Abstract
Helicobacter pylori causes gastric ulcer diseases and gastric adenocarcinoma in humans. Not much is known regarding DNA replication in H.pylori that is important for cell survival. Here we report the cloning, expression and characterization of H.pylori DnaB (HpDnaB) helicase both in vitro and in vivo. Among the DnaB homologs, only Escherichia coli DnaB has been studied extensively. HpDnaB showed strong 5′ to 3′ helicase and ATPase activity. Interestingly, H.pylori does not have an obvious DnaC homolog which is essential for DnaB loading on the E.coli chromosomal DNA replication origin (oriC). However, HpDnaB can functionally complement the E.coli DnaB temperature-sensitive mutant at the non-permissive temperature, confirming that HpDnaB is a true replicative helicase. Escherichia coli DnaC co-eluted in the same fraction with HpDnaB following gel filtration analysis suggesting that these proteins might physically interact with each other. It is possible that a functional DnaC homolog is present in H.pylori. The complete characterization of H.pylori DnaB helicase will also help the comparative analysis of DnaB helicases among bacteria.
INTRODUCTION
Helicobacter pylori, a Gram-negative, spiral bacterial pathogen is considered to be the causative agent for the induction of chronic gastritis, gastroduodenal ulcer diseases, gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (1–4). Approximately half of the world’s population is infected with H.pylori. The bacterium was first isolated and cultured from gastric biopsy samples by Marshall and Warren in 1984 (5,6). Although a lot of effort has been made to understand the major virulence factors secreted by the bacteria and the underlying mechanisms which cause the disease, the basic biology of the bacteria is not completely understood.
DNA replication is a fundamental process that takes care of the faithful duplication of the genome in each cell division cycle. Central to this process are the events that take place at the replication origin. In bacteria, the initiator protein DnaA binds specifically to the DnaA boxes. The Escherichia coli oriC region contains four DnaA boxes which allow binding of DnaA proteins leading to the unwinding of the adjacent AT-rich region. This is followed by the entry of a helicase complex (DnaB6–DnaC6) at the origin, with the concomitant loading of other proteins required to form the replisome (7,8).
Thorough in silico analysis of the genome sequence of two unrelated isolates of H.pylori strains 26695 and J99 suggests that the basic mechanism of chromosomal DNA replication may likely be similar to that found in other eubacteria, although experimental evidence to support this hypothesis is not available (9,10).
There are many dissimilarities when compared with the E.coli genome, like the absence of the recF gene, the presence of the dnaA gene ∼600 kb away from the dnaN–gyrB genes and most importantly the absence of the dnaC gene, which codes for DnaC protein, required to load DnaB helicase to the origin. DnaC is essential in E.coli. It is possible that a functional DnaC counterpart is present in H.pylori, though it is not very obvious from the sequence analysis. Alternatively, HpDnaB can independently be loaded on oriC without the presence of DnaC. Therefore, it is important to find out first whether HpDnaB is functionally a true DnaB homolog of E.coli DnaB.
Recently, H.pylori DnaA protein has been purified and characterized (11). HpDnaA contains four domains like other DnaA proteins. The C-terminal domain of HpDnaA has been shown to be responsible for DNA binding. The putative oriC region in H.pylori containing five DnaA binding sites has also been mapped to be located upstream of the dnaA gene (11).
DnaB is considered to be a true multifunctional enzyme since it interacts with a number of other proteins during replication of DNA. The enzymatic activities of DnaB includes, but is not limited to, helicase activity, ATP hydrolysis and DNA binding (8). Escherichia coli DnaB interacts with DnaG primase (12), DNA polymerase III holoenzyme (13) and the helicase loader DnaC (8). It is considered to be the major replicative helicase in most eubacteria. However, among these helicases only E.coli DnaB has been characterized extensively both in vitro and in vivo. Apart from E.coli, some of the bacterial species whose DnaB homologs have been reported so far include Thermus aquaticus (14), Bacillus stearothermophilus (15), Pseudo monas putida and Pseudomonas aeruginosa (16).
