Abstract
Studies of cellular and tissue dynamics benefit greatly from tools that can control protein activity with specificity and precise timing in living systems. We describe here a new approach to confer allosteric regulation specifically on the catalytic activity of kinases. A highly conserved portion of the kinase catalytic domain is modified with a small protein insert that inactivates catalytic activity, but does not affect other protein interactions. Catalytic activity is restored by addition of rapamycin or non-immunosuppresive analogs (Fig. 1A). We demonstrate the approach by specifically activating focal adhesion kinase (FAK) within minutes in living cells, thereby demonstrating a novel role for FAK in regulation of membrane dynamics. Molecular modeling and mutagenesis indicate that the protein insert reduces activity by increasing the flexibility of the catalytic domain. Drug binding restores activity by increasing rigidity. Successful regulation of Src and p38 suggest that modification of this highly conserved site will be applicable to other kinases.
Recent novel methods for regulation of kinases with precise timing in living cells include induced dimerization, subcellular localization, proteolytic degradation or chemical rescue from an inactivating mutation1–4. Engineered allosteric regulation as well shows great promise for precise control of protein activity 5–7. Nonetheless, important challenges remain in that existing methods are limited to specific targets, inactivate rather than activate kinases, and/or do not enable regulation of a particular domain within the target. We describe here a new method to activate specifically the catalytic domain within a multidomain protein kinase, using FAK as a model. FAK has been implicated in a wide variety of cell behaviors, including proliferation, apoptosis, migration and tumorigenesis8–11. It is a multidomain protein that functions as both a scaffold and a kinase11 and relatively little is known about the specific role of its catalytic activity. It therefore served as a good test of the new method, which enabled us to specifically dissect the function of FAK kinase activity without affecting scaffolding functions, controlling it with a temporal resolution of 1–2 minutes.
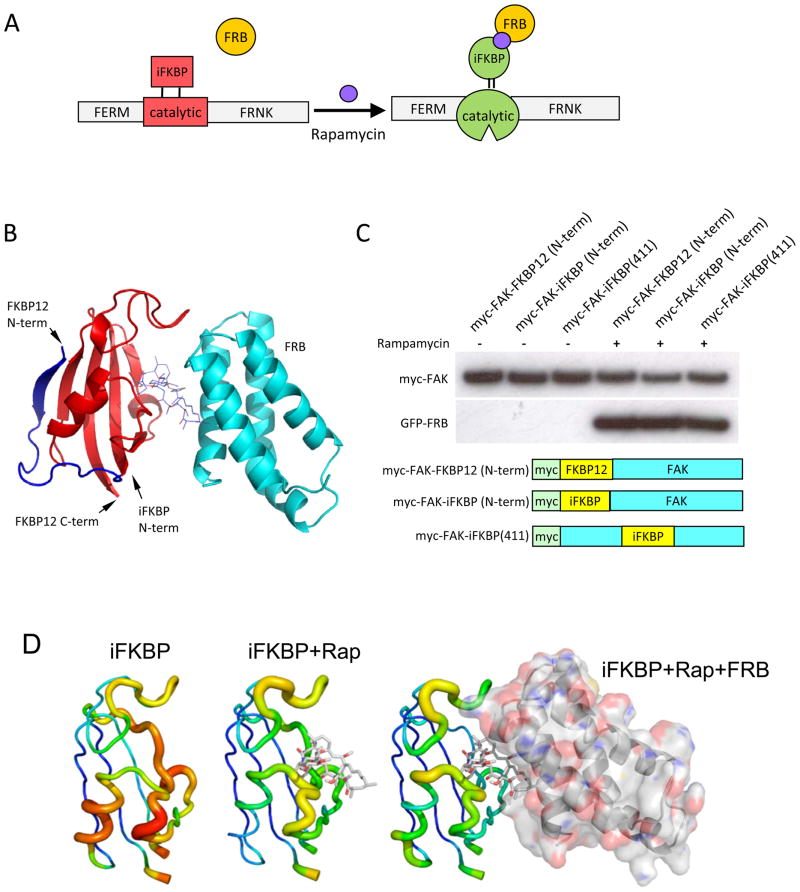
To allosterically regulate FAK’s catalytic activity we used a portion of the small protein FKBP12; a previous study showed that ligand binding to FKBP12 greatly increased its conformational rigidity 12, suggesting that insertion of FKBP12 near the catalytic site of kinases could be used to control the conformational mobility of the kinase active site. It was, however, unclear that FKBP12 could be inserted into the middle of the kinase sequence without severely disrupting kinase structure or FKBP12 binding interactions. We therefore experimented with truncated forms of FKBP12, leading to an FKBP12 derivative named iFKBP (insertable FKBP, Fig. 1B). In iFKBP, the N and C termini are positioned near one another for minimal perturbation of kinase secondary structure (Fig 1B). Co-immunoprecipitation experiments showed that iFKBP interacts with rapamycin and FKBP12-Rapamycin Binding domain (FRB) as efficiently as does wild type FKBP12, even when inserted in the middle of the FAK molecule (Fig. 1C, Supplementary Fig. S1). Molecular dynamics studies of iFKBP indicated that its conformational fluctuation is reduced by interaction with rapamycin or by rapamycin-induced heterodimerization with FRB (Fig. 1D and Supplementary Fig. S2). Changes in conformational fluctuations were especially pronounced at the N and C termini where iFKBP would be linked to FAK, suggesting that the effects of rapamycin/FRB binding could be communicated to FAK.
