and 2 and residues 5 and 6, which are one helix turn apart) making contacts with four bases from the top and bottom strands. According to this recognition code, the residues in PrrAC involved in recognising the core consensus motif (GCG) would be R171, R172, Q175 and R176 as represented in Figure 5. These arginines were proposed above as being important for specific recognition; furthermore, arginines contact guanine bases almost exclusively, with the possibility of forming two hydrogen bonds (35). Q175 is in a good position on the recognition helix to have contact with bases, but without specificity. Glutamines are able to bind bases almost indifferently (35).
Figure 5.
Proposed contacts between PrrAC and the GCG conserved motif. Pattern of contacts suggested between the residues proposed to be in contact with bases and the GCG motif according to the contact code (35). The residues on the recognition helix are represented between the two strands of DNA. The lines represent observed contacts in HTH/DNA complexes (35). The continuous lines represent the contacts with the best chemical and stereochemical scores and the dashed lines the possible contacts, considering the length of the amino acid side chains. The best scores contacts were used to make the model in Figure 6. Arginines bind almost exclusively guanine bases, so make highly specific protein/DNA contacts, whereas glutamines are observed to be able to make contacts with any base.
A model of the complex was built, in which two PrrAC domains were fitted to bind two GCG inverted repeats (the bases corresponding to these positions in 434CRO-bound DNA) separated by 6 bp (Fig. 6). The orientations of the R171, R172, Q175 and R176 side chains are at a distance compatible with the contacts proposed in Figure 5. DNA and PrrAC have been manually positioned to minimize van der Waals contacts and reflect titration results without any attempt at energy minimisation. The arrangement of the PrrAC molecules on the model places their N-termini 30 Å apart (S140-S140 Cα), and presents no dimer interface. The distance between the two N-termini of the effector domain is similar to the distance of ∼30 Å observed between the two C-termini of phosphorylated FixJN dimer (5), and would not be incompatible with PrrAN adopting the same dimer conformation. The inter-domain linker of about 10 residues would furthermore allow some flexibility in the relative position of the two effector domains bound on the two DNA half-sites, and therefore allow variability in the separation of the GCG repeats.
Figure 6.
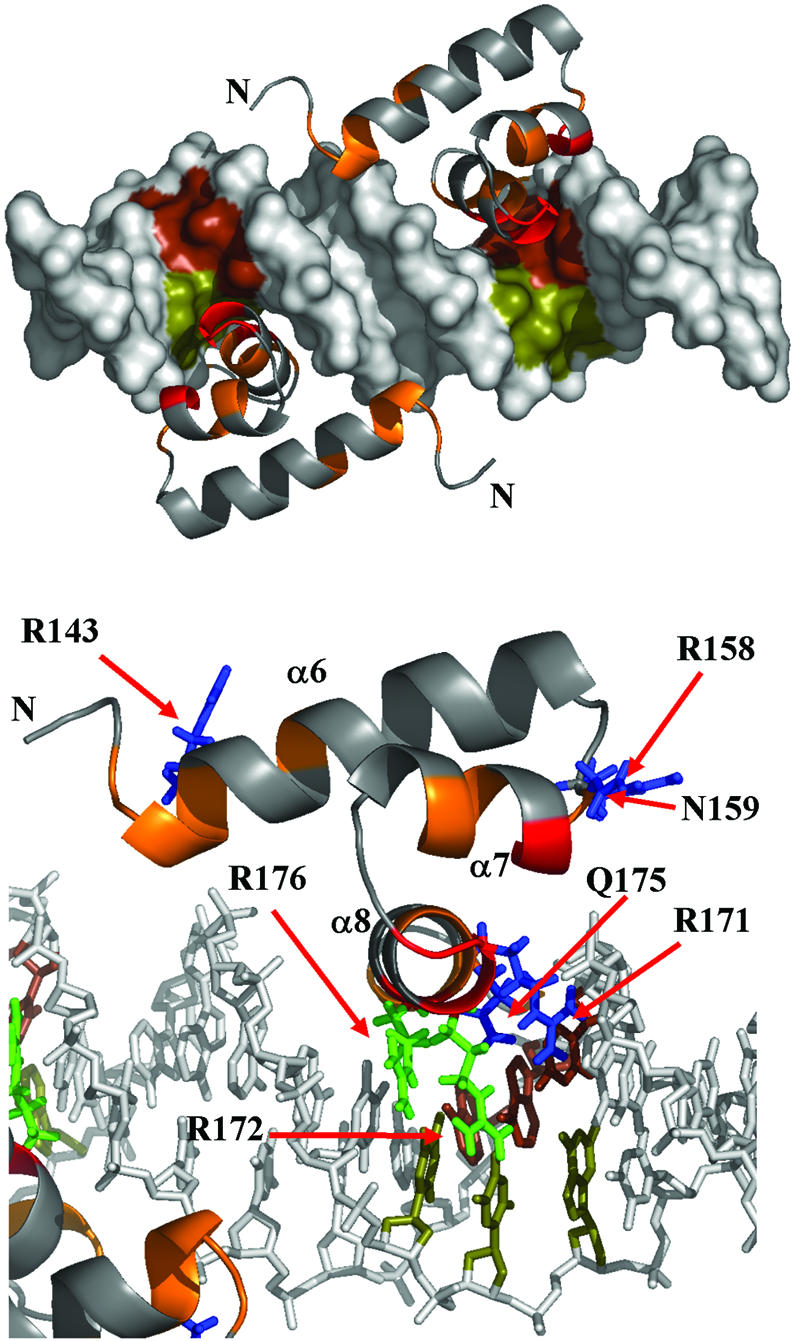
Model of two PrrAC monomers bound to a 20 bp DNA fragment. (Top) A view of both monomers. (Bottom) A detail of PrrAC/DNA interactions. PrrAC monomers were fitted to respect the protein/DNA contacts predicted in Figure 5, the DNA titrations and the DNA and protein surfaces. R171, R172, Q175 and R176 side chain positions are at a distance where the predicted contacts would be possible. The recognised GCG inverted repeats have been coloured by DNA strand (brown and olive). The PrrAC colour code is as in Figure 4. R166 and R181 have been omitted for clarity.