A putative E.coli DnaB homolog has been identified by in silico analysis of the H.pylori (strain 26695) genome database (9). The open reading frame (ORF) HP1362 has been found to be 32% identical and 57% similar to its E.coli counterpart suggesting that this could be a putative DnaB homolog in H.pylori. However, there is no experimental evidence to support this hypothesis.
In an effort to understand the replication initiation processes in H.pylori, we cloned, purified and characterized the putative DnaB homolog of H.pylori strain 26695 (ORF HP1362). Although H.pylori lacks an obvious DnaC homolog, we found that HpDnaB fulfils the characteristics of the E.coli DnaB both in vivo and in vitro. Unlike E.coli DnaB, HpDnaB can use UTP as energy source, Ca2+ as a co-factor and untailed substrate for the helicase activity.
MATERIALS AND METHODS
Bacterial strains
The E.coli strains and plasmids used in this work are listed in Table 1. Escherichia coli strains were grown in LB media (supplemented with 100 µg/ml ampicillin or 50 µg/ml kanamycin wherever needed) either at 37 or 30°C, as per the requirement.
Table 1. Bacterial strains and plasmids used in this work.
| Strain/plasmid | Genotype/relevant characteristics | Reference |
|---|---|---|
| Escherichia coli DH5α | supE44, hsdR17, recA1, endA1, gyrA96, thi-1, relA1 | Molecular cloning, Sambrook et al. (18) |
| DJ58 (E.coli dnaBts) | Temperature-sensitive mutant of E.coli | Gift from Dr Dhruba K. Chattoraj |
| pET28a | T7, his, kanR | Novagen |
| pET28a HpDnaB (wild-type and mutant) | pET28a derivative containing 1.5 kb of H.pylori DnaB | This work |
| pBR322 | tetR, ampR | Bolivar et al. (17) |
| BL21 (DE3) | F– ompT hsdSB (rB– mB) gal dcm (DE3) | Novagen |
| pdnaBC | Plasmid expressing E.coli DnaB and DnaC (wild type) | Gift from Dr Dhruba K. Chattoraj |
| Helicobacter pylori 26695 | ATCC™ 700392D |
DNA manipulations
Helicobacter pylori dnaB gene was amplified by polymerase chain reaction (PCR) using Pfu DNA polymerase (Stratagene) and H.pylori strain 26695 genomic DNA (obtained from ATCC™) as template with the following primers: 5′-GCGGATCCATGGATCATTTAAAGCATTTGC-3′ and 5′-GCGGATCCTCAAGTTGTAACTATATCATAA-3′.
The PCR-amplified DNA (1.5 kb) was cloned in expression vector pET28a (Novagen) at the BamHI site and subsequently sequenced. For glutathione-S-transferase (GST) fusion protein, the PCR product was cloned into pGEX2T (Amersham Pharmacia) at the BamHI site. Escherichia coli dnaC was PCR amplified using genomic DNA of E.coli strain K12 with the following primers: 5′-GCGGATCCATGAAAA ACGTTGGCGACCTG-3′ and 5′-GCGGATCCATACTCTT TACCTGTTACCCG-3′. The PCR-amplified product was cloned into the BamHI site of pET28a and subsequently sequenced.
Mutation in the ATP binding site of HpDnaB (ARPSMGKT) was introduced by PCR using the following mutagenic oligonucleotides: 5′-GTCATTATAGGGGCAT GCCCGTCTATGGGTAAA-3′ and 5′-TTTACCCATAGA CGGGCATGCCCCTATAATGAC-3′.
Mutagenesis was carried out on a pET28aHpDnaB DNA template using a Stratagene site-directed mutagenesis kit, as directed by the vendor. This mutation introduces a cysteine (C) residue at position 204 instead of arginine (R) and created a SphI site in the DNA.