Fig. 1.
Design and generation of RapR-FAK. (A) Schematic representation of the approach used to regulate the catalytic activity of FAK. A fragment of FKBP is inserted at a position in the catalytic domain where it abrogates catalytic activity. Binding to rapamycin and FRB restores activity. (B) The truncated fragment of human FKBP12 (amino acids Thr22 through Glu108) inserted into the kinase domain. Blue and red, full length FKBP12; red, proposed structure of the inserted fragment. The FKBP12 is shown in complex with rapamycin and FRB (cyan). (C)Immunoblot analysis of iFKBP interaction with rapamycin and FRB. Myc-tagged FKBP12 and iFKBP constructs were immunoprecipitated from cells treated for 1 hour with either 200 nMrapamycin or ethanol (solvent control). Co-immunoprecipitation of co-expressed GFP-FRB was detected using anti-GFP antibody. (D) Changes in the molecular dynamics of iFKBP upon binding to rapamycin and FRB. Warmer colors and thicker backbone indicate increasing root mean square fluctuation (RMSF).
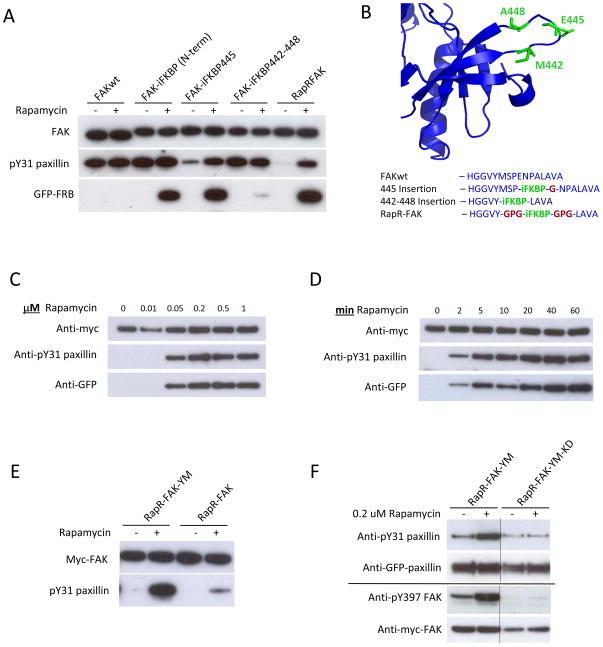
Optimization of the insertion site and the linkers connecting iFKBP to FAK led to a version of FAK that was susceptible to regulation by rapamycin-induced FRB binding. With insertion of iFKBP at Glu445 (FAK-iFKBP445 construct), FAK catalytic activity was dramatically reduced. Rapamycin-induced binding to FRB restored activity (Fig. 2A). Treatment with rapamycin/FRB did not affect the activity of wild-type FAK (FAKwt) or a construct with iFKBP attached to the FAK N-terminus, demonstrating that regulation of catalytic activity is dependent on specific placement of the insert in the catalytic subunit. To optimize regulation of FAK by rapamycin, several modifications were introduced into the regions where iFKBP was connected to FAK. iFKBP was positioned within the FAK loop Met442-Ala448, between two β-strands in the N-terminal lobe of the FAK catalytic domain (Fig. 2B). Replacing FAK residues Met442-Ala448 with iFKBP, using no linkers, negated the effect of iFKBP on FAK activity and dramatically reduced interaction with rapamycin and FRB (Figs. 2A and 2B, construct FAK-iFKBP 442–448). Computational analysis revealed that the construct without linkers is locked in a distorted conformation that prevents ligand binding (Supplementary Fig. S3). In contrast, introduction of short linkers to connect iFKBP with the β-strands of the FAK catalytic domain led to the optimized structure used henceforth (named RapR-FAK, for Rapamycin-regulated FAK). In RapR-FAK, activity in the absence of rapamycin was significantly lower than that of FAK-iFKBP445 (Fig. 2A). Rapmycin-induced FRB binding restored activity to near wild type level. Activation of RapR-FAK catalytic activity was achieved in living cells within 2 minutes and with 50 nM rapamycin(Fig. 2C and D). Activation was also achieved by treatment with rapamycin alone, without co-expression of FRB (Supplementary Fig. S4). However, this required significantly higher concentrations of rapamycin (up to 4 μM), so the remaining studies described here were carried out using rapamycin-induced FRB binding. Computational analysis indicated that rapamycin alone does not stabilize iFKBP to the same extent as rapmycin/FRB (Fig. 1D and Supplementary Fig. S2).
