Abstract
In recent years microRNAs have become recognized as pervasive, versatile agents of gene regulation. Some widely embraced rules involving Watson-Crick hybridization of microRNAs with mRNAs have generated great interest as scientists envision potential RNA cargoes for gene therapy and other experimental systems. However, while researchers ardently seek simplifying principles, nature seems very uncooperative. This article reviews some small RNA mechanisms that potentially regulate genes and which are not covered by previous microRNAs characterizations. In addition, we report here results of fluorescence microscopy experiments to directly demonstrate nuclear importation of small RNAs equal in length to typical mature microRNAs, implying that gene regulation at the locus of transcription might be possible.
1. Introduction
“When we add to the truth, we subtract from it.” This saying, attributed to the Talmud, is certainly exemplified by microRNA (miRNA) research. For the present and foreseeable future, the arrival rate of new miRNA phenomena and layers of complexity exceeds and will exceed the departure rate of solved problems.
Current research points to miRNA roles in the general management and fine-scale control of protein synthesis (Baek et al., 2008; Selbach et al., 2008), with implications regarding cancer (Lujambio et al., 2008), immune response (Stern-Ginossar et al., 2008), viral immunoevasion (Umbach et al., 2008), apoptosis (Yamakuchi et al., 2008), cell cycle control (Cloonan et al., 2008; Chivukula and Mendell, 2008), and stem cell differentiation (Li Z et al., 2008). Gene management by miRNAs and other noncoding RNAs can employ alteration of transcription rates, RNA stability, translational efficiency, and methylation of chromatin. Furthermore, proteins can return the favor by controlling miRNA biogenesis (Chang et al., 2007), suggesting a vast world of complex gene expression regulation suitable for anyone seeking a really hard network control problem.
miRNA gene regulation is conventionally regarded as targeting 3′ untranslated regions (3′UTRs) of mRNAs and inhibiting gene expression. However, a recent report (Tay et al., 2008) identifies targets occurring throughout some mRNAs; in particular, mouse transcription factors Nanog, Pou5f1 (formerly called Oct4) and Sox2 display many naturally occurring miRNA targets in their amino acid coding sequences. The seed region of an miRNA is defined as the sequence of nucleotides in positions 2 through 8 from the 5′ terminus. Popular web engines use bioinformatic complementarity of the seed with 3′UTRs and additional rules such as conservation to provide a first approximation of miRNA-mRNA targeting. However, some experimentally verified targets (Tay et al., 2008) do not contain the miRNA seed, some span exon-exon junctions, and some are not conserved across human and rhesus genomes.
miRNAs are generally considered post-transcriptional inhibitors of translation or accelerants of mRNA degradation, that is, downregulators of gene expression. However, recent reports (Li et al., 2006; Janowski et al., 2007; Place et al., 2008) have further increased the potential complexity of miRNA regulation by describing instances in which miRNAs or transfected double-stranded RNAs (dsRNAs) designed as if they were short interfering RNAs (siRNAs) can upregulate gene expression. Evidently several genes–genes intensively studied in other contexts–can be upregulated by siRNA targeting of their promoter regions. This means targeting chromosomal dsDNA in the nucleus, using target selection algorithms designed for ssRNA messages in the cytoplasm. Puzzles implied by the observations of upregulation include:
Given that the effects are actually on transcription (Janowski et al., 2007), how is ectopic siRNA (dsRNA with TT 3′overhangs) imported into the nucleus?
To what degree is nuclear import of RNA sequence-specific?
Into what form is ectopic dsRNA processed or complexed in order to interact with a chromosomal promoter region?
Does promoter dsDNA somehow open and hybridize sequence-specifically with imported ssRNA?
How can observed shifting by only a few bases change activation into deactivation, or change a strong effect such as 18-fold upregulation into no effect (Janowski et al., 2006)?
If cryptic noncoding antisense transcripts from promoter regions are the actual targets (as suggested by Schwartz et al. (2008)), then what function do they have in routine gene expression?
Do endogenous miRNAs or siRNAs exist that target promoter regions and upregulate transcription?
The purpose of this paper is to provide some background information that might support resolution of the puzzles.