For the complementation assay, wild-type and mutated dnaB genes were released from respective pET28a recombinant clones by using XbaI (followed by end-filling using Klenow polymerase) and EcoRI and cloned into pBR322 (downstream of the Bla-P2 promoter) (17) at HindIII (followed by end-filling using Klenow polymerase) and EcoRI sites. Escherichia coli DJ58 (dnaBts) strain was transformed with the recombinant positive clones.
Purification of His-tagged HpDnaB (Wt), His-tagged HpDnaB (Mut), GST-HpDnaB (Wt) and His-tagged E.coli DnaC; gel filtration chromatography and western blotting
Escherichia coli strain BL21 (DE3) (Novagen) harboring pET28aHpDnaB (Wt), pET28a HpDnaB (Mut) or pET28aEcDnaC were grown at 37°C in LB media containing 50 µg/ml kanamycin. The bacterial cultures were induced for the expression of the recombinant proteins using 1 mM IPTG. His-tagged proteins were purified using Ni-NTA agarose (Qiagen) beads essentially following the protocol supplied by the vendor. Fractions containing HpDnaB (Wt) or HpDnaB (Mut) or EcDnaC were pooled, dialyzed against buffer A [50 mM Tris–Cl (pH 7.4), 1 mM EDTA, 10 mM βME, 100 µM PMSF, 10% glycerol] and loaded on a 1 ml Q–Sepharose column (Amersham) that had been previously equilibrated with buffer A with 50 mM NaCl (buffer B). The column was connected to a Duo-Flow protein purification system (Bio-Rad). After loading the protein, the column was washed with the same buffer until OD280 reached baseline and proteins were eluted by applying a linear gradient of 50 mM–700 mM NaCl in buffer A. All the fractions were checked for HpDnaB (∼55 kDa) on 10% SDS–PAGE. Positive fractions were pooled and dialyzed against buffer A with 100 mM NaCl and stored at –80°C. GST-HpDnaB was purified using agarose immobilized GST beads (Sigma-Aldrich) using the instructions supplied by the vendor.
The molecular masses of the native wild-type or mutated HpDnaB were determined by subjecting them to size- exclusion chromatography on a Bio-Sil SEC 250-5 column (7.8 × 300 mm; Bio-Rad, Hercules, CA) in buffer B supplemented with 2 mM MgCl2 and 2 mM ATP. The column was previously calibrated against Bio-Rad molecular weight standards.
Protein concentrations were determined by the Bradford method (Bio-Rad kit) as per the instructions of the vendor with BSA as a standard.
Western blot analysis was carried out following standard procedures (18).
Helicase activity
The following oligonucleotides were used for the helicase activity: 5′-TCGGTACCCGGGGATCCT-3′ (18mer); 5′-CCCAGTCACGACGTTGTAAAACG-3′ (23mer); 5′-TCG AGCTCGGTACCCGGGGATCCTCTAGAG-3′ (30mer); 5′- AATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTC G-3′ (36mer); 5′-CCAAAACCCAGTCACGACGTTGTAA AACG-3′ (5′ tailed 23mer); and 5′-CCCAGTCACGAC GTTGTAAAACGTGCCGG-3′ (3′ tailed 23mer). The oligonucleotides were radiolabeled at the 5′ end and were allowed to anneal to either M13mp18 or M13mp19 as described earlier (19), with minor modifications. Briefly, 100 ng of the respective labeled oligonucleotides were annealed to 1 µg of M13mp19 or M13mp18 single-stranded DNA (ssDNA) in 40 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 50 mM NaCl and 1 mM DTT and purified by passing through a 1 ml Sepharose CL-4B (Amersham) spin column.