In the model, side chains affected by both specific and non-specific binding are in good position to contact the phosphate backbone (R143, R158 and N159). Furthermore, side chains affected only by specific binding, or affected differently between pucR and pucL, are in position to make specific contacts with bases with the exception of R166 and R181, whose conformation does not allow contact with DNA bases without drastic changes in backbone angles (Fig. 4). The R166 side chain has a well-defined position in the structures having medium and long-range NOE restraints with α6 and α7 residues (R151, I152, M155, E162 and T163) and is packed against the hydrophobic core. This residue could be affected by a conformational change (or a change in local dynamics) occurring only when a specific DNA sequence is bound. This residue might be reporting a conformational change important in signalling the correct DNA binding to the N-terminal domain or to a σ-RNA Polymerase subunit.
DISCUSSION
The Prr/Reg system has been extensively studied, both from genetic and biochemical approaches. The way PrrA is activated upon phosphorylation, a phenomenon still difficult to characterise in RRs, is still unclear. We report here the structure of the effector domain of PrrA, which belongs to the abundant family of three-helix bundle HTH-containing domains but is not closely homologous to any characterised RR effector domain or DNA-binding domain. Upon phosphorylation, PrrA forms a dimer and must undergo an inter-domain rearrangement, weakening inter-domain interactions, to allow dimerisation and correct orientation of PrrAC for efficient DNA binding.
Alignment of the known sequences bound by PrrA/RegA allowed a refinement of the PrrA consensus sequence showing the presence of two inverted GCGNC repeats, which are poorly conserved and present different spacings between the two half-sites. An interesting characteristic shared by the sequences is the presence of pyrimidine–purine steps, particularly flexible in DNA. It suggests that the recognition of PrrA/RegA proteins involves also structural features of the DNA. Analysis of the two promoter sequences cycAP2 and puc using the program bend.it (36) shows a pronounced bendability about 10 bases apart at the two half-sites and predicts an intrinsic curvature of the DNA centred between the two half-sites in the A/T-rich region (data not shown). PrrA and homologues might not only recognise a sequence, which is poorly conserved and often asymmetric, but also the curvature of promoter DNA and its bendability at the binding sites. These calculations support, on two promoter DNA sequences demonstrated in this study to be bound specifically by PrrAC, the hypothesis of an indirect read-out of the DNA sequence, adding to the specific recognition elements.