2. Materials and methods
2.1. RNAi mechanisms
Experimentally manipulated, conventional RNA interference (RNAi) is synthetic, sequence-specific suppression of gene expression by introduction of double-stranded RNAs (dsRNAs) into cells. Introduction means the RNA is treated in some way to allow passage through the cell membrane and into the cytoplasm, and interference usually means accelerated degradation or translational inhibition of a specific mRNA. In RNAi of human cells, the dsRNA is then processed by DICER ribonuclease into small interfering RNAs (siRNAs) which are complexed with argonaute (Ago) proteins, forming an RNA induced silencing complex (RISC). The RISC-associated siRNA can hybridize with a complementary section of an mRNA message, causing argonaute-mediated cleavage of the mRNA or otherwise interfering with translation. There are multiple variations on the siRNA theme with modified nucleic acids and other chemical modifications to enhance stability, selectivity, and specificity, as well as reduce toxicity. Alternatively, when the antisense strand of the siRNA is somehow imported into the nucleus, it can target chromatin of a specific promoter region (Morris KV, 2008). Such siRNA causes epigenetic revision of local histone code and chromatin marks, leading to heterochromatization of the targeted gene. As pointed out by Morris, epigenetic modifications could be much more persistent that conventional siRNA targeting mRNAs.
Several web-based engines, some with proprietary algorithms, are available for selection of optimal siRNA targets within an mRNA sequence for custom RNAi. Alternatively, several vendors guarantee effectiveness of their stock or made-to-order siRNA products. A guarantee means generally that some fraction such as two-thirds of products purchased are effective, and effectiveness means downregulation of mRNA by at least a certain fraction such as 50%. Regarding the usefulness of the algorithms, it should be noted that random siRNA targeting also has significant impact on gene expression in about half of tries, but typically, design and synthesis in a lab would start with comparison of several algorithm outputs. Eventually, successful trials of siRNA pharmaceuticals will require that siRNA treatments strongly downregulate targeted genes (selectivity) and only do so in targeted cells (specificity) (Krützfeldt et al., 2005; Kumar et al., 2008). There is already a rich literature on siRNA drug design dealing with these notions.
Regarding transcriptional silencing (so RNAi in the nucleus applied to chromosomal dsDNA), Weinberg et al. (2006) measured histone methylation at targeted promoters. They reported that siRNA treatment by the antisense strand alone can increase both H3K9 and H3K27 methylation of the promoter of a targeted gene, EF1A, and that this increase is dependent on nuclear specific delivery of the siRNA.
Janowski et al. (2006) showed that AGO1 and AGO2 connect pathways for mRNA silencing with pathways for recognition of chromosomal DNA. Complements to transcription start sites or upstream regions in gene promoters called synthetic antigene RNAs (agRNAs) inhibited gene transcription. Such silencing occurs in the nucleus, requires high promoter activity, and does not necessarily require histone modification. The researchers reported that AGO1 and AGO2 proteins associated with promoter DNA in cells treated with agRNAs, and that inhibiting expression of AGO1 or AGO2 reversed transcriptional and post-transcriptional silencing. Their data indicated key linkages and important mechanistic distinctions between transcriptional and post-transcriptional silencing pathways in mammalian cells.
2.2 Sources of endogenous dsRNAs
Aside from pre-miRNAs, there might be many additional sources of RNA hairpins that contribute to gene regulation. Pervasive transcription of human and mouse genomes (Katayama, 2005; ENCODE, 2007) yields a potentially huge supply of noncoding RNAs, including dsRNAs that could enter the miRNA pathway to emerge both as mRNA inhibitors and as activators of promoters. Inverted repeat structures, bidirectional transcription, and antisense transcripts from various loci are sources of dsRNAs. Okamura and Lai (2008) have reviewed recent research regarding endo-siRNA, meaning processing of diverse hairpins (other than pre-miRNAs) that do not generally conform to pre-miRNA sequence statistics (Zhang et al., 2006). The stems could become RISC-mounted exact complements of mRNAs, promoting cleavage. In particular, such endo-siRNA may be responsible for control of transposable element transcripts in mouse oocytes (Puschendorf et al., 2006; Tam et al., 2008). That is, transposon-derived hairpin sequence elements may contribute to the metabolism of mouse maternal transcripts through a Dicer-dependent pathway. Thus RNAi triggered by antisense transcripts may modulate human L1 retrotransposition efficiently and economically (Yang and Kazazian, 2006). Naturally occurring dsRNAs might also regulate protein-coding transcripts (Watanabe et al., 2008).