For determination of polarity of unwinding (3′-5′ or 5′-3′), 5′-end-labeled 36mer oligonucleotides (5′-AATTCGAGCTC GGTACCCGGGGATCCTCTAGAGTCG-3′) were annealed to M13mp19 DNA, digested with SmaI and purified by passing through a 1 ml Sepharose CL-4B spin column. For 5′-3′ polarity determination, 36mer oligonucleotides were annealed to M13mp19 DNA followed by 3′-end-labeling by Klenow polymerase, digested with SmaI, purified as before and used for the helicase assay.
The helicase assay was carried out in 10 µl of reaction buffer containing 20 mM Tris–HCl (pH 7.5), 8 mM DTT, 1.5 mM MgCl2, 2 mM ATP, 80 µg/ml BSA, 10 mM KCl, 4% (w/v) sucrose, ∼2000 c.p.m. of labeled partial duplex DNA and purified HpDnaB (Wt) or HpDnaB (Mut) (1.8 pmol or as indicated in the Figure legends) at 37°C for 30 min. Reactions were stopped by adding 0.25% SDS, 15 mM EDTA, 5% glycerol and products were separated on 10% non-denaturing PAGE. The gel was dried and analyzed by a phosphorimager (Fujifilm-BAS 1800) or autoradiographed.
ATP hydrolysis assay
The ATPase activity of HpDnaB was measured in a reaction mixture (10 µl) containing 20 mM Tris–HCl (pH 8.0), 1 mM MgCl2, 100 mM KCl, 8 mM DTT, 4% sucrose, 80 µg/ml BSA, 1 mM ATP, 3.4 fmol of [γ-32P]ATP and the required amount of DnaB, as indicated in the Figure legends. The reaction mixtures were incubated at 37°C for 30 min and the reactions were stopped by putting the tubes on ice. Released inorganic Pi was separated by thin-layer chromatography (TLC) on a polyethylenemine cellulose strip (Sigma-Aldrich) in 0.5 M LiCl and 1 M formic acid at room temperature for 1 h. The TLC plate was dried, autoradiographed and analyzed by a phosphorimager (Fujifilm-BAS-1800) for quantitation.
Nucleotide binding assay
UV-mediated cross-linking of [α-32P]dATP. The cross-linking mixtures, in a final volume of 20 µl, contained 20 mM HEPES buffer (pH 7.5), 10% glycerol, 0.1 mM DTT, 10 µCi of [α-32P]dATP (3000 Ci/mmol) and 1 µg of wild-type or mutated HpDnaB. The mixtures were incubated for 10 min on ice, followed by UV (254 nm) irradiation at a distance of 5 cm (Stratagene) for 30 min at 4°C. After termination of UV exposure, 0.8 µl of 100 mM dATP and 20 µg of BSA were added to reaction mixtures. Proteins were precipitated by trichloroacetic acid, washed with acetone containing 0.5% HCl and then twice with acetone. Proteins were separated by SDS–PAGE and the labeled protein was visualized by a phosphoimager.
Complementation assay
Complementarity of HpDnaB with its E.coli counterpart was tested by transforming E.coli strain DJ58 (dnaBts), a conditional lethal mutant for the E.coli dnaB allele, with recombinant plasmids containing HpDnaB, E.coli DnaB and DnaC (wild type) or pBR322 alone, followed by checking the survival of E.coli colonies at the non-permissive temperature in the presence of ampicillin. Escherichia coli plasmid pdnaBC was used as a positive control for complementation analysis.