Binding of PrrAC to specific, non-specific and half-site DNA sequences showed which areas of the domain are involved in binding DNA and what residues are involved in specific DNA binding. The HTH is highly involved in binding, forming a continuous binding surface (Fig. 4), both with bases on the major groove floor but also with the phosphate backbone. More unexpected is the involvement of the beginning of α6 in unspecific binding, probably with the phosphate backbone, helping the HTH in binding DNA.
The model presented in this study agrees with established dimeric HTH/DNA interactions (32); binding occurs with dyad symmetry on two adjacent major grooves through their recognition helix. Whereas Suzuki and Yagi’s prediction of recognition helix/DNA-specific contacts in Figure 5 fits well with the model presented, it does not fully explain both the results from the titration experiments and the consensus sequence. It is likely that PrrA recognition involves more residues than predicted, at the C-terminal end of the domain. K180, in the model, is in an ideal position for the recognition of the last cytosine (the G on the opposite strand) of the GCGNC full consensus sequence to contribute to a more effective binding when this element of the consensus sequence is present. R181, which is affected by specific DNA binding but cannot easily make contacts with bases, could be affected because of K180 binding, or contact the phosphate backbone. Finally, the backbone of R184, the C-terminal residue of PrrA, undergoes chemical shift changes during puc and cycAP2 titrations and could be involved in additional specific or non-specific binding.
The alignment of the sequences bound by PrrA suggests a more versatile mode of DNA binding than typical HTH proteins. First, the two DNA consensus half-sites are not symmetrical, and rarely are both of the half-sites conserved (Fig. 2). This has implications for the binding of a PrrA dimer on DNA. Because PrrA is able to bind specific and less specific sequences, a tight binding on one consensus-like half-site could be enough to allow a rather non-specific binding on the second half-site. The cooperativity of the binding as a dimer would then compensate the lack of conservation. The different spacing between the half-sites is also uncommon amongst proteins recognising specific DNA targets for gene regulation. Different spacings imply not only that the two monomers will be at different distance apart, but also that their orientation on the double helix will be different. The inter-domain linker as well as the dimer interface would have to adapt to the various sequences, affecting the cooperativity of binding and also the affinity for different DNA targets. The structure of the DNA itself should undergo variable deformations in terms of bending, compression of the grooves and even maybe local B- to A-DNA transitions. Furthermore, even if a symmetrical binding of the effector domain is likely, it cannot be ruled out, for the long half-sites spacings, that binding might be possible in a tandem arrangement.
This ability to bind differently spaced, poorly conserved sequences but with some similar structural features might be an advantage for PrrA activity as a global regulator, controlling many different genes and highly conserved in several, even non-related, organisms. The investigation of how the full-length PrrA dimer binds DNA targets with drastically different half-site spacings, and the influence on DNA local structure, remain to be investigated. It could provide important information on the strategies a transcription activator can adopt to bind different DNA sequences and its influence on the gene regulation exerted by the PrrA family of RRs in bacteria.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Andrea Hounslow, Jeremy Craven and Laszlo Hosszu for assistance with the NMR, Chris Potter for supplying plasmids, and Peter Henderson for helpful discussions. We acknowledge the provision of FELIX software by Accelrys. C.L. and M.P.W. are members of the Krebs Institute and the North of England Structural Biology Centre, which are funded by the Biotechnology and Biological Sciences Research Council (BBSRC). We thank the Wellcome Trust and BBSRC for instrument grants, and the BBSRC for funding (grant 24/B12958).