It has been hypothesized (Jeffries, 2006) that large, well-formed hairpins from intronic Alu sequences adjacent to their complements could serve as substrates to produce multiple siRNA fragments. Processing through an miRNA pathway could yield cleavage or translational inhibition of mRNAs having Alu sequences in their UTRs or introns. Such Alu hairpins hypothetically could be produced from a large number of introns. For example, the gene encoding pro-apoptotic PAWR could generate such byproduct hairpins from three loci in one of its introns, leading to downregulation of genes such as anti-apoptotic BACL4 with a 3′UTR Alu. In this manner, initiation of apoptosis could automatically inhibit anti-apoptosis response in a cell. In fact, a naturally occurring, miRNA instance of this type of feed forward control has been reported (Barik, 2008) for AATK, a gene essential for promoting neuronal differentiation. Transcription of AATK yields miR-338 as an intronic byproduct, and miR-338 silences a family of genes that are negative regulators of neuronal differentiation.
However, only a small subset of hypothetical hairpins might be selected for siRNA production, since long dsRNAs (Stein et al., 2005) in mammalian cells induce an antiviral response mediated by interferon (IFN) that leads to general inhibition of protein synthesis and nonspecific degradation of mRNAs.
2.3 Nuclear import
That miRNAs might be naturally imported into HeLa nuclei was suggested by Hwang et al. (2007). They observed that miR-29b, tagged with a fluorophore, localized in HeLa nuclei. Experiments included using confocal microscopy (so that images are virtual sections that show what is in, not on, nuclei). Hwang et al. found that other small ssRNAs when modified to include AGUGUU at 3′ terminus also entered the nucleus. The same authors later submitted a patent application (Hwang et al., patent application, 2007) covering the pattern AGUGUU and seven other nucleotide (nt) patterns (UGUGUU, ACUGUU, AGAGUU, AGUCUU, AGUGAU, AGUGUA, AGNGUN) as distinguished sequences for enabling nuclear import. They demonstrated as well that several similar sequences were not imported. However, the complexity of nuclear import mechanisms might not be consistent with such a concise list.
The mechanism of nuclear import of miR-29b is unknown. Possibly AGUGUU is an RNA sequence that binds sequence-specifically to a protein already subject to nuclear import. Clues include a report by Guang et al (2008) on factors essential for RNA interference (RNAi) in nuclei of Caenorhabditis elegans. A cytoplasmic Argonaute protein NRDE-3 with a bipartite nuclear localization sequence (NLS) binds exclusively to endogenous siRNAs generated from mRNA templates; binding of siRNAs to NRDE-3 is required to promote relocation of NRDE-3 from the cytoplasm to the nucleus. In mutant worms defective in endogenous siRNA production, NRDE-3 becomes associated with exogenous siRNAs, suggesting a mechanism adaptable to various RNA sequences. The NRDE-3 associated RNA is recruited by importin proteins and conveyed via the nuclear pore complex to and into the nucleus where the RNA might associate with nascent transcripts.
2.4 Extraordinary extensions of known siRNA and miRNA capabilities
Li LC et al. (2006) reported that 21 nt dsRNA (with 3′ overhang = TT) targeted (exact match) to sequences selected by conventional siRNA algorithms could have the effect of upregulation of CDH1 (alias E-cadherin) gene expression. The effect was discerned down to application of ~5 nM. Indeed, the intersection of outputs of several public, web-based siRNA target selection algorithms includes the specified region. Upregulation was AGO2-dependent and did not induce an IFN response. dsRNAs targeting nearby regions instead resulted in slight downregulation, indicating the effect is sequence specific. Shortening the dsRNA to 16 nt or extending it to 26 nt also abrogated enhanced expression. The researchers noted that while RNAi by siRNA transfection typically lasts 5–7 days, observed enhancement persisted for more than 10 days. Chromatin immunoprecipitation analysis revealed epigenetic changes that might be inheritable through mitosis, providing a possible explanation for persistence.
Also studied by Li LC et al. (2006) with similar results were genes CDKN1A (alias p21WAF1/CIP1) and VEGF. Increases from 2- to 10-fold in mRNA and protein levels were variously detected. However, tests with genes ATR, PTEN, and APC did not produce strong upregulation. It would be of interest to understand this difference in susceptibility to upregulation.