RESULTS
Cloning and purification
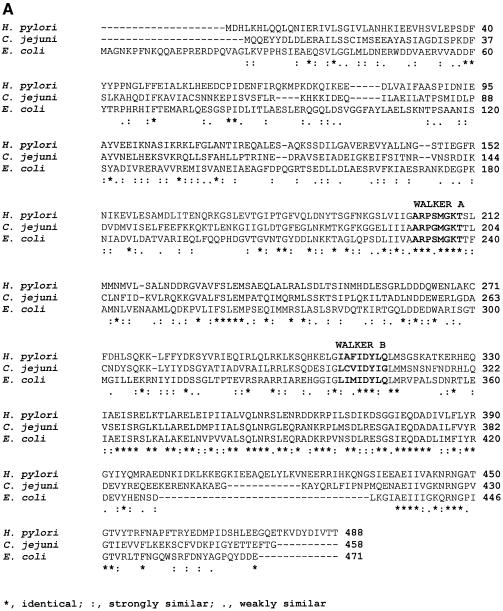
In the H.pylori genomic database, an ORF HP1362 was annotated as the putative H.pylori DnaB homolog. When the amino acid sequence of the gene was aligned with its E.coli and Campylobacter jejuni counterparts, it was found that H.pylori DnaB is ∼32% identical and 57% homologous to E.coli DnaB (Fig. 1A). Expression of the dnaB gene in H.pylori was confirmed by western blotting using anti-DnaB (E.coli) antibody (Fig. 1B).
Figure 1.
Primary sequence analysis of HpDnaB and in vivo expression. (A) Protein sequence alignment of homologous DnaB proteins. Comparison of the H.pylori, C.jejuni and E.coli DnaB primary structures. The amino acid sequences of the respective proteins were aligned by CLUSTALW multiple alignment program. ‘*’ indicates the residues which are identical, ‘:’ indicates the residues which are strongly similar and ‘.’ indicates the residues which are weakly similar. Nucleotide binding (Walker A) and hydrolysis (Walker B) motifs are also shown. (B) In vivo expression of DnaB in H.pylori. Western blot of H.pylori and E.coli cell extract using a polyclonal antisera against DnaB (E.coli). Lane 1 contains 1.0 µg of purified E.coli DnaB, lanes 2 and 3 contain cell extract of H.pylori (5.0 and 10.0 µg, respectively), lanes 4 and 5 contain cell extract of E.coli (5.0 and 10.0 µg, respectively) and lane 6 contains 100 ng of purified GST-tagged DnaB of H.pylori.
In order to clone the H.pylori dnaB gene (HP1362; strain 26695) (9), the 1.5 kb DNA fragment representing the ORF was amplified from H.pylori strain 26695 genomic DNA by PCR. The amplified product was subsequently cloned into E.coli expression vector pET28a (Novagen) and sequenced. The cloned gene when expressed in the E.coli strain BL21 (DE3) produced a ∼55 kDa polypeptide that matches with the calculated molecular weight of the His-tagged gene product (Fig. 2A). The protein was subsequently purified using an Ni-NTA agarose (Qiagen) affinity matrix (Fig. 2B). The peak fractions were found to be >90% pure. The protein was further purified on a Mono-Q ion-exchange column to near homogeneity as shown in Figure 2C.
Figure 2.
Purification of HpDnaB. (A) Induction of expression of His6-HpDnaB protein in E.coli strain BL21(DE3). (B) Purification of His6-HpDnaB by Ni-NTA resin. The fractions which contain the purified proteins are indicated on the top. (C) Purification of His6-HpDnaB using an ion-exchange Mono-Q column. The fractions containing DnaB after Ni-NTA purification were pooled and passed through the Mono-Q column. The fraction numbers are indicated on the top.
In vitro helicase activity of H.pylori DnaB
DnaB, a hexameric helicase is essential for replication of the bacterial chromosome, some bacterial phage genomes (like bacteriophage lambda and bacteriophage P2) and plasmid DNA (8,20,21). In order to test helicase activity of the purified HpDnaB, unwinding of a partial duplex DNA by the protein was monitored as described earlier (22). As shown in Figure 3A and B, the peak fractions from the Mono-Q column exhibited the protein concentration and ATP-dependent helicase activity, converting >80% of the partial duplex substrate to product. We also checked the effect of ATP concentration on helicase activity (Fig. 3B). HpDnaB helicase activity was stimulated at low ATP concentrations, reached a peak at 2 mM ATP concentration, followed by a decrease in the activity at higher concentrations. Since the Mg2+ ion concentration was not compensated with increasing ATP concentration, it is possible that Mg2+ ions become limiting with increasing ATP concentration, affecting both ATP hydrolysis and helicase activity.