PDB no. 1umq
REFERENCES
- 1.Swem L.R., Elsen,S., Bird,T.H., Swem,D.L., Koch,H.G., Myllykallio,H., Daldal,F. and Bauer,C.E. (2001) The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J. Mol. Biol., 309, 121–138. [DOI] [PubMed] [Google Scholar]
- 2.Robinson V.L., Buckler,D.R. and Stock,A.M. (2000) A tale of two components: a novel kinase and a regulatory switch. Nature Struct. Biol., 7, 626–633. [DOI] [PubMed] [Google Scholar]
- 3.Masuda S., Matsumoto,Y., Nagashima,K.V., Shimada,K., Inoue,K., Bauer,C.E. and Matsuura,K. (1999) Structural and functional analyses of photosynthetic regulatory genes regA and regB from Rhodovulum sulfidophilum, Roseobacter denitrificans and Rhodobacter capsulatus. J. Bacteriol., 181, 4205–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emmerich R., Hennecke,H. and Fischer,H.M. (2000) Evidence for a functional similarity between the two-component regulatory systems RegSR, ActSR and RegBA (PrrBA) in alpha-Proteobacteria. Arch. Microbiol., 174, 307–313. [DOI] [PubMed] [Google Scholar]
- 5.Birck C., Mourey,L., Gouet,P., Fabry,B., Schumacher,J., Rousseau,P., Kahn,D. and Samama,J.P. (1999) Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure Fold Des., 7, 1505–1515. [DOI] [PubMed] [Google Scholar]
- 6.Volkman B.F., Nohaile,M.J., Amy,N.K., Kustu,S. and Wemmer,D.E. (1995) Three-dimensional solution structure of the N-terminal receiver domain of NTRC. Biochemistry, 34, 1413–1424. [DOI] [PubMed] [Google Scholar]
- 7.Emmerich R., Strehler,P., Hennecke,H. and Fischer,H.M. (2000) An imperfect inverted repeat is critical for DNA binding of the response regulator RegR of Bradyrhizobium japonicum. Nucleic Acids Res., 28, 4166–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maris A.E., Sawaya,M.R., Kaczor-Grzeskowiak,M., Jarvis,M.R., Bearson,S.M., Kopka,M.L., Schroder,I., Gunsalus,R.P. and Dickerson,R.E. (2002) Dimerization allows DNA target site recognition by the NarL response regulator. Nature Struct. Biol., 9, 771–778. [DOI] [PubMed] [Google Scholar]
- 9.Blanco A.G., Sola,M., Gomis-Ruth,F.X. and Coll,M. (2002) Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure, 10, 701–713. [DOI] [PubMed] [Google Scholar]
- 10.Zhao H., Msadek,T., Zapf,J., Madhusudan, Hoch,J.A. and Varughese,K.I. (2002) DNA complexed structure of the key transcription factor initiating development in sporulating bacteria. Structure, 10, 1041–1050. [DOI] [PubMed] [Google Scholar]
- 11.Potter C.A., Ward,A., Laguri,C., Williamson,M.P., Henderson,P.J. and Phillips-Jones,M.K. (2002) Expression, purification and characterisation of full-length histidine protein kinase RegB from Rhodobacter sphaeroides. J. Mol. Biol., 320, 201–213. [DOI] [PubMed] [Google Scholar]
- 12.Yan D., Cho,H.S., Hastings,C.A., Igo,M.M., Lee,S.Y., Pelton,J.G., Stewart,V., Wemmer,D.E. and Kustu,S. (1999) Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc. Natl Acad. Sci. USA, 96, 14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornilescu G., Delaglio,F. and Bax,A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR, 13, 289–302. [DOI] [PubMed] [Google Scholar]
- 14.Brunger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.S., Kuszewski,J., Nilges,M., Pannu,N.S., Read,R.J., Rice,L.M., Simonson,T. and Warren,G.L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 15.Linge J.P., Williams,M.A., Spronk,C.A., Bonvin,A.M. and Nilges,M. (2003) Refinement of protein structures in explicit solvent. Proteins, 50, 496–506. [DOI] [PubMed] [Google Scholar]
- 16.Linge J.P., Habeck,M., Rieping,W. and Nilges,M. (2003) ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics, 19, 315–316. [DOI] [PubMed] [Google Scholar]
- 17.Du S., Bird,T.H. and Bauer,C.E. (1998) DNA binding characteristics of RegA. A constitutively active anaerobic activator of photosynthesis gene expression in Rhodobacter capsulatus. J. Biol. Chem., 273, 18509–18513. [DOI] [PubMed] [Google Scholar]