In a subsequent paper (Place et al., 2008) by researchers also affiliated with the Dahiya lab, gene promoters were scanned for sequences complementary to known miRNAs seeds. An miR-373-3p target site was predicted in the promoter of CDH1. Transfection of miR-373-3p and its pre-miRNA into a human prostate cancer cell line induced CDH1 expression (but slightly mutated sequences did not) in a DICER-dependent manner and concomitant with enrichment of Pol II at the promoter. The miR-373-3p target site is further upstream from the siRNA target used by Li LC et al. (2006) and near the 5′ end of an Alu with + orientation. In Fig 1 the seed target is AGCACTT within the blue boundary:
Fig 1.
How miR-373-3p might hybridize with a hypothetical transcript from an Alu repeat in the promoter of CDH1. The seed region is within the blue boundary.
The significance of location in an Alu is that so many Alus appear in gene promoters, suggesting vast potential for analogous effects on other genes. Searching ~14,000 human promoter regions (up to 5000 bases upstream of transcription start) has revealed heavy Alu enrichment (Polak and Domany, 2006). The distribution is highly dependent upon gene function. For example, Alus are dense in promoters of RNA processing genes (about 5 Alus per 5 kb region) and relatively sparse in CNS development genes (about 2 per 5 kb, which is close to random distribution of one million Alus). Furthermore, several miRNAs are similar to miR-373 and might share targets, including miR-20a,b, -93, -17-5p, -106a,b, -372, -302a,b,c,d (Smalheiser and Torvik, 2006). Presumably almost all of the putative interactions are not in effect due to epigenetic or other factors, but it would be strange if miR-373 upregulation of CDH1 were the only case in nature.
The Corey lab has also done much to substantiate the case for upregulation by siRNAs and miRNAs (Janowski et al., 2007). They reported identification of multiple duplex RNAs complementary to the promoter of PR (progesterone receptor) that increase expression of PR mRNA and protein after transfection into cultured T47D or MCF7 human breast cancer cells. The dsRNA or ssRNA sequences that induced upregulation of PR included a 19 nt exact complement to a target sequence in the PR promoter plus a TT tail. Upregulation of PR was discovered by growing T47D cells in serum-stripped medium lacking hormones, thus having reduced expression of PR. Adding a particular dsRNA rescued normal PR expression. The researchers wrote, “It is possible that the activating RNAs bind directly to DNA, but is also possible that they bind to rare RNA transcripts that initiate upstream from the TSS [transcription start site] or bind to antisense transcripts; our data are consistent with either mechanism.” Interestingly, shifting the target sequence only two nts nullified the effect in some instances. In a follow-up study, the same lab showed that, indeed, the actual targets for “promoter-directed” siRNAs which either increased or decreased PR transcription are antisense transcripts initiating elsewhere but overlapping the PR transcription unit (Schwartz 2008). The siRNAs affecting transcription were dubbed “antigene RNAs” (agRNAs). Their ability to affect gene expression requires Argonaute proteins which target the agRNAs to the antisense transcript. The AGO-agRNA-antisense RNA complex then recruits other factors, such as hnRNP-k and heterochromatin protein 1, that then alter gene expression.
As another instance of miRNA upregulation of translation, Vasudevan et al. (2007) and Vasudevan et al. (2008) connected cell cycle phases with regulatory effects. They showed in HEK293 cells (Human Embryonic Kidney) that miRNAs including let-7 induced translation downregulation of target mRNAs in proliferating cells but upregulation in quiescent cells. They suggested that miRNAs generally have the potential to regulate translation in a manner determined by cell cycle phase.
Cloonen et al. (2008) studied the cluster of miRNAs known as miR-17-92 (Woods et al., 2007). They found it to be a cell cycle regulated locus. Ectopic expression of a member miRNA (miR-17-5p) was sufficient to drive a proliferative signal in HEK cells in a mechanism acting specifically at the G1/S cell cycle boundary when pro-proliferative mRNAs were upregulated through secondary effects. Thus the connection of cell cycle with miRNA signals includes a network of direct and indirect, up- and downregulation effects.
3. Results
3.1 Additional study of AGUGUU
Returning to the important observations of Hwang et al. (2007) on AGUGUU, our own investigation revealed the following. miRNAs miR-29b and miR-29a are defined as
miR-29b 5′-UAGCACCAUUUGAAAUCAGUGUU-3′
miR-29a 5′-UAGCACCAUCUGAAAUCGGUU -3′
For mitotic HeLa cells, Hwang et al. (2007) found using northern blots that the nuclear/cytoplasmic distribution of miR-29b was 71/29 while that of miR-29a was 42/58. Confocal microscopy results with fluorophore-tagged 29a and 29b siRNAs showed a localization consistent with the endogenous miR-29 RNAs. In additional tests with synthetic siRNAs, sequences ending in AGUGUU were found to have the strongest nuclear concentrations. Thus the researchers concluded, “These RNAs may prove useful for the manipulation of nuclear steps in gene expression,” a belief reflected in their patent activities.
We transfected into HeLa cells selected RNA sequences tagged with fluorescein (FAM). Transfection employed the DeliverX protocol (Panomics, Fremont CA) using a short amphipathic peptide that forms stable nanoparticles with nucleic acids that enter cells independent of endosomal pathways (Morris MC et al., 2008). Small ssRNA species were obtained from IDT (Coralville IA).
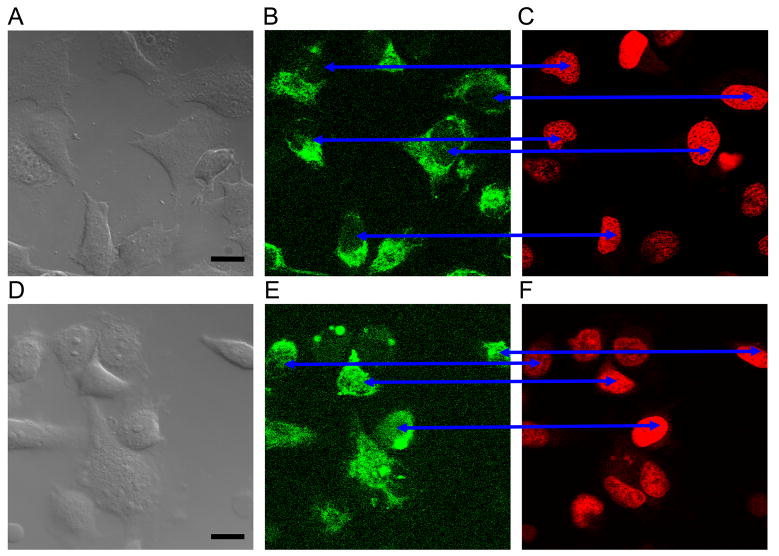
We first applied tagged sequences equal to or derived from the miR-29b sequence; the derived sequence had AGUGUU omitted from the 3′ terminus. Results are shown in Fig 2.
Fig 2.
Agent with AGUGUU penetrated some HeLa nuclei while agent without did not. Images A,D are DIC micrographs with bar = 10 um; images B,E are FAM emission confocal micrographs; images C,F are DRAQ5™ emission confocal micrographs. Cells in the upper three images were transfected with FAM+UAGCACCAUUUGAAAUC; no nuclei have FAM fluorescence. Cells in the lower three images were transfected with FAM+UAGCACCAUUUGAAAUCAGAGUU; most nuclei have at least some FAM fluorescence (blue arrows). Here and also in Figs 3 and 5 the brightness and contrast settings for each fluorescence channel are identical. The confocal images are from a ~400 nm slice of the sample. Cells were alive. The images were obtained 3 to 4 hours post-treatment, hence nuclear import was not always related to a particular cell cycle phase, including the mitosis phase. Scale bar = 10 μm.
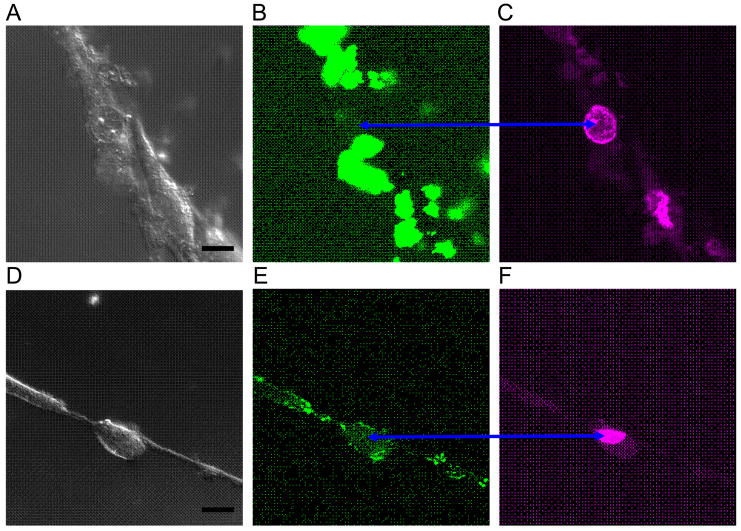
Next we treated human neural progenitors derived from olfactory epithelium. The agent was not so readily transfected, but nuclear penetration of agent with AGUGUU was observed as shown in Fig 3.
Fig 3.
Agent with AGUGUU penetrated some olfactory epithelium neural progenitor nuclei while agent without did not. Images A,D are DIC micrographs with bar = 10 um; images B,E are FAM emission confocal micrographs; images C,F are DRAQ5™ emission confocal micrographs. Cells in the upper three images were transfected with FAM+UAGCACCAUUUGAAAUC; nuclei have no FAM fluorescence, even at high excitation power. The cell in the lower three images was transfected with FAM+UAGCACCAUUUGAAAUCAGAGUU and has some FAM fluorescence (blue arrows) from its nucleus.
We also reviewed the discoveries of Place et al. (2008) on miR-373. Fig 4 shows the predicted structure of the pre-miRNA; miR-373-5p is in the blue boundary and miR-373-3p is in the red. Underlined is subsequence AGUGCUU, suggesting a relationship with the AGUGUU sequence studied above.
Fig 4.
The two miRNAs from the pre-miR-373 hairpin. The observation of miR-373 regulatory activity by Place et al, (2008) might be due to the inclusion of AGUGCUU, as it is similar to the sequence AGUGUU associated with nuclear import by Hwang et al. (2007).
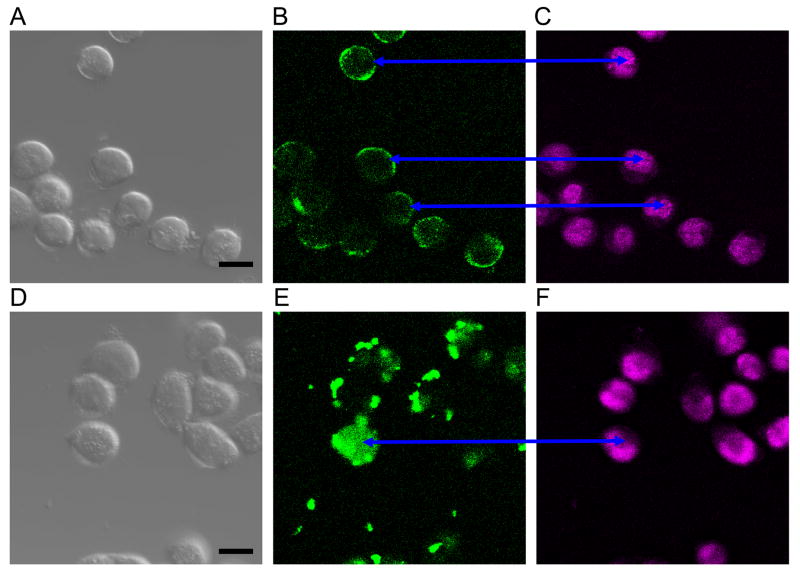
Again transfection of agents was used to study nuclear import. The two agents were FAM+adenine+mature miR-373-5p versus FAM+miR-373-3p mature. Results are shown in Fig 5. Interestingly the agent with AGUGCUU penetrated some nuclei while the -5p agent did not.
Fig 5.
Agent with miR-373-3p penetrated ~10% HeLa nuclei while agent with miR-373-5p penetrated none in our observations. Images A,D are DIC micrographs with bar = 10 um; images B,E are FAM emission confocal micrographs; images C,F are DRAQ5™ emission confocal micrographs. Cells in the upper three images were transfected with FAM+UAGCACCAUUUGAAAUC; no nuclei have FAM fluorescence. Cells in the lower three images were transfected with FAM+UAGCACCAUUUGAAAUCAGAGUU; most nuclei have at least some FAM fluorescence (blue arrows).
4. Discussion
We have suggested or described several speculative miRNA mechanisms for which recent evidence exists. Clearly there is much to keep miRNA researchers busy. We note that our own evidence is based upon confocal microscopy, but care should be taken in interpretation since identical cells in the one experiment are capable of very nonidentical, stochastic phenomena (Raj and van Oudenaarden, 2008). Nuclear assays are needed that corroborate and investigate putative nuclear miRNA import mechanisms such as the possible connection suggested in Fig 4 between AGUGUU in miR-29b (Hwang et al., 2007) and AGUGCUU in miR-373-3p (Place et al., 2008). Much additional work is needed to clarify nuclear import mechanisms for small ssRNAs..
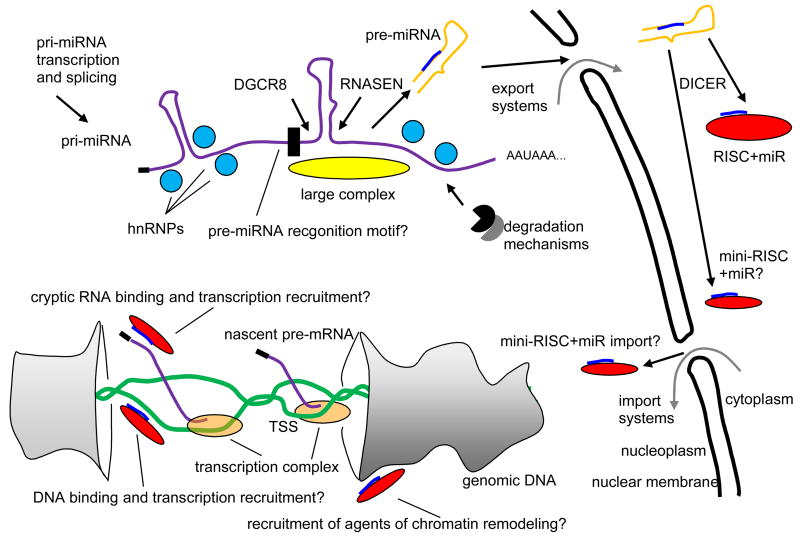
However, a diagram that organizes the potential interactions and flows is shown in Fig 6. While a number of nuclear roles for mi- and siRNAs have been reported, so far only human Ago2 and C. elegans NRDE-3, both members of the Argonaute family of RNA-binding proteins, have been found responsible for RNA nuclear localization (Guang 2008, Ohrt 2008). However, it is unknown how these proteins with their bound RNAs are imported. AGO2 might or might not be related to the up- or downregulation phenomena involving promoter regions described above.
Fig 6.
Mechanisms, largely speculative or surmised, suggested in this paper. A primary miRNA transcript (pri-miRNA), possibly from an intron, is chaperoned into folded RNA (purple) by heterogeneous ribonucleoprotein particles (hnRNPs) (light blue), countering RNA degradation mechanisms. Possibly a large complex (Gregory et al., 2004) (yellow) or a sequential or geometric recognition motif (black) guide recruitment of protein products of genes DGCR8 and RNASEN, yielding excision of a precursor RNA hairpin (pre-miRNA) (orange with blue subsequence that becomes the mature miRNA). The pre-miRNA is exported. Protein product of the DICER gene selects an ssRNA section of the hairpin stem, the mature miRNA, for mounting on an RNA induced silencing complex (RISC) (red). Evidence exits (Ohrt et al., 2008) that a single protein product of AGO2 enables re-import of some miRNAs or other short ssRNAs, possibly as a “mini-RISC.” Back in the nucleus the miRNA might engage open genomic DNA (green) or some cryptic noncoding RNA to enhance or suppress recruitment of transcription complex to the transcription start site (TSS) of a gene. Recruitment of agents of chromatin remodeling could be part of the mechanisms.
Strategies exist for advancement against the bewildering complexity of miRNA mechanics. A good example is that of Fedorov and Karpilow (2008), who have addressed the question: what miRNAs and proteins determine differentiation lineages of stem cells? Suppose a phenotypic outcome is detected. A screen strategy described in their technical note employs transfection in a propagation medium with a library of miRNA inhibitors followed by substitution with differentiation medium and phenotype monitoring. The same is done with a matched library of miRNA mimics. Transfection hits are selected on the basis of dosage dependency and induction of opposite effects from inhibitor and mimic pairs. Then a bioinformatic search interrogates 3′UTR tables for logically related genes. It might be advisable to apply loose versions of targeting algorithms that allow a single gap on one side in the seed region. Finally, a rescue strategy for implicated miRNA-protein pairs enforces inhibition of the miRNA, then checks for loss of phenotype, then inducts an exogenous protein source, and finally checks for rescue of phenotype. Many variations on such strategies become possible. In the particular case of mesenchymal stem cell osteogenesis studied by Fedorov and Karpilow, the outcome was selection of just three miRNAs; interestingly, two have very similar seed regions (hsa-miR-489 = GUGACAUCACA... and hsa-miR-148b = UCAGUGCAUCACA...), suggesting closely related targeting. Thus there are examples in which careful experimental design, modern screening products, and judicious use of bioinformatics enable researchers to bridge the yawning gap between what they know and what they would like to know.