Figure 3.
Effect of protein amount, ATP, time and KCl on DnaB helicase activity. Annealed substrates (18mer radiolabeled oligo annealed to M13mp19 ssDNA, as described in the Materials and Methods) were used in all four cases. (A) The helicase reaction was carried out using increasing amounts of purified protein (0.9 pmol to 9.0 pmol). Annealed and unwound oligos were separated by non-denaturing PAGE. (B) The helicase activity of HpDnaB was further characterized by using 1.8 pmol (∼100 ng) of purified HpDnaB in the presence of increasing concentrations of ATP. (C) HpDnaB helicase activity varying the time of incubation of reaction or (D) increasing KCl concentration. Percent unwinding activity of HpDnaB for each reaction was also plotted.
The in vitro helicase activity was also dependent on the time of incubation. In the presence of 1.8 pmol (∼100 ng) protein ∼50% of the substrate was converted to product in 10 min (Fig. 3C). We also tested the effect of KCl concentration on helicase activity. Helicase activity was not affected up to 25 mM concentration. However, increasing the KCl concentration beyond that point inhibited the helicase activity (Fig. 3D).
HpDnaB unwinding activity ranges from ∼45% to nearly 100% in different sets of experiments (Fig. 3A–D) under the same experimental conditions. This discrepancy in enzyme activity in different sets of experiments could be due to the variation in the enzyme activity in the different batches of enzyme purification.
Like other DnaB from different bacterial sources, HpDnaB was also found to be dependent on nucleoside triphosphate for helicase activity and ATP, GTP and UTP were found to be preferred compared with other NTPs and dNTPs (Fig. 4A). However, when ADP and ATPγS were included in the reaction mixture instead of ATP, no helicase activity was detected (Fig. 4B), suggesting nucleoside triphosphate hydrolysis is required.
Figure 4.
Effect of nucleotides and divalent cations on DnaB helicase activity. (A) The helicase reaction was carried out using 1.8 pmol (∼100 ng) of purified protein in the presence of a 5 mM concentration of various nucleotides, as indicated on the top. (B) The effect of ATP analogs was tested on HpDnaB helicase activity. The substrates used in both cases were 18mer radiolabeled oligos (as described in the Materials and Methods) annealed to M13mp19 ssDNA.
Mutation in the ATP binding motif of DnaB affects helicase activity
To further test that the unwinding activity shown by HpDnaB was not due to the presence of any other minor contaminating proteins and the activity required NTP binding, a mutation was introduced in the Walker A motif (R→C at position 204) and the mutated protein was analyzed for helicase activity.
His-tagged mutant HpDnaB was purified following the wild-type HpDnaB purification protocol (Fig. 5A) and tested for helicase activity using the same 23mer partial duplex substrate.
Figure 5.
Helicase and nucleotide binding activity of HpDnaB (Wt) and HpDnaB (Mut). (A) Purification of wild-type and mutated HpDnaB proteins. (B) The effect of wild-type and mutated HpDnaB on helicase activity. (C) HpDnaB (Wt) but not HpDnaB (Mut) binds to radiolabeled nucleotide in the presence of UV light. One microgram of the wild-type or mutated DnaB was incubated with [α-32P]dATP and was further cross-linked using UV, as described in Materials and Methods. (D) Gel showing the presence of proteins in all the lanes of Figure 6C.
No DNA unwinding activity was detected even at the highest protein amount tested, 3.6 pmol (∼200 ng), whereas 1.8 pmol (∼100 ng) of wild-type HpDnaB showed a strong helicase activity (Fig. 5B). This indicates that the helicase activity shown here is solely due to HpDnaB and not due to the presence of any other contaminating protein and also reconfirms that the helicase activity is dependent on ATP. In order to test the difference in nucleotide binding of wild-type and mutated HpDnaB, both wild-type and mutated HpDnaB proteins were incubated with [α32-P]dATP and subjected to UV cross-linking analysis. Unlike wild-type protein, no ATP binding was detected in the case of mutated HpDnaB (Fig. 5C and D).
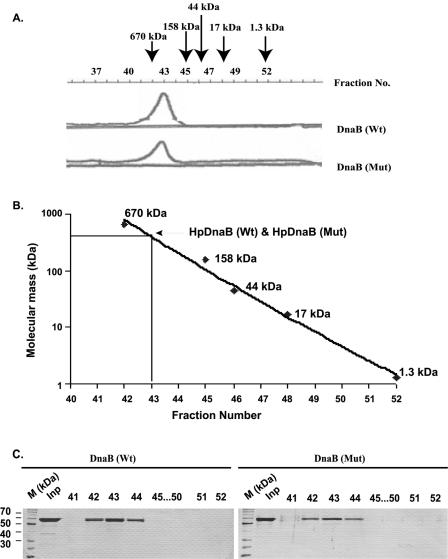
In solution, HpDnaB exists as a hexamer
Wild-type and mutated HpDnaB protein were subjected to size-exclusion chromatography on a calibrated Bio-Sil SEC 250-5 column. In each case, a single protein peak (Fig. 6A) was observed. Molecular mass standards were also subjected to gel filtration chromatography under the same conditions. A standard curve was obtained by plotting the molecular mass of the standards (in logarithmic scale) against the fraction number. From this plot, the native molecular mass of HpDnaB (Wt) and HpDnaB (Mut) was estimated to be ∼360 kDa (Fig. 6B). SDS–PAGE followed by silver staining of the peak fractions revealed the presence of a ∼55 kDa protein band that corresponds to His-tagged HpDnaB (Wt) and HpDnaB (Mut) (Fig. 6C).
Figure 6.
Size-exclusion chromatography of wild-type and mutated DnaB. (A) Wild-type or mutated DnaB was passed through a Bio-Sil SEC 250-5 column and 0.2 ml fractions were collected in each case. The fraction numbers and the elution pattern of the proteins are shown. The elution pattern of the gel filtration standards is marked on the top as followed: thyroglobulin (670 kDa), bovine gamma globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa) and vitamin B-12 (1.35 kDa). (B) Estimation of molecular masses of HpDnaB (Wt) and HpDnaB (Mut). Molecular masses of the standards were plotted in logarithmic scale against the fraction numbers. Molecular masses of HpDnaB (Wt) and HpDnaB (Mut) were deduced from the plot. (C) SDS–PAGE followed by Coomassie stain of the gel filtration fractions.
HpDnaB shows ATPase activity
All the helicases tested so far, have been shown to require NTPase activity for helicase action (23,24). It has already been shown that a non-hydrolysable ATP analog could not support unwinding activity (Fig. 4B). Therefore, HpDnaB was tested for associated ATPase activity. To test ATPase activity, increasing amounts of HpDnaB were incubated with [γ-32P]ATP and released free phosphate from labeled ATP was monitored following TLC.
Wild-type HpDnaB showed strong ATPase activity as evidenced by the release of more free phosphate with increasing amounts of HpDnaB (Fig. 7A). However, mutated HpDnaB failed to show any ATPase activity, even at the highest amount of protein tested (Fig. 7A), suggesting that mutation in the Walker A motif also affects ATP hydrolysis, which is expected since mutated HpDnaB cannot bind ATP. The HpDnaB-associated ATPase activity was also observed to be stimulated by ssDNA (Fig. 7B), suggesting that HpDnaB has DNA-dependent helicase activity.
Figure 7.
Characterization of ATPase activity of HpDnaB (Wt) and HpDnaB (Mut). (A) ATPase activity is enhanced with increasing amounts of HpDnaB (Wt). HpