Abstract
The development and maintenance of most tissues and organs requires the presence of multipotent and unipotent stem cells that have the ability of self-renewal as well as of generating committed, further differentiated cell types. The transcription factor Sox2 is essential for embryonic development and maintains pluripotency and self-renewal in embryonic stem cells. It is expressed in immature osteoblasts/osteoprogenitors in vitro and in vivo and is induced by FGF signaling, which stimulates osteoblast proliferation and inhibits differentiation. Sox2 overexpression can by itself inhibit osteoblast differentiation. To elucidate its role in the osteoblastic lineage, we generated mice with an osteoblast-specific, Cre-mediated knockout of Sox2. These mice are small and osteopenic, and mosaic for Sox2 inactivation. However, culturing calvarial osteoblasts from the mutant mice for 2-3 passages failed to yield any Sox2 null cells. Inactivation of the Sox2 gene by Cre-mediated excision in cultured osteoblasts showed that Sox2 null cells could not survive repeated passage in culture, could not form colonies, and arrested their growth with a senescent phenotype. Additionally, expression of Sox2 specific shRNAs in independent osteoblastic cell lines suppressed their proliferative ability. Osteoblasts capable of forming “osteospheres” are greatly enriched in Sox2 expression. These data identify a novel role for Sox2 in the maintenance of self-renewal in the osteoblastic lineage.
Introduction
The development and maintenance of all tissues and organs is orchestrated by a complex interplay of signaling molecules and transcription factors, whose expression governs cell fate determination, proliferation and differentiation. Signaling by the FGF family of growth factors and cognate receptors plays an important role in skeletal development by affecting the proliferation and differentiation of chondrocytes and osteoblasts, the two major cell types responsible for bone formation. Several human skeletal disorders are caused by activating mutations in FGF receptors (FGFR) (1-3). In the osteoblastic lineage, FGF stimulates the proliferation of immature cells and inhibits their differentiation (4-7). By studying the mechanisms underlying the FGF response of osteoblasts, we found that FGF signaling antagonizes Wnt-induced transcription, a process which promotes osteoblast differentiation and function (8, 9). In these cells, treatment with exogenous FGF or the presence of activated FGFR2 strongly induces the expression of the transcription factor Sox2. Sox2 contributes to the inhibition of Wnt signaling by FGF by binding to β-catenin, and its overexpression can by itself inhibit osteoblast differentiation (8, 9).
The transcription factor Sox2 is a member of the SRY-related, HMG box family, that plays a critical role in embryonic development and in maintaining the pluripotency and self-renewal of embryonic stem (ES) cells (10-12). It was originally identified as a binding partner of the transcription factor Oct-4 in ES and embryonal carcinoma (EC) cells, where it is crucial for the expression of FGF-4 (13), and was later shown to regulate the expression of many other ES cell-specific genes. It has also been recently shown to be a critical factor for the reprogramming of somatic cells into “induced” pluripotent stem cells (iPS) (14).
Our knowledge about the origins, fate and life span of cells along the osteoblast lineage is limited. Primitive osteoprogenitors arise from multipotent mesenchymal stem cells that commit and then differentiate to preosteoblasts and mature osteoblasts. While the transcription factors Runx2, and Osterix (OSX) have been shown to be essential for commitment to the osteoblastic lineage, few factors that play a defined role in early osteoblast lineage expansion have been identified (15-19).
To elucidate the role of Sox2 in osteogenesis and in osteoblasts self-renewal and differentiation, we have generated a conditional Cre-mediated knockout of Sox2 in the osteoblast lineage. These mice are highly mosaic for Sox2 inactivation, but are smaller and show decreased trabecular bone density compared to their wild-type littermates. By culturing calvarial osteoblasts from these mutant mice we were unable to recover any viable Sox2 null cells. Excision of the Sox2 gene in primary or immortalized osteoblasts in culture produced Sox2 null cells that did not survive passaging in culture, could not form colonies, and arrested their growth with a senescent phenotype. These and other results identify a previously unknown role for this transcription factor in maintaining self-renewal of the osteoblastic lineage and suggest that Sox2 expression is required to maintain osteoprogenitor cells in a multi-or unipotent stem like proliferative state.
Results and Discussion
Generation of SOX2 Conditional Knockout Mice
Since the Sox2 KO is lethal in early embryonic development (10), to precisely define the role of Sox2 in osteoblast proliferation and differentiation we created a conditional knock-out (CKO) of the Sox2 gene in the osteoblastic lineage utilizing mice with a Sox2flox/geo genotype (hereafter referred to as Sox2flox/-). In these mice, one of the two Sox2 alleles, is bracketed by loxP sequences, providing a target for the Cre-recombinase, while the other is replaced by the β-gal (geo) gene, that inactivates the Sox2 gene (Fig. 1A) (20) These mice, which are phenotypically normal, were crossed with transgenic mice expressing the Cre-recombinase under the control of the 2.3kb collagen I promoter, which is specifically expressed in the osteoblastic lineage, to elicit Sox2 excision in Cre-bearing Sox2-floxed F2 offspring (Fig. 1B) (21). The resulting Sox2 CKO mice were initially difficult to study because they died at birth with esophageal atresia, most likely due to aberrant expression of the Cre-recombinase in the esophagus and trachea and the essential role of Sox2 in the development of the foregut (22). However this phenotype has low penetrance, and at least 50% of the pups survive to adulthood. These mice are smaller than their wildtype littermates and are mostly sterile (Fig.1C). MicroCT analysis reveals that mutant mice have reduced bone density with defects in trabecular bone. Bone mineral density and bone volume are reduced, while trabecular pattern factor, a parameter of micro-CT analysis that is increased in poor quality bone (23), is higher in the Sox2 CKO mice (Fig. 1D-G). Cortical bone thickness is also reduced as is the density of parietal bones in the skull (not shown). Interestingly, these effects are clearly detected even though these mice are highly mosaic for Sox2 inactivation. At birth, >50% of the calvarial osteoblasts appear to have maintained the Sox2 gene. While in principle this could be due to inefficient expression or activity of the Cre recombinase, this seemed unlikely, since, if anything, the collagen I Cre mice appear to express Cre more uniformly and less tissue specifically than expected. This finding suggested that Sox2 expressing cells have a growth advantage over Sox2 null osteoblasts, a notion supported by the experiments described below.
Figure 1. Osteoblast-specific Sox2 conditional knock-out mice are smaller and have low bone density.
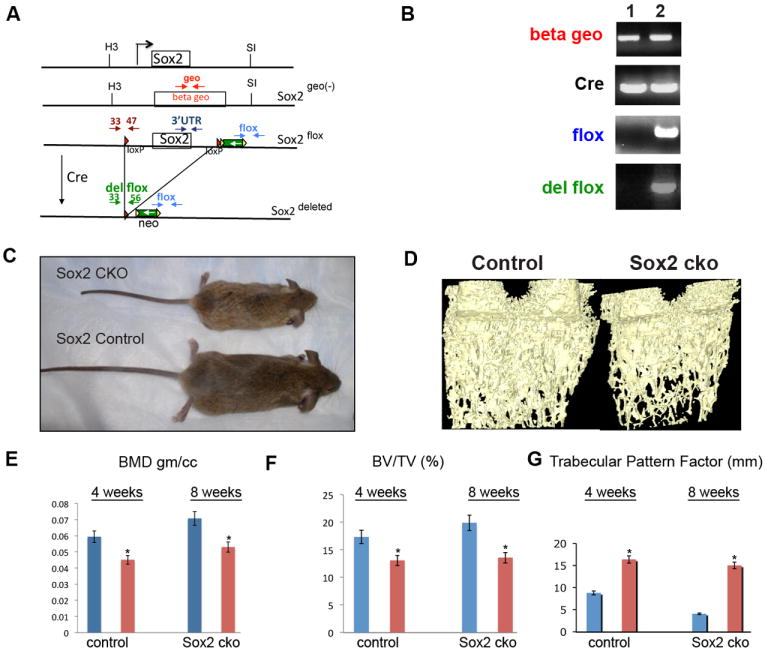
(A) Schematic of Sox2, Sox2geo (-), Sox2flox and Sox2del alleles. Map shows the location of diagnostic primer pairs in the Sox2 alleles. Beta geo is the beta geo gene in the Sox2 null allele Sox2geo(-); green box is the neo gene; red triangles are the loxP sites. H3, HindIII; S1, SalI. The results obtained within the diagnostic PCR primer pairs depicted in this figure using DNA from Sox2flox/- osteoblasts or Sox2flox/- osteoblasts subjected to Cre excision are shown in Supplementary Figure 1A.
(B) PCR analysis of calvarial DNA. DNA from littermates was isolated using standard techniques and PCR was performed with the indicated primers. The flox primer pair detects the deleted and undeleted flox alleles. The del flox primer pair detects only the deleted Sox2 allele after Cre recombination. 1. Sox2+/geo;Cre (control), 2. Sox2flox/geo;Cre (cko)
(C) Four week old Sox2 cko (Sox2flox/-; Cre) and control (Sox2+/geo;Cre) littermate.
(D) Reconstructed image from Micro cT analysis of 4-week-old distal femurs from Sox2 cko and control littermate.
(E) Quantification of average bone mineral density, (F) bone volume per tissue volume (BV/TV), and (G) trabecular pattern factors obtained from micro-CT analysis of 4 and 8 week femurs of Sox2 cko and control littermate animals.
Sox2 inactivation impairs osteoblast proliferation
To analyze the phenotype of Sox2 null osteoblasts, we generated cultures of calvarial osteoblasts from the mutant mice. PCR analysis of DNA from freshly cultured cells showed that deletion of the floxed Sox2 allele had occurred. However, after culturing osteoblast pools from the P1 calvaria of the mutant Sox2flox/-;Cre mice for 3-4 passages, the deleted Sox2 allele could no longer be detected by PCR analysis, while it was maintained in cultures from Sox2flox/+;Cre mice, which carry a wild type Sox2 allele that cannot be excised by the Cre recombinase, suggesting that Sox2 deletion is incompatible with osteoblast survival (Fig. 2A). To directly determine whether Sox2 inactivation was deleterious to cell survival, we infected freshly cultured primary Sox2flox/- osteoblast populations with a Cre-expressing adenovirus (also expressing GFP linked to Cre by an IRES) to induce Sox2 excision. We could show that Sox2 excision had taken place in a majority of the cells by PCR analysis, but again, after 2-3 passages in culture Sox2-negative cells could no longer be detected (Fig. 2B). To better quantitate these experiments, we performed colony assays. We infected independent cultures of primary osteoblasts derived from Sox2flox/- mice with the Cre-expressing adenovirus and the corresponding control adenovirus expressing only GFP. While cells infected in the GFP-virus yielded a very similar number of colonies, Cre-virus infection drastically reduced the colony forming ability of these cultures. In contrast, cultures of primary osteoblasts from Sox2flox/+ mice were completely resistant to the effect of Cre-virus infection (Fig. 2C). Together these results strongly suggested that Sox2 inactivation abolished the ability of osteoblasts to sustain proliferation in culture
Figure 2. Sox2 inactivation impairs proliferation of primary osteoblasts.
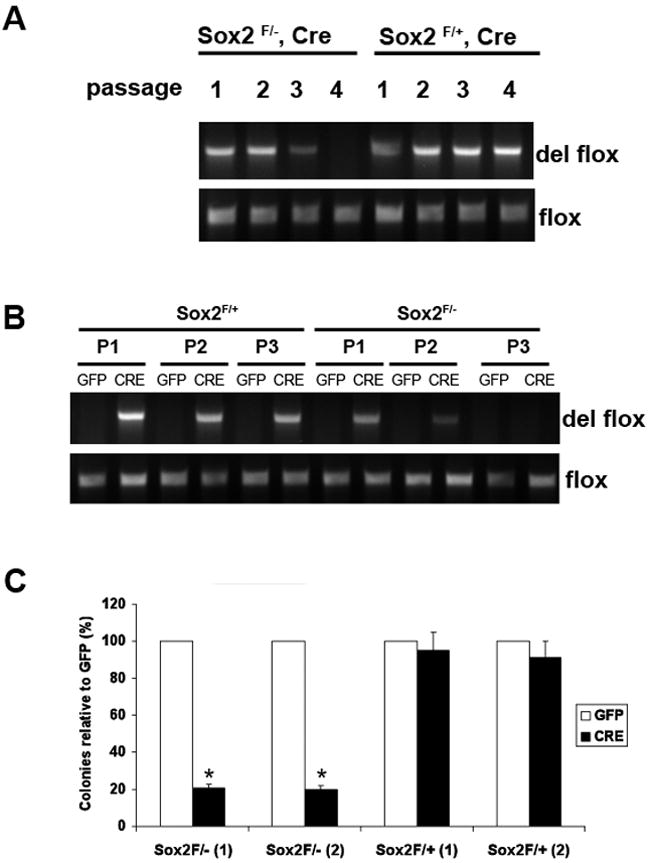
(A) PCR analysis of primary osteoblast DNA. Primary calvarial osteoblasts were prepared from Sox2flox/-;Cre (cko) mice and Sox2flox/+;Cre (control). Upon confluency, cells were passaged and DNA was extracted at each passage. The deleted flox and the flox allele were detected by PCR using specific primers to assess persistence of Sox2-null cells. Numbers indicate passage number following isolation.
(B) PCR analysis of GFP- and Cre-virus infected primary osteoblast DNA. Primary osteoblasts from Sox2 flox/+ (control) and Sox2 flox/- (cko) mice were infected with GFP or Cre-adenovirus and passaged after infection. DNA was extracted from each passage and the deleted flox allele was detected by PCR using specific primers to check for persistence of Sox2-null cells.
(C) Colony Assay. Primary calvarial osteoblasts from Sox2 flox/+ (control) and Sox2flox/- (cko) mice were infected with either GFP or Cre-adenovirus for 72 hours and plated in triplicate in 6-well plates. Colonies were counted after crystal violet staining. Percent of colonies obtained in Cre infection are plotted as a percentage of the colonies in the corresponding GFP infection in two independent Sox2 flox/- and Sox2 flox/+ osteoblast pools [(1) and (2)]. * = p<0.05. GFP Adenovirus infection of Sox2flox/- or Sox2flox/+ cells produced equivalent number of colonies (~120 colonies/plate).
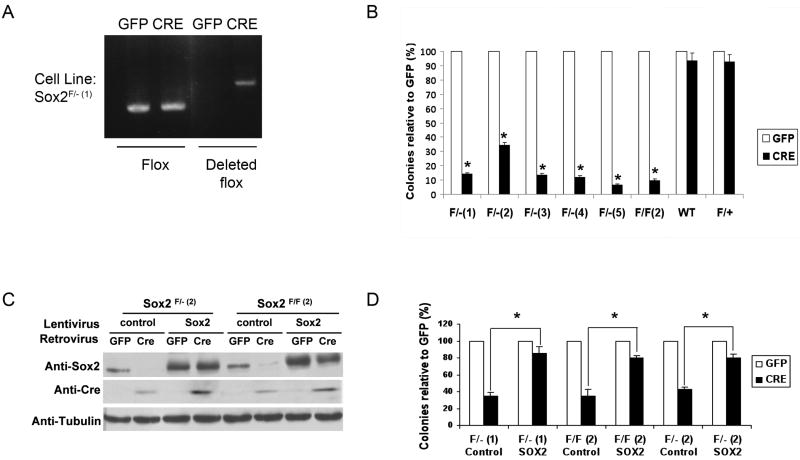
To facilitate biochemical and genetic experiments that are difficult to perform in primary cultures that have a limited life span, we created several clonal lines of Sox2flox/- osteoblasts by immortalizing them with Polyoma large T antigen (PyLT). PyLT inactivates the Rb proteins, but not p53. As previously reported, PyLT immortalized osteoblasts are still capable of differentiating into mature osteoblasts in culture, although this process is slightly slower than in primary osteoblasts (6). Several independent clones of immortalized Sox2flox/- cells were infected with a Cre-expressing retrovirus that also carries the GFP gene. Infection of such cell lines leads to deletion of Sox2 gene and reduction in Sox2 protein level (Fig 3A, C). As observed in primary osteoblasts, however, subsequent passaging of the Cre virus infected cells led to disappearance of the deleted floxed allele, indicating that cells in which Sox2 excision had occurred were incapable of survival in culture. We therefore performed a colony assay where either GFP- or Cre-GFP infected cells were plated and the number of colonies formed determined after 7 days. As shown in Fig. 3B, the number of colonies obtained was drastically reduced (70-90%) in Cre-infected cells with respect to those produced by a control retroviral infection (no Cre), and all surviving clones were GFP negative, representing cells that escaped retrovirus infection. Infection of wild type or Sox2flox/+ osteoblasts with the Cre-virus had no deleterious effect.
Figure 3. Transgenic expression of Sox2 rescues the lethality caused by Sox2 deletion in immortalized osteoblasts.
(A) PCR analysis of immortalized osteoblast DNA. Representative immortalized Sox2-floxed osteoblast clone Sox2 flox/-(1) (referred to as Sox2F/-(1)) was infected with GFP or Cre-retrovirus for 72 hours. DNA was extracted and the deleted flox and flox bands were detected by PCR using specific primers to determine efficiency of in vitro Sox2 deletion by Cre-dependent excision.
(B) Colony Assay. Immortalized Sox2-floxed or wild type osteoblasts were infected with either GFP or Cre-retrovirus for 72 hours and plated in triplicate in 6-well plates. Colonies were counted after crystal violet staining. Percent of colonies obtained in Cre infection are plotted as a percentage of the colonies in the corresponding GFP infection in five independent Sox2 F/- lines (F/-1-5), one Sox2 F/F line F/F (2), Sox2 F/+ and wild type (WT) cells. * = p<0.05
(C) Western analysis. Two Sox2-floxed osteoblasts clones Sox2 F/-(2) and Sox2 F/F(2) were infected with either control or Sox2-expressing lentivirus for 72 hours. Following infection, they were re-infected with GFP or Cre-retrovirus for 72 hours. Sox2 and CRE protein were detected by immunoblotting using antibodies against Sox2 and Cre. γ-tubulin was used as a loading control.
(D) Colony Assay. Sox2-floxed osteoblasts [F/F(2)) or (F/-(1) and F/-(2)] were infected as described in 3C and plated in 6-well plates in triplicate. Colonies were counted after 10 days. Note that all colonies surviving Cre-virus infection in the control cells were GFP negative (therefore not expressing Cre), while most of the colonies in the lenti-Sox2 expressing cells were GFP positive. * = p<0.02
Several groups have reported that Cre expression can cause non-specific DNA-damage effects in cell culture (24). Despite the lack of any effects of the Cre recombinase in wildtype and Sox2flox/+ osteoblasts, we wanted to rule out the possibility that the observed cell lethality following Cre-virus expression might be due to the toxic effects of Cre. We thus isolated fibroblasts (MEF) from mouse embryos and infected them with the same Cre and GFP retroviruses. Infection of Sox2flox/- fibroblasts, which do not express Sox2, produced excision of the Sox2 gene, but Sox2 null fibroblasts could be easily maintained in culture (Supplementary Fig. 1B). To conclusively demonstrate that the decrease in colony formation induced by Cre-mediated excision in Sox2flox/- osteoblasts was due only to Sox2 inactivation, we introduced into several clones of immortalized Sox2flox/- or Sox2flox/flox cells a “transgenic” copy of the Sox2 gene using lentivirus-mediated transduction (Fig. 3C). Infection with the Cre-virus led to excision of the endogenous Sox2 gene but no significant inhibition of colony formation was observed, showing that constitutive expression of Sox2 abolishes the detrimental effects of the inactivation of the endogenous Sox2 gene (Figure 3D). These experiments therefore show that the inability of Sox2 floxed osteoblasts to proliferate following Cre-virus infection is due to the lack of Sox2 expression and not to Cre-mediated cell damage.
Inactivation of Sox 2 in cultured osteoblasts causes a senescent-like growth arrest
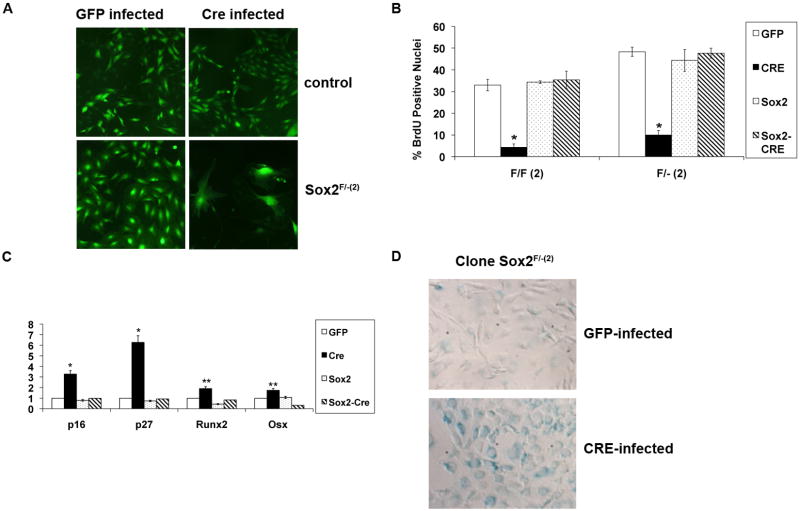
Examination of Sox2 depleted cell populations 6-7 days after infection showed extensive morphological changes with the appearance of large cells with a “flat”, senescent-like morphology, and substantial inhibition of proliferation. (Figure 4A). DNA synthesis was greatly decreased by 48 hours after Cre-virus infection (Figure 4B), but no evidence of increased apoptosis, as measured by TUNEL assay or caspase activation, was observed. As expected, Sox2 transduced cells experienced no reduction in DNA synthesis. Since Sox2 overexpression inhibits differentiation in immortalized osteoblasts (8) as well as in primary cells (Supplementary Fig. 2), we tested the expression of several markers of osteoblast differentiation, including alkaline phosphatase (ALP), Runx2 and OSX expression (17) We did not observe any increased expression of ALP by 7 days after Cre-infection, and only a minor increase in the expression of Runx2 and OSX RNAs. Instead, we noticed a clear increase in the expression of cdk inhibitors p16 and p27 (Fig. 4C). Cdk inhibitors expression is elevated in senescent cells (25), and this together with the cell morphology of Cre-infected osteoblasts made us assay for the presence of senescence-associated- β-galactosidase (SABG) that is considered to be a hallmark of senescence (26). Cre-infected osteoblasts stained positive for SABG while Sox2- transduced cells did not, indicating that loss of Sox2 might produce a phenotype akin to senescence in osteoblasts (Figure 4D). It is relevant to mention here that the endogenous β-gal from the Sox2 null allele is not expressed in these cells, probably because of the deletion of a ~2kb DNA region (20) that appears to contain osteoblast-specific regulatory elements.
Figure 4. Inactivation of Sox2 in immortalized osteoblasts causes a senescent-like growth arrest with p16 and p27 upregulation.
(A) Morphology of colonies. Live colonies of GFP and Cre-infected control and Sox2F/-(2) immortalized osteoblast clones were examined under a fluorescence microscope (10X magnification) 1 week after infection.
(B) BrdU incorporation of GFP and Cre-infected Sox2-floxed osteoblasts Two independent clones (Sox2 F/-(2) and Sox2 F/F(2)) were infected as described in Figure 3C. BrdU incorporation 48 hours after infection was assessed by labeling cells with 4 μg/ml BrdU for 3 hours and staining with an anti-BrdU antibody. GFP/BrdU-positive cells were counted using a fluorescence microscope. At least 10 independent fields were counted and each experiment was repeated at least twice. * = p<0.01
(C) Gene expression analysis Sox2F/F(2) was infected as described in Figure 3C and mRNA was extracted. Gene expression of p16, p27, Osterix and Runx2 was analyzed after 72 hours of infection by qRT-PCR using specific primers. All values are normalized to actin as an internal control and are expressed relative to GFP-infected control cells. * = p<0.01, **=p<0.05
(D) Senescence-associated β-galactosidase (SABG) expression. SABG activity in Sox2-floxed osteoblast clone Sox2 F/-(2) was determined after 96 hours of infection. Slides were mounted in DAKO mounting medium and photographed under 40X magnification.
A threshold of Sox2 is necessary to maintain osteoblast survival
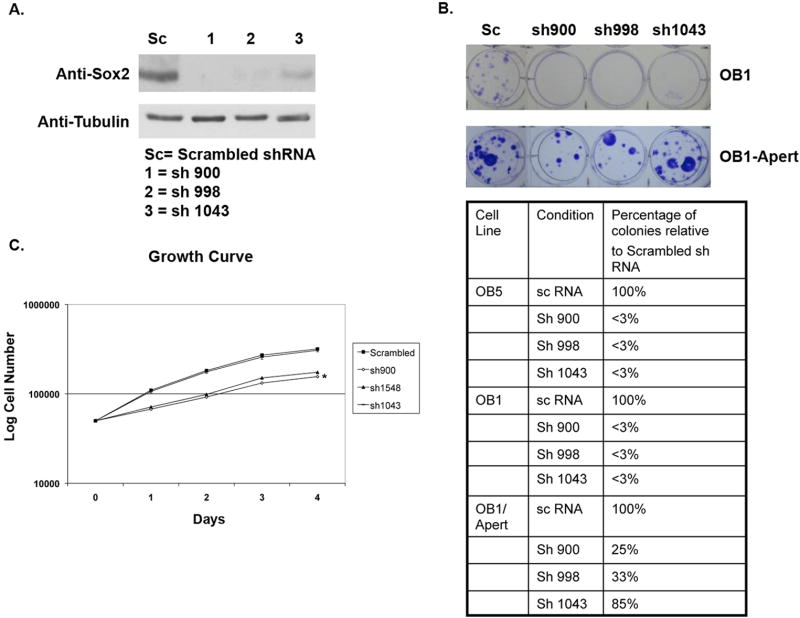
While our findings with osteoblasts from Sox2 mutant mice highlighted the role of Sox2 in maintaining proliferation potential in osteoblasts, all of our experiments had been performed with osteoblasts from mutant mice carrying a floxed Sox2 allele, and it was of interest to be able to extend these observations to independent osteoblastic cell lines. Furthermore, we wished to rule out the effect of excision of any Sox2 flanking sequences such as those that may encode regulatory microRNAs. To address these questions, we used two immortalized osteoblast cell lines that our laboratory had previously characterized: OB1, a PyLT immortalized immature osteoblast line, and OB5, a spontaneously immortalized cell line derived from p53+/- calvaria (6, 27). We also tested OB1-Apert cells, an OB1-derived clone carrying the activated FGFR2 bearing the S252W mutation found in Apert syndrome, that express high levels of Sox2, at least 10-fold more than the parental OB1 cells (8). We infected these cell lines with pBabe-hygro retrovirus expressing three shRNAs directed against murine Sox2, and a scrambled shRNA control and selected for Hygromycin resistant cell clones. These three shRNAs were found to differentially reduce Sox2 expression in the OB1-Apert cells (Figure 5A), ranging from 60 to 90% inhibition of Sox2 protein expression. As seen in Figure 5B, infection of OB1 or OB5 cells with Sox2 shRNA vectors completely abrogated colony formation. On the other hand, OB1-Apert cells were still capable of forming colonies, with an efficiency ranging between 25-85% of the colonies produced by the cells expressing the scrambled control shRNA (Fig. 5B). The differential ability of the shRNAs to inhibit Sox2 protein expression was reflected in the ability to form colonies, as the shRNA (1043) that caused the least reduction in Sox2 levels produced the highest number of colonies. The OB1-Apert cells expressing Sox2 shRNA 900 and 998 that could form colonies grew considerably more slowly than the parental cells (Fig. 5C), while the growth of cells expressing shRNA 1043 was not affected, as expected. The proliferation of NIH3T3 mouse fibroblasts, that do not express Sox2, was unaffected by Sox2 shRNAs as infection with all Sox2-specific shRNA viruses resulted in a number of hygromycin resistant colonies essentially identical to that produced by the scrambled shRNA control (not shown). These results demonstrate that the requirement for Sox2 expression is not limited to osteoblasts from Sox2 mutant mice and suggest that a threshold level of Sox2 expression is necessary to maintain osteoblast self-renewal.
Figure 5. A threshold of Sox2 is required to maintain osteoblast survival.
(A) Western Blot for Sox2 expression. OB1-Apert cells were infected with pBabe-Hygro retrovirus expressing the indicated shRNAs against Sox2 or scrambled (sc) shRNA. Pools of cells were selected in Hygromycin B and protein was extracted. Sox2 expression was determined by immunoblotting.
(B) Colony formation by Sox2 shRNA-expressing osteoblasts. OB5, OB1 and OB1-Apert cells infected with pBabe-Hygro retro-virus expressing either scrambled (sc) or sh900, sh998 and sh1043 were plated in triplicate and selected in Hygromycin B for 7-10 days followed by staining with Crystal Violet. Pictures of representative plates of OB1 and OB1-Apert cells expressing shRNA are shown and quantified in adjoining Table
(C) Growth of Sox2 shRNA-expressing OB1-Apert osteoblasts. OB1-Apert cells (0.1 × 106 cells/well) stably expressing either scrambled (sc) or sh900, sh998 or sh1043 were plated in triplicate and growth of cells assessed by counting cells in a Z1 Beckman Coulter counter. * = p<0.05
Sphere-forming osteoblasts are enriched for Sox2 expression
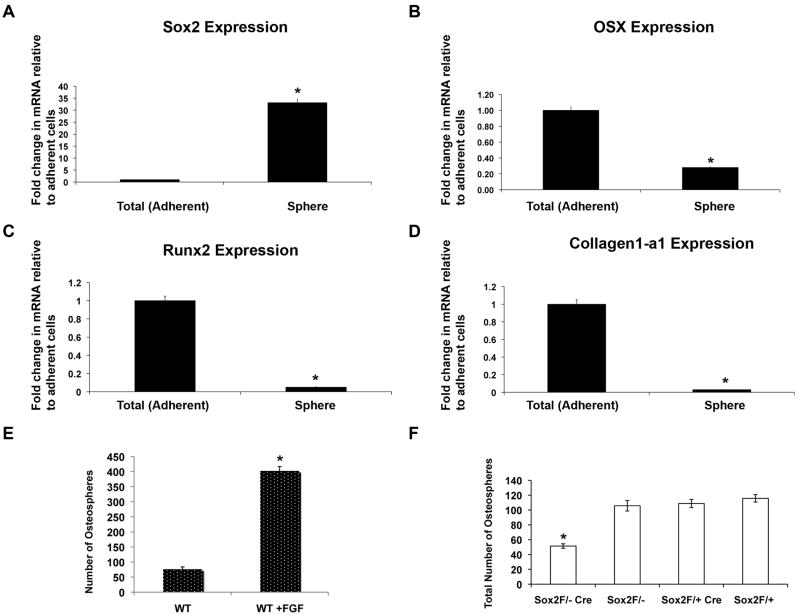
The results presented so far demonstrate that Sox2 is essential for osteoblast self-renewal and suggest that its expression may mark a population of uncommitted or unipotent osteoblasts. To investigate this hypothesis we have used a technique that has been successful in identifying such stem-like cells in the nervous system, mammary gland, skin and tumor tissues, i.e. the ability of these cells to form “spheres” of unattached cells in suspension culture. Indeed, a recent report has shown that calvarial osteoblasts are capable of forming “osteospheres” in suspension, representing progenitor/multipotent cell populations (28). In line with this observation, we found that osteoblasts isolated from calvaria of P1 pups are capable of forming osteospheres. We found that Sox2 expression is greatly enriched in the osteosphere fractions as compared to the total adherent cell population (Fig. 6A). On the other hand, the expression of OSX, Runx2, and ColI, markers of committed osteoblasts, was down-regulated in the spheres, confirming that they indeed represent a precursor cell population (Fig. 6B-D). Treatment with FGF, that induces Sox2 expression, increased osteosphere formation by primary osteoblasts 5-6 fold (Fig. 6E). As expected, we also found that osteoblasts made from calvaria of Sox2flox/-;Cre mice, that are a mosaic of Sox2 positive and Sox2 null cells, show reduced osteosphere forming ability as compared to the osteoblasts isolated from control littermate mice, indicating that Sox2 is necessary to maintain a pool of osteoprogenitor cells (Figure 6F).
Figure 6. Sox2 is enriched in osteospheres and is required for osteosphere formation.
(A-D) Osteospheres from osteoblasts derived from P1 calvaria of wild type mice were collected by straining through a 40 μM strainer and RNA was extracted from the spheres. Gene expression of Sox2 (A), Osterix (B), Runx2 (C), and Collagen1-a1 (D) was analyzed by qRT-PCR using specific primers. All values are normalized to actin as an internal control and are expressed relative to total (adherent) cells in each case. * = p<0.05
(E) Primary calvarial osteoblasts were plated in suspension culture in the presence or absence of 10 ng/ml recombinant FGF1. Spheres were counted after 7 days. * = p<0.05
(F) Primary P1 calvarial osteoblasts from control (Sox2 flox/+;Cre, Sox2flox/- and Sox2flox/+) and cko (Sox2flox/-;Cre) littermates were plated in triplicate in 6 cm Corning ultra-low attachment plates. Total number of osteospheres was counted after 7 days. * = p < 0.05
Together these results support the notion that, while Sox2 expression is necessary for the survival of the entire osteoblast population, high levels of Sox2 correlate with the ability to form osteospheres, and mark a subpopulation of immature, progenitor cells.
In this report, we described the novel finding that the transcription factor Sox2 plays an essential role in the expansion of the osteoblastic lineage. Progenitor stem-like cells are responsible for the development and maintenance of most tissues and organs. Osteoblast populations in culture represent a mixture of osteoprogenitor cells and immature osteoblasts, and proliferation in culture selects for more immature multi-or unipotent stem-like cells (28) (29). Our data show quite clearly that inhibition of Sox2 expression, whether by excision of the endogenous Sox2 gene or by RNA interference, abolishes the ability of both primary and immortalized osteoblasts to proliferate. These conclusions are also supported by the observation that mice we have generated with a conditional knock-out of Sox2 in the osteoblastic lineage have reduced bone density, in spite of being highly mosaic for Sox2 inactivation. While a full description of the phenotype of these mice is beyond the scope of this report, clearly this phenotype is less severe that what could have been expected from the behavior of Sox2 null osteoblasts in culture. A likely reason for this is that only ~50% of the osteoblasts in the skull (and presumably also in other bones) have lost the Sox2 gene, probably because the collagen I promoter-driven Cre used in these experiments is expressed rather late during osteoblastic lineage development (21, 30, 31), or is inefficient, allowing cells escaping Cre-mediated Sox2 excision to proliferate at the expense of Sox2 null cells. Another possible explanation is that Sox2’s function could be redundant with that of other Sox factors in vivo, such as Sox4, whose inactivation has been also shown to impair bone development (32). However, such redundancy does not seem to occur in cell cultures, and would not account for the mosaicism of the Sox2 CKO mice.
The results presented here strengthen the notion that Sox2 expression is required for the maintenance of a stem cell progenitor-like state in several tissues. Indeed a similar requirement for Sox2 expression has been reported for neural and retinal progenitor cells (20, 33, 34). Our results also demonstrate that Sox2 inactivation causes growth-arrest by inducing a senescent-like phenotype. Runx2 deficient osteoblasts have been reported to bypass senescense (35), but we found that the level of Runx2 in Sox2-null cells is not appreciably altered. Sox2 null cells do not appear to be in a terminally differentiated osteocyte-like state as DMP1, a marker of osteocytes is not expressed in Sox2 null cells (data not shown). This suggests that Sox2 inactivation targets specific osteoblast genes that may be important for their proliferation. Although we have showed that Sox2 can interfere with Wnt/β-catenin signaling by binding to β-catenin, such an effect requires relatively high levels of Sox2 such as those induced by FGF (8, 9), while the basal levels of Sox2 expression in osteoblasts are quite low. We therefore do not think the main role of Sox2 in the osteoblast lineage is, in the absence of exogenous Wnt stimulation, that of inhibiting Wnt signaling. Rather we interpret the ability of Sox2 overexpression to inhibit differentiation as reflecting its function in maintaining osteoblasts in an immature, unipotent state. This is further supported by our observation that Sox2 expresssion is higher in osteosphere-forming cells as well as by the finding that Sox2 expression is enhanced in osteogenic cells in which differentiation is impaired (36). While further work will be required to conclusively answer these questions, it is interesting to note that forced Sox2 expression in bone marrow derived-mesenchymal stem cells (MSC) has been reported to promote their expansion and ability to differentiate into the osteoblastic lineage (37). Furthermore, several groups have described an anabolic effect of FGF treatment in vivo on bone formation, a finding that still awaits a mechanistic explanation (38-40). MSCs persist in adult organisms and are thought to contribute to the replacement of osteoblasts in bone turnover and fracture healing. It is tempting to speculate that the bone anabolic effect of FGF in vivo results from its ability to induce Sox2 and expand the pool of osteoprogenitor cells or of MSCs that commit to the osteoblastic lineage. If this hypothesis is correct, compounds that mimic or induce Sox2 expression in the osteoblastic lineage could be useful bone anabolic agents.
Materials and Methods
Generation of Sox2-floxed mice
The Sox2-floxed mice have been recently described elsewhere (20). Primers used for PCR genotyping of such mice are available upon request.
Analysis of skeletal phenotype of mutant mice
Mice were euthanized by carbon dioxide narcosis and defleshed in 2% KOH for 7 days. Quantitative micro-computed tomography (micro-CT) analysis was conducted at the Center for Bone and Periodontal Research, McGill University, Montreal, Quebec, Canada. Data acquisition and analysis was on a SkyScan T-1072 microtomograph using software supplied by SkyScan.
Cell culture
Osteoblasts from P1 calvaria were isolated by serial collagenase digestion and immortalized as described (6). Mouse embryonic fibroblasts (MEFS) were isolated from E16.5 embryos. All cells were maintained in DMEM supplemented with 10% fetal bovine serum.
Viral vectors
For primary cells, an adenovirus expressing either eGFP (control) or Cre (linked to an IRES-GFP to detect expression) was obtained from the Vector Development Laboratory at Baylor College of Medicine, Houston, TX, and used at an MOI of 50 in 2% FBS containing DMEM for 2 hours. For immortalized cells and MEFs, a retrovirus either encoding eGFP (MSCV-eGFP) or CRE (MSCV-CRE-eGFP) was used at an MOI of 10 to infect cells in the presence of 8 μg/ml Polybrene for 24 hours. For Sox2 lentiviral transduction experiments, a lentivirus, expressing full-length human Sox2 was used at an MOI of 10 for infection in the presence of 8 μg/ml Polybrene for 24 hours.
Colony Assay
GFP or Cre-GFP virus infected osteoblasts were plated at a density of 500-5000 cells/well in 6-well plates. GFP-positive colonies were visualized and counted under a fluorescence microscope after 7-10 days. Colonies were fixed in 20% methanol for 15 minutes at room temperature, and counted after staining with 0.5% crystal violet in 20% methanol.
Detection of Senescence-Associated β–Galactosidase (SABG) Activity
Cells were plated on coverslips and infected with GFP or CRE retrovirus, and recovered for 96 hours. SABG activity was detected at pH 6.0 as described (26).
Gene expression analysis by quantitative real-time RT-PCR and Western Blotting
mRNA was prepared using Trizol Reagent (Invitrogen, USA). Reverse transcription and real-time PCR analysis was carried out as previously described (8) using specific primers. Actin was used as a normalization control. Antibodies used in the study were Sox2 (Cell Signaling, Danvers, MA), Cre (Chemicon, Temecula, CA) and γ-tubulin (Sigma, St. Louis, MO)
DNA Synthesis Assay
BrdU incorporation assay was carried out on cultured cells as previously described (8).
Colony formation assay with Sox2 shRNA
shRNAs against murine Sox2 were designed using the RNAi oligo retriever program (http://katahdin.cshl.org:9331/RNAi/html/rnai.html). For each target sequence, a U6 promoter/hairpin cassette was generated by a PCR-based strategy, and then cloned in the Bam HI/Eco RI sites of retroviral vector pBabe which carries resistance to hygromycin. The shRNAs used in this study are as follows: Sox2 1043 - AAA AAC ATA CCG GGA ATG CCC TGC TGC GAG TAG G, Sox2 900 - AAA AAC ATT GAG GCC CAG GTG CTG CGG GTA GCC C and Sox2 998 - AAA AAC AAA CCG TTC ATG TAG GTC TGC GAG CTG G.
Osteosphere Assay
Osteosphere assay was carried out as described in Gutierrez et al (28). Briefly, 1 × 105 cells were plated in triplicate in 6 cm Corning ultra-low attachment plates for 7 days. Osteospheres were collected by sieving through a 40 μM strainer and RNA was extracted from the spheres for gene exprerssion analysis. Spheres were counted in each plate under a dissecting microscope.
Supplementary Material
Acknowledgments
We wish to thank Cuauhtémoc Castillo, Tom Tao and Jeff Kraynak for technical assistance, Dr. Greg Holmes for help with the Sox2 cko mice, and Dr. Eva Hernando for helpful discussion. We also wish to thank Dr. Gerard Karsenty, Columbia University for the Collagen I-Cre mice and Drs. Dan Littman and Derya Unutmaz from NYU School of Medicine for the GFP/Cre-EGFP viruses and the Sox2 lentivirus respectively. This work was supported by PHS Grant AR0151358 from NIAMS.
References
- 1.Ornitz DM, Marie PJ. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002 Jun 15;16(12):1446–1465. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- 2.Muenke M, Schell U. Fibroblast-growth-factor receptor mutations in human skeletal disorders. Trends Genet. 1995 Aug;11(8):308–313. doi: 10.1016/s0168-9525(00)89088-5. [DOI] [PubMed] [Google Scholar]
- 3.Marie PJ, Coffin JD, Hurley MM. FGF and FGFR signaling in chondrodysplasias and craniosynostosis. J Cell Biochem. 2005 Dec 1;96(5):888–896. doi: 10.1002/jcb.20582. [DOI] [PubMed] [Google Scholar]
- 4.Tang KT, Capparelli C, Stein JL, Stein GS, Lian JB, Huber AC, et al. Acidic fibroblast growth factor inhibits osteoblast differentiation in vitro: altered expression of collagenase, cell growth-related, and mineralization-associated genes. J Cell Biochem. 1996 Apr;61(1):152–166. doi: 10.1002/(sici)1097-4644(19960401)61:1<152::aid-jcb16>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 5.Debiais F, Hott M, Graulet AM, Marie PJ. The effects of fibroblast growth factor-2 on human neonatal calvaria osteoblastic cells are differentiation stage specific. J Bone Miner Res. 1998 Apr;13(4):645–654. doi: 10.1359/jbmr.1998.13.4.645. [DOI] [PubMed] [Google Scholar]
- 6.Mansukhani A, Bellosta P, Sahni M, Basilico C. Signaling by fibroblast growth factors (FGF) and fibroblast growth factor receptor 2 (FGFR2)-activating mutations blocks mineralization and induces apoptosis in osteoblasts. J Cell Biol. 2000 Jun 12;149(6):1297–1308. doi: 10.1083/jcb.149.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalajzic I, Kalajzic Z, Hurley MM, Lichtler AC, Rowe DW. Stage specific inhibition of osteoblast lineage differentiation by FGF2 and noggin. J Cell Biochem. 2003 Apr 15;88(6):1168–1176. doi: 10.1002/jcb.10459. [DOI] [PubMed] [Google Scholar]
- 8.Mansukhani A, Ambrosetti D, Holmes G, Cornivelli L, Basilico C. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J Cell Biol. 2005 Mar 28;168(7):1065–1076. doi: 10.1083/jcb.200409182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambrosetti D, Holmes G, Mansukhani A, Basilico C. Fibroblast growth factor signaling uses multiple mechanisms to inhibit Wnt-induced transcription in osteoblasts. Mol Cell Biol. 2008 Aug;28(15):4759–4771. doi: 10.1128/MCB.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003 Jan 1;17(1):126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lefebvre V, Dumitriu B, Penzo-Mendez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39(12):2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niwa H. How is pluripotency determined and maintained? Development. 2007 Feb;134(4):635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 13.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995 Nov 1;9(21):2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006 Aug 25;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997 May 30;89(5):755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 16.Ducy P, Schinke T, Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000 Sep 1;289(5484):1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 17.Aubin J, Triffit J. Mesenchymal Cells and Osteoblast Differentiation. In: Bilezikian J, Raisz LG, Rodan GA, editors. Principles of Bone Biology. Vol. 1. Academic Press; New York: 2002. pp. 59–82. [Google Scholar]
- 18.Nakashima K, Zhou X, Kunkel G, Zhang ZP, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor Osterix is required for osteoblast differentiation and bone formation. Cell. 2002 Jan 11;108(1):17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 19.Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, et al. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006 Jun;7(1-2):1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- 20.Favaro R, Valotta M, Ferri AL, Latorre E, Mariani J, Giachino C, et al. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci. 2009 Oct;12(10):1248–1256. doi: 10.1038/nn.2397. [DOI] [PubMed] [Google Scholar]
- 21.Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002 Jun;224(2):245–251. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- 22.Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007 Jul;134(13):2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn M, Vogel M, Pompesius-Kempa M, Delling G. Trabecular bone pattern factor--a new parameter for simple quantification of bone microarchitecture. Bone. 1992;13(4):327–330. doi: 10.1016/8756-3282(92)90078-b. [DOI] [PubMed] [Google Scholar]
- 24.Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci U S A. 2001 Jul 31;98(16):9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003 Jun 13;113(6):703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 26.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995 Sep 26;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raucci A, Bellosta P, Grassi R, Basilico C, Mansukhani A. Osteoblast proliferation or differentiation is regulated by relative strengths of opposing signaling pathways. J Cell Physiol. 2008 May;215(2):442–451. doi: 10.1002/jcp.21323. [DOI] [PubMed] [Google Scholar]
- 28.Gutierrez GM, Kong E, Sabbagh Y, Brown NE, Lee JS, Demay MB, et al. Impaired bone development and increased mesenchymal progenitor cells in calvaria of RB1-/-mice. Proc Natl Acad Sci U S A. 2008 Nov 25;105(47):18402–18407. doi: 10.1073/pnas.0805925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coussens AK, Hughes IP, Wilkinson CR, Morris CP, Anderson PJ, Powell BC, et al. Identification of genes differentially expressed by prematurely fused human sutures using a novel in vivo - in vitro approach. Differentiation. 2008 May;76(5):531–545. doi: 10.1111/j.1432-0436.2007.00244.x. [DOI] [PubMed] [Google Scholar]
- 30.Liu F, Woitge HW, Braut A, Kronenberg MS, Lichtler AC, Mina M, et al. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 2004 Sep;48(7):645–653. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- 31.Dacic S, Kalajzic I, Visnjic D, Lichtler AC, Rowe DW. Col1a1-driven transgenic markers of osteoblast lineage progression. J Bone Miner Res. 2001 Jul;16(7):1228–1236. doi: 10.1359/jbmr.2001.16.7.1228. [DOI] [PubMed] [Google Scholar]
- 32.Nissen-Meyer LSH, Jemtland R, Gautvik VT, Pedersen ME, Paro R, Fortunati D, et al. Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. Journal of Cell Science. 2007 Aug 15;120(16):2785–2795. doi: 10.1242/jcs.003855. [DOI] [PubMed] [Google Scholar]
- 33.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003 Aug 28;39(5):749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 34.Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, et al. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006 May 1;20(9):1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaidi SK, Pande S, Pratap J, Gaur T, Grigoriu S, Ali SA, et al. Runx2 deficiency and defective subnuclear targeting bypass senescence to promote immortalization and tumorigenic potential. Proc Natl Acad Sci U S A. 2007 Dec 11;104(50):19861–19866. doi: 10.1073/pnas.0709650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muraglia A, Perera M, Verardo S, Liu Y, Cancedda R, Quarto R, et al. DLX5 overexpression impairs osteogenic differentiation of human bone marrow stromal cells. Eur J Cell Biol. 2008 Oct;87(10):751–761. doi: 10.1016/j.ejcb.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Go MJ, Takenaka C, Ohgushi H. Forced expression of Sox2 or Nanog in human bone marrow derived mesenchymal stem cells maintains their expansion and differentiation capabilities. Exp Cell Res. 2008 Mar 10;314(5):1147–1154. doi: 10.1016/j.yexcr.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura T, Hanada K, Tamura M, Shibanushi T, Nigi H, Tagawa M, et al. Stimulation of endosteal bone formation by systemic injections of recombinant basic fibroblast growth factor in rats. Endocrinology. 1995 Mar;136(3):1276–1284. doi: 10.1210/endo.136.3.7867582. [DOI] [PubMed] [Google Scholar]
- 39.Fromigue O, Modrowski D, Marie PJ. Growth factors and bone formation in osteoporosis: roles for fibroblast growth factor and transforming growth factor beta. Curr Pharm Des. 2004;10(21):2593–2603. doi: 10.2174/1381612043383773. [DOI] [PubMed] [Google Scholar]
- 40.Dunstan CR, Boyce R, Boyce BF, Garrett IR, Izbicka E, Burgess WH, et al. Systemic administration of acidic fibroblast growth factor (FGF-1) prevents bone loss and increases new bone formation in ovariectomized rats. J Bone Miner Res. 1999 Jun;14(6):953–959. doi: 10.1359/jbmr.1999.14.6.953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.