Acknowledgments
Confocal micrographs were generated by our colleague Dr. Michael Chua of the Michael Hooker Microscopy Facility, University of North Carolina at Chapel Hill. The research reflected in this paper was supported in part by Stanley Medical Research Foundation grant 08R-1978, NIH grant 5P01ES014635-02, and NIH grant 2R01GM066940-05A1. The content is solely the responsibility of the authors, not the funding institutions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64 – 71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Edgar R. Reannotation of array probes at NCBI’s GEO database. Nat Methods. 2008;5:117. doi: 10.1038/nmeth0208-117b. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745 – 52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivukula RR, Mendell JT. Circular reasoning: microRNAs and cell-cycle control. Trends Biochem Sci. 2008 Sep 5; doi: 10.1016/j.tibs.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Brown MK, Steptoe AL, Wani S, Chan WL, Forrest AR, Kolle G, Gabrielli B, Grimmond SM. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9:R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799 – 816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov Y, Karpilow J. Successful use of Thermo Scientific Dharmacon® miRIDIAN microRNA mimic and inhibitor libraries identify miRNAs involved in human mesenchymal stem cell osteogenesis. Thermo Scientific Dharmacon application note. 2008 http://www.dharmacon.com/docs/miRNA_library_hMSC_app_note.pdf.
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235 – 40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537 – 41. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97 – 100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. Nucleotide motifs providing localization elements and methods of use. World Intellectual Property Organization 2007 WO 2007/149521 A2.
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787 – 92. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:136 – 7. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- Jeffries CD, Perkins DO, Jarstfer M. Systematic discovery of the grammar of translational inhibition by RNA hairpins. J Theor Biol. 2006;241:205 – 15. doi: 10.1016/j.jtbi.2005.11.027. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engström PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564 – 6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685 – 9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, Laouar A, Yao J, Haridas V, Habiro K, Yang YG, Jeong JH, Lee KY, Kim YH, Kim SW, Peipp M, Fey GH, Manjunath N, Shultz LD, Lee SK, Shankar P. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577 – 86. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103:17337 – 42. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hassan MQ, Volinia S, Wijnen AJ, Stein JL, Croce CM, Lian JB, Stein GS. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci U S A. 2008;105:13906 – 11. doi: 10.1073/pnas.0804438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556 – 61. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV. RNA-mediated transcriptional gene silencing in human cells. Curr Top Microbiol Immunol. 2008;320:211 – 24. doi: 10.1007/978-3-540-75157-1_10. [DOI] [PubMed] [Google Scholar]
- Morris MC, Deshayes S, Heitz F, Divita G. Cell-penetrating peptides: from molecular mechanisms to therapeutics. Biol Cell. 2008;100:201 – 17. doi: 10.1042/BC20070116. [DOI] [PubMed] [Google Scholar]
- Ohrt T, Mütze J, Staroske W, Weinmann L, Höck J, Crell K, Meister G, Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439 – 49. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673 – 8. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608 – 13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006;7:133. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschendorf M, Stein P, Oakeley EJ, Schultz RM, Peters AH, Svoboda P. Abundant transcripts from retrotransposons are unstable in fully grown mouse oocytes. Biochem Biophys Res Commun. 2006;347:36 – 43. doi: 10.1016/j.bbrc.2006.06.106. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842 – 8. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58 –63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532 – 6. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Stein P, Zeng F, Pan H, Schultz RM. Absence of non-specific effects of RNA interference triggered by long double-stranded RNA in mouse oocytes. Dev Biol. 2005;286:464 – 71. doi: 10.1016/j.ydbio.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065 – 73. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534 – 8. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124 – 8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. MicroRNA biogenesis: drosha can’t cut it without a partner. Curr Biol. 2005 Jan 26;15(2):R61–4. doi: 10.1016/j.cub.2004.12.057. [DOI] [PubMed] [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780 – 3. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931 – 4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545 – 9. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539 – 43. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, Riggs AD, Rossi JJ, Morris KV. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256 – 62. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130 – 4. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421 – 6. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763 – 71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA. Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci. 2006;63:246 – 54. doi: 10.1007/s00018-005-5467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]