- 18.Karls R.K., Wolf,J.R. and Donohue,T.J. (1999) Activation of the cycA P2 promoter for the Rhodobacter sphaeroides cytochrome c2 gene by the photosynthesis response regulator. Mol. Microbiol., 34, 822–835. [DOI] [PubMed] [Google Scholar]
- 19.Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- 20.Kelley L.A., MacCallum,R.M. and Sternberg,M.J. (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol., 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 21.Kostrewa D., Granzin,J., Koch,C., Choe,H.W., Raghunathan,S., Wolf,W., Labahn,J., Kahmann,R. and Saenger,W. (1991) Three-dimensional structure of the E. coli DNA-binding protein FIS. Nature, 349, 178–180. [DOI] [PubMed] [Google Scholar]
- 22.Pelton J.G., Kustu,S. and Wemmer,D.E. (1999) Solution structure of the DNA-binding domain of NtrC with three alanine substitutions. J. Mol. Biol., 292, 1095–1110. [DOI] [PubMed] [Google Scholar]
- 23.Comolli J.C., Carl,A.J., Hall,C. and Donohue,T. (2002) Transcriptional activation of the Rhodobacter sphaeroides cytochrome c(2) gene P2 promoter by the response regulator PrrA. J. Bacteriol., 184, 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubbs J.M. and Tabita,F.R. (2003) Interactions of the cbbII promoter-operator region with CbbR and RegA (PrrA) regulators indicate distinct mechanisms to control expression of the two cbb operons of Rhodobacter sphaeroides. J. Biol. Chem., 278, 16443–16450. [DOI] [PubMed] [Google Scholar]
- 25.Swem D.L. and Bauer,C.E. (2002) Coordination of ubiquinol oxidase and cytochrome cbb(3) oxidase expression by multiple regulators in Rhodobacter capsulatus. J. Bacteriol., 184, 2815–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elsen S., Dischert,W., Colbeau,A. and Bauer,C.E. (2000) Expression of uptake hydrogenase and molybdenum nitrogenase in Rhodobacter capsulatus is coregulated by the RegB-RegA two-component regulatory system. J. Bacteriol., 182, 2831–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubbs J.M., Bird,T.H., Bauer,C.E. and Tabita,F.R. (2000) Interaction of CbbR and RegA* transcription regulators with the Rhodobacter sphaeroides cbbI promoter-operator region. J. Biol. Chem., 275, 19224–19230. [DOI] [PubMed] [Google Scholar]
- 28.Poirot O., O’Toole,E. and Notredame,C. (2003) Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res., 31, 3503–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickerson R.E. (1998) DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res., 26, 1906–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson W.K., Gorin,A.A., Lu,X.J., Hock,L.M. and Zhurkin,V.B. (1998) DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc. Natl Acad. Sci. USA, 95, 11163–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickerson R.E. and Chiu,T.K. (1997) Helix bending as a factor in protein/DNA recognition. Biopolymers, 44, 361–403. [DOI] [PubMed] [Google Scholar]
- 32.Harrison S.C. and Aggarwal,A.K. (1990) DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem., 59, 933–969. [DOI] [PubMed] [Google Scholar]
- 33.Mondragon A. and Harrison,S.C. (1991) The phage 434 Cro/OR1 complex at 2.5 Å resolution. J. Mol. Biol., 219, 321–334. [DOI] [PubMed] [Google Scholar]
- 34.Tzou W.S. and Hwang,M.J. (1999) Modeling helix-turn-helix protein-induced DNA bending with knowledge-based distance restraints. Biophys. J., 77, 1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki M. and Yagi,N. (1994) DNA recognition code of transcription factors in the helix-turn-helix, probe helix, hormone receptor and zinc finger families. Proc. Natl Acad. Sci. USA, 91, 12357–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vlahovicek K., Kajan,L. and Pongor,S. (2003) DNA analysis servers: plot.it, bend.it, model.it and IS. Nucleic Acids Res., 31, 3686–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laskowski R.A., Rullmannn,J.A., MacArthur,M.W., Kaptein,R. and Thornton,J.M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR, 8, 477–486. [DOI] [PubMed] [Google Scholar]