Fig. 2.
Development and biochemical characterization of RapR-FAK. (A) Rapamycin regulation of FAK variants with iFKBP inserted at different positions. HEK293T cells co-expressing myc-tagged FAK constructs and GFP-FRB were treated for one hour with either 200 nMrapamycin or ethanol (solvent control). The activity of immunoprecipitated FAK variants was tested using the N-terminal fragment of paxillin as a substrate. (B) Sites of iFKBP insertion (green) and connecting linkers (red). (C, D) HEK293T cells co-expressing RapR-FAK and FRB were treated with the indicated amount of rapamycin for 1 hour or with 200 nMrapamycin for the indicated period of time. The kinase was immunoprecipitated and its activity tested as described above. (E) FAK Y180A and M183A mutations were introduced to eliminate autoinhibitory interactions, thereby generating RapR-FAK-YM, which was tested as in A. (F) HEK293T cells co-expressing Cherry-FRB, GFP-paxillin and either myc-tagged RapR-FAK-YM or its kinase-inactive mutant (RapR-FAK-YM-KD) were treated with rapamycin or ethanol (solvent control) for 1 hour. GFP-paxillin was immunoprecipitated and its phosphorylation was assessed using anti-phospho-Tyr31 antibody. Autophosphorylation of FAK on Tyr397 was analyzed using total cell lysate.
iFKBP-mediated FAK regulation was designed to specifically control catalytic activity without perturbing other FAK functions. Thus, it was important to test the effects of iFKBP insertion on normal FAK binding interactions and FAK regulation. FAK catalytic activity is regulated by an autoinhibitory interaction between the N-terminal FERM domain and the catalytic domain 13. Two amino acids known to be involved in this interaction were mutated to alanines (Y180A and M183A, previously described 13) to test if RapR-FAK remains regulated by autoinhibition. When activated by rapamycin, the Y180A/M183A construct (RapR-FAK-YM) demonstrates significantly higher activity than RapR-FAK (Fig. 2E), consistent with published results for constitutively active FAK 13 and demonstrating that RapR-FAK is still regulated by autoinhibition. Importantly, RapR-FAK-YM is regulated solely by rapamycin, and not by endogenous mechanisms. To confirm that RapR-FAK phosphorylates substrates in a rapamycin-dependent manner in cells, phosphorylation of two known FAK substrates was tested before and after addition of rapamycin. Upon activation of RapR-FAK-YM, phosphorylation of paxillin on residue Tyr31 and autophosphorylation of FAK on residue Tyr397 are significantly increased (Fig. 2F). A control construct lacking catalytic activity (RapR-FAK-YM-KD, with additional mutation D546R) failed to demonstrate any change in phosphorylation. RapR-FAK and wtFAK showed similar binding to paxillin and Src in co-immunoprecipitation assays (Supplementary Fig. S5), indicating that introduction of iFKBP into the catalytic domain of FAK does not affect interaction with binding partners. Also, iFKBP insertion did not perturb the intracellular distribution of RapR-FAK; its localization was identical to that of wild-type FAK (Supplementary Fig. S6). Activation of RapR-FAK was accompanied by translocation of fluorescently labeled FRB into focal adhesions and co-localization with fluorescent RapR-FAK (Supplementary Fig. S7). The translocation of fluorescent FRB into adhesions served as a useful marker of FAK activation in live cells. Overall we conclude that RapR-FAK enables robust and specific activation of FAK catalytic activity in living cells without perturbation of other FAK properties.
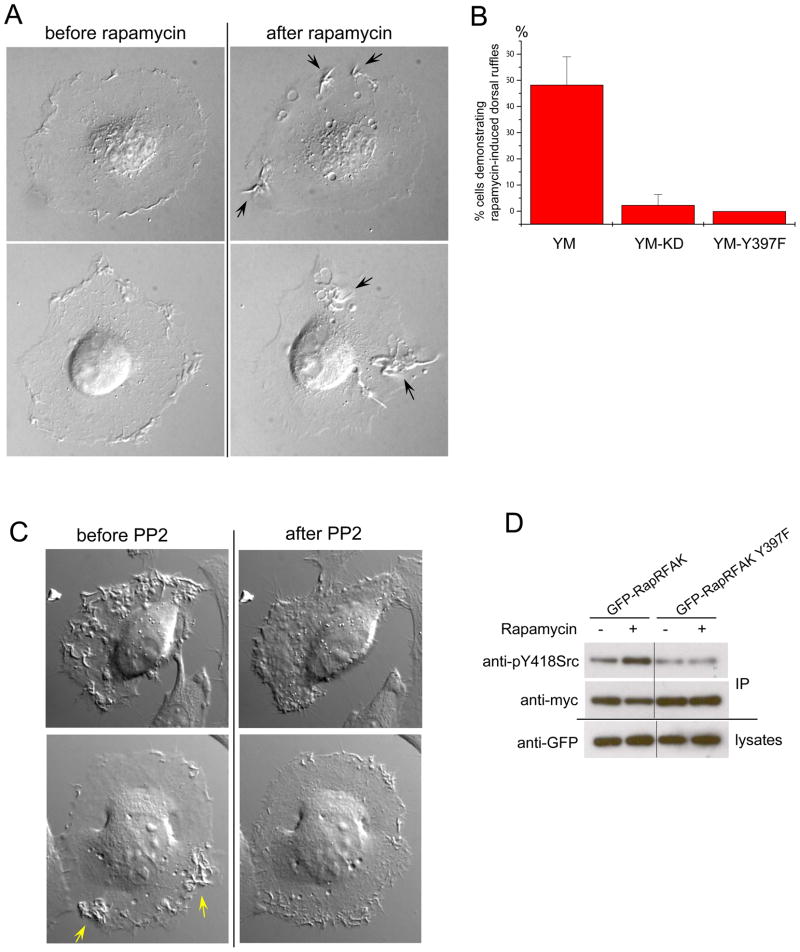
FAK is known to be overexpressed and activated in human tumors 14–16, but the specific role of its catalytic activity remains unclear. To identify processes affected specifically by FAK catalytic activity, we examined the activation of RapR-FAK-YM in HeLa cells. The Y180A/M183A mutant was used to ensure the regulation of RapR-FAK by rapamycin only, and exclude modulation by endogenous upstream factors. Consistent with previous reports showing that catalytic activity is not required for FAK’s role in growth factor-stimulated motility17, activation of RapR-FAK-YM did not significantly affect cell movement (Supplementary Fig 8). However, we did observe a distinct effect on membrane dynamics. HeLa cells normally show small peripheral ruffles that remain near the cell border. Upon addition of rapamycin, the extent of ruffling greatly increased, and very large and dynamic ruffles appeared across the dorsal surface (Fig. 3A, B, Supplementary movie S1, 36/64 analyzed cells). In control studies, cells expressing similar levels of catalytically inactive RapR-FAK-YM-KD showed no change in normal ruffling activity (Fig. 3B, 34/35 analyzed cells). RapR-FAK was localized within these ruffles (Supplementary Fig. S9). Importantly, wild-type FAK was also detected in ruffles stimulated by RapR-FAK and in those produced by PDGF treatment (Supplementary Fig. S10, S11, S12), indicating that dorsal ruffles are not an artifact of RapR-FAK mislocalization. Furthermore, FAK-null fibroblasts failed to produce PDGF-stimulated dorsal ruffles (158 cells analyzed), whereas 50% of control fibroblasts expressing FAK (59 out of 118 analyzed) exhibited distinct dorsal ruffling under the same stimulation conditions. These data implicate FAK catalytic activity in the regulation of dorsal membrane protrusions.
Fig. 3.
Activation of FAK catalytic activity initiates large dorsal ruffles via the activation of Src. (A)Rapamycin treatment of HeLa cells co-expressing RapR-FAK-YM and FRB caused formation of large dorsal ruffles. (B) HeLa cells expressing either GFP-RapR-FAK-YM (YM, 64 cells), GFP-RapR-FAK kinase-dead mutant (YM-KD, 35 cells) or GFP-tagged Y397F mutant (YM-Y397F, 47 cells) were scored for ruffle induction by rapamycin. No dorsal ruffles were seen before rapamycin addition. (C) Inhibition of Src family kinases eliminated the FAK-induced ruffles. Cells co-expressing GFP-RapR-FAK-YM and Cherry-FRB were treated with rapamycin for 1 hour and imaged before and after addition of Src family kinase inhibitor PP2. PP2 addition stopped dorsal protrusion in all cells analyzed (16 cells). (D) Activation of FAK leads to activation of Src. HeLa cells co-expressing myc-tagged Src, Cherry-FRB and either GFP-RapR-FAK-YM or its Y397F mutant were treated with rapamycin for 1 hour. Src was immunoprecipitated using anti-myc antibody and its phosphorylation on Tyr418 was assessed by immunoblotting.
Published work has demonstrated that FAK autophosphorylation of Tyr397 plays an important role in FAK-mediated signaling, and that Tyr397 phosphorylation level correlates with FAK activation18. Because autophosphorylation of FAK on Tyr397 creates a binding site for Src family kinases 18, 19 it has been proposed that interaction of FAK with Src leads to Src activation 18. Furthermore, Src is involved in the formation of dorsal protrusions stimulated by platelet derived growth factor (PDGF) 20. Together these observations led us to hypothesize that the FAK-stimulated formation of dorsal protrusions occurs via activation of Src. In our studies, mutation of Tyr397 to phenylalanine in RapR-FAK completely abolished the formation of dorsal protrusions (Fig. 3B). To test the potential role of Src, cells were treated with PP2, an inhibitor of Src family kinases, after stimulation of RapR-FAK-YM. This abrogated the FAK-induced dorsal ruffling (Fig 3C, Supplementary movie S2). In contrast, control compound PP3, an inactive PP2 stereoisomer, or imatinib, an inhibitor of Abl kinase, had no effect (data not shown). Phosphorylation of Src Tyr418 (Tyr416 in avian Src) is known to occur upon Src activation 21, 22. Rapamycin addition to cells transfected with RapR-FAK-YM led to increased Src Tyr418 phosphorylation, while cells expressing RapR-FAK-YM with an additional mutation that abolishes Src binding (Y397F mutation) showed no effect (Fig. 3D). Together, these data directly demonstrate that FAK catalytic activity stimulates Src, and that this in turn leads to dorsal protrusions. Dorsal protrusions play an important role in the invasive migration of cells into extracellular matrix 23 and enhanced FAK expression in tumor cells is associated with cell invasiveness. Our data suggests a mechanism whereby FAK overexpression contributes to cancer.
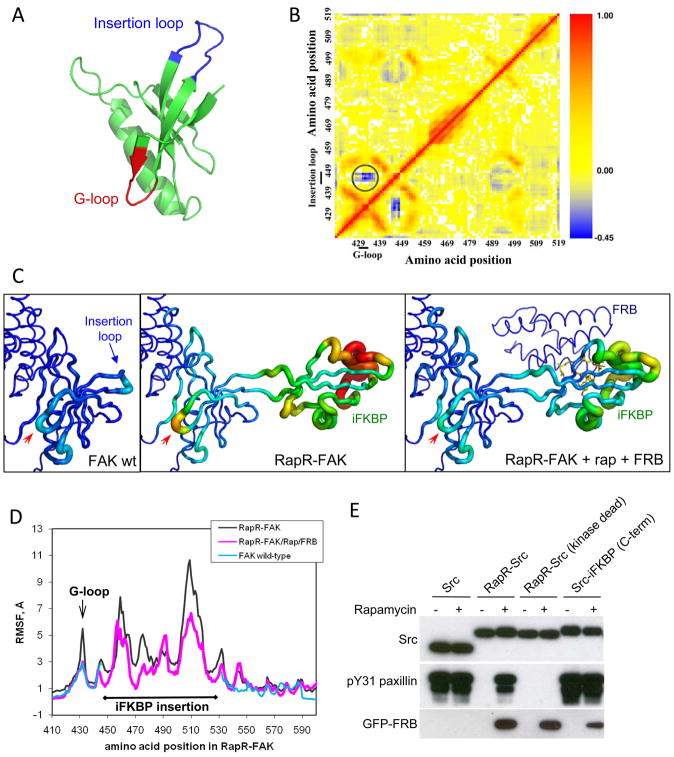
In order to understand the molecular mechanism of RapR-FAK allosteric regulation and explore the generalizability of the approach, we performed molecular dynamics simulations24, 25. Combined with the biochemical data, the computational analysis indicated a mechanism for iFKBP-mediated regulation. The point where iFKBP is inserted connects via a β strand to FAK’s Gly loop (G-loop), a structural feature critical for positioning the ATP phosphate groups in the catalytic site (Fig 4A) 26. Molecular dynamics analysis indicated that the conformational mobility of the G-loop is correlated with that of the FAK region where iFKBP is inserted (the ‘insertion loop’, Fig. 4B), suggesting that the dynamics of the insertion loop could affect the dynamics of the G-loop and thereby change the catalytic activity. Comparison of wt FAK and RapR-FAK dynamics indicated that the amplitude of G-loop conformational dynamics is dramatically increased when iFKBP is inserted in the catalytic domain. These dynamics decreased back to wild-type levels upon binding to rapamycin/FRB (Fig. 4C, D, supplementary movie S3). Based on this analysis, we postulate that the effectiveness of the G-loop in the phosphate transfer reaction is reduced due to greater conformational flexibility produced by insertion of iFKBP. Interaction with rapamycin and FRB stabilizes the G loop to rescue FAK catalytic activity. Molecular dynamics analysis was consistent with empirical measurements; dynamics analysis of the FAK-iFKBP445 variant suggested that its longer linkers decreased coupling between the iFKBP insert and G-loop dynamics (Supplementary Fig. S13), resulting in the less effective FAK inhibition observed in biochemical studies (Fig. 2A, FAK-iFKBP445 construct). In contrast, insertion of iFKBP without any linkers restricted the structural dynamics of iFKBP, consistent with the observed minimal effects on catalytic activity (Supplementary Fig. S13, FAK-iFKBP442-8 construct). In summary, computational analysis indicates that the allosteric modulation of RapR-FAK activity results from dynamic coupling of the optimized iFKBP insertion and the kinase G-loop, highly conserved structural features in all known kinases 26.
Fig. 4.
Mechanism of regulation by iFKBP; Src regulation. (A) The portion of the FAK catalytic domain targeted for insertion of iFKBP (blue) and the G-loop (red). (B) Dynamic correlation analysis of the wild type FAK catalytic domain (red, positive correlation; blue, negative correlation). The circled region indicates strong negative correlation between the movement of the insertion loop and the G-loop. (C) Tube representation depicting changes in the dynamics of the FAK catalytic domain’s N-terminal lobe, based on molecular dynamics simulations. Warmer colors and thicker backbone correspond to higher RMSF values, reflecting the degree of free movement within the structure. The red arrows points to the G-loop. (D) Root mean square fluctuation (RMSF) of amino acids in FAK and RapR-FAK(arrow indicates G-loop). The break in the wild type FAK graph corresponds to the iFKBP insert in RapR-FAK. (E) Regulation of Src kinase by insertion of iFKBP. HEK293T cells co-expressing the indicated myc-tagged Src construct and GFP-FRB were treated with either 200 nMrapamycin or ethanol solvent control. The kinase activity of immunoprecipitatedSrc was tested as in 2A.
Because the mechanism of allosteric regulation is based on coupling of highly conserved structural elements, the RapR approach may well be applicable to other kinases. We explored this by inserting iFKBP into a tyrosine kinase, Src, and into a serine/threonine kinase, p38, at a site analogous to that used in FAK (Gly288 in Src, Lys45 in p38α) (Supplementary Fig. S14). In both Src and p38, insertion of iFKBP strongly inhibited activity, and activity was rescued by interaction with rapamycin/FRB (Fig. 4E, Supplementary Fig. S15). Treatment with rapamycin did not affect wild-type Src or control Src constructs in which iFKBP was added to the C-terminus, nor did it have any significant effect on wild-type p38. In molecular dynamics simulations, Src showed the same coupling between iFKBP and the Gly-loop that was observed for FAK (Supplementary Fig. S16, S17). These data suggest that the iFKBP cassette can be used for allosteric regulation of a wide variety of both tyrosine and serine/threonine kinases.
Although we saw no effects of rapamycin in the absence of RapR kinases, we were concerned that some potential studies could be complicated by the known immunosuppressive effects of rapamycin. We therefore tested the ability to regulate RapR kinases using known non-immunosuppresive analogs of Rapamycin, iRap and AP21967. Both compounds regulated RapR-FAK activity at concentrations comparable to those reported previously for dimerization of proteins in living cells 27 (Supplementary Fig. S18). Importantly, AP21967 and a similar analog of rapamycin (C20-MaRap) have been successfully used for experiments in animals 28, 29, indicating that the RapR method can be applied in live animal studies. The F36V mutant of FKBP, which interacts tightly with the Shield 1 compound 4, could potentially eliminate the requirement for FRB and minimize effects on endogenous FKBP12 function.
In summary, we describe a protein modification to confer rapamycin sensitivity specifically on the catalytic activity of kinases. The approach is based upon highly conserved regions in both serine/threonine and tyrosine kinases, promising broad applicability. It requires only addition of a small protein insert into the conserved portion of the catalytic subunit, and can be used with non-immunosuppresive rapamycin analogs suitable for in vivo studies. The approach combines the temporal resolution of small molecule inhibitors with the absolute specificity of genetic approaches, and enables allosteric regulation of a single domain in a multidomain protein. A mechanistic model based on molecular dynamics, and application to analogous sites in FAK, Src and p38α, indicate that rapamycin exerts its effect by modulating the conformational flexibility of the conserved catalytic subunit. By selectively activating FAK catalytic activity in living cells, we directly demonstrated that FAK catalysis activates Src to trigger large dorsal protrusions, a potential mechanism explaining how overexpression and activation of FAK contributes to tumor progression.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturebiotechnology/.
Supplementary Material
Acknowledgments
We thank Jonathan Edwards, Daniel Dominguez and Vishal Rao for help with construction and testing of RapR-Src and RapR-p38α constructs, Betsy Clarke for her design of figures, and are grateful to the UNC Cancer Research Fund and the National Institutes of Health for funding (GM64346 and GM057464 to KMH; GM080742 and GM080742-03S1 to N.V.D).
Footnotes
Supplementary Information is linked to the online version of the paper at http://www.nature.com/naturebiotechnology/.
Author Contributions A.V.K initiated the project, developed and validated regulation of RapR-kinases, and performed the studies of FAK biological function. F.D. performed molecular modeling of FKBP12 variants, RapR-FAK and RapR-Src. P.K. performed biochemical characterization of RapR-p38. N.V.D. coordinated molecular dynamics studies. K.M.H. coordinated the study and wrote the final version of the manuscript, based on contributions from all authors.
Author Information Reprints and permissions information is available at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–24. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 2.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 3.Qiao Y, Molina H, Pandey A, Zhang J, Cole PA. Chemical rescue of a mutant enzyme in living cells. Science. 2006;311:1293–7. doi: 10.1126/science.1122224. [DOI] [PubMed] [Google Scholar]
- 4.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tucker CL, Fields S. A yeast sensor of ligand binding. Nat Biotechnol. 2001;19:1042–6. doi: 10.1038/nbt1101-1042. [DOI] [PubMed] [Google Scholar]
- 6.Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc Natl Acad Sci U S A. 2005;102:11224–9. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radley TL, Markowska AI, Bettinger BT, Ha JH, Loh SN. Allosteric switching by mutually exclusive folding of protein domains. J Mol Biol. 2003;332:529–36. doi: 10.1016/s0022-2836(03)00925-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev. 2009;28:35–49. doi: 10.1007/s10555-008-9165-4. [DOI] [PubMed] [Google Scholar]
- 9.Gabarra-Niecko V, Schaller MD, Dunty JM. FAK regulates biological processes important for the pathogenesis of cancer. Cancer Metastasis Rev. 2003;22:359–74. doi: 10.1023/a:1023725029589. [DOI] [PubMed] [Google Scholar]
- 10.Tilghman RW, Parsons JT. Focal adhesion kinase as a regulator of cell tension in the progression of cancer. Semin Cancer Biol. 2008;18:45–52. doi: 10.1016/j.semcancer.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlaepfer DD, Mitra SK, Ilic D. Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim Biophys Acta. 2004;1692:77–102. doi: 10.1016/j.bbamcr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Marquis-Omer D, et al. Stabilization of the FK506 binding protein by ligand binding. Biochem Biophys Res Commun. 1991;179:741–8. doi: 10.1016/0006-291x(91)91879-h. [DOI] [PubMed] [Google Scholar]
- 13.Lietha D, et al. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 2007;129:1177–87. doi: 10.1016/j.cell.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golubovskaya VM, Kweh FA, Cance WG. Focal adhesion kinase and cancer. Histol Histopathol. 2009;24:503–10. doi: 10.14670/HH-24.503. [DOI] [PubMed] [Google Scholar]
- 15.Chatzizacharias NA, Kouraklis GP, Theocharis SE. Clinical significance of FAK expression in human neoplasia. Histol Histopathol. 2008;23:629–50. doi: 10.14670/HH-23.629. [DOI] [PubMed] [Google Scholar]
- 16.Sood AK, et al. Biological significance of focal adhesion kinase in ovarian cancer: role in migration and invasion. Am J Pathol. 2004;165:1087–95. doi: 10.1016/S0002-9440(10)63370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sieg DJ, et al. FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol. 2000;2:249–56. doi: 10.1038/35010517. [DOI] [PubMed] [Google Scholar]
- 18.Schaller MD, et al. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol Cell Biol. 1994;14:1680–8. doi: 10.1128/mcb.14.3.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing Z, et al. Direct interaction of v-Src with the focal adhesion kinase mediated by the Src SH2 domain. Mol Biol Cell. 1994;5:413–21. doi: 10.1091/mbc.5.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veracini L, et al. Two distinct pools of Src family tyrosine kinases regulate PDGF-induced DNA synthesis and actin dorsal ruffles. J Cell Sci. 2006;119:2921–34. doi: 10.1242/jcs.03015. [DOI] [PubMed] [Google Scholar]
- 21.Smart JE, et al. Characterization of sites for tyrosine phosphorylation in the transforming protein of Rous sarcoma virus (pp60v-src) and its normal cellular homologue (pp60c-src) Proc Natl Acad Sci U S A. 1981;78:6013–7. doi: 10.1073/pnas.78.10.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Playford MP, Schaller MD. The interplay between Src and integrins in normal and tumor biology. Oncogene. 2004;23:7928–46. doi: 10.1038/sj.onc.1208080. [DOI] [PubMed] [Google Scholar]
- 23.Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell. 2003;5:595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- 24.Ding F, Dokholyan NV. Dynamical roles of metal ions and the disulfide bond in Cu, Zn superoxide dismutase folding and aggregation. Proc Natl Acad Sci U S A. 2008;105:19696–701. doi: 10.1073/pnas.0803266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding F, Tsao D, Nie H, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–8. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krupa A, Preethi G, Srinivasan N. Structural modes of stabilization of permissive phosphorylation sites in protein kinases: distinct strategies in Ser/Thr and Tyr kinases. J Mol Biol. 2004;339:1025–39. doi: 10.1016/j.jmb.2004.04.043. [DOI] [PubMed] [Google Scholar]
- 27.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–8. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stankunas K, et al. Conditional protein alleles using knockin mice and a chemical inducer of dimerization. Mol Cell. 2003;12:1615–24. doi: 10.1016/s1097-2765(03)00491-x. [DOI] [PubMed] [Google Scholar]
- 29.Vogel R, Mammeri H, Mallet J. Lentiviral vectors mediate nonimmunosuppressive rapamycin analog-induced production of secreted therapeutic factors in the brain: regulation at the level of transcription and exocytosis. Hum Gene Ther. 2008;19:167–78. doi: 10.1089/hum.2007.125. [DOI] [PubMed] [Google Scholar]
- 30.Cai X, et al. Spatial and temporal regulation of focal adhesion kinase activity in living cells. Mol Cell Biol. 2008;28:201–14. doi: 10.1128/MCB.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerwins P, Blank JL, Johnson GL. Cloning of a novel mitogen-activated protein kinase kinase kinase, MEKK4, that selectively regulates the c-Jun amino terminal kinase pathway. J Biol Chem. 1997;272:8288–95. doi: 10.1074/jbc.272.13.8288. [DOI] [PubMed] [Google Scholar]
- 32.Sharma S, Ding F, Dokholyan NV. Multiscale modeling of nucleosome dynamics. Biophys J. 2007;92:1457–70. doi: 10.1529/biophysj.106.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teotico DG, et al. Active nuclear receptors exhibit highly correlated AF-2 domain motions. PLoS Comput Biol. 2008;4:e1000111. doi: 10.1371/journal.pcbi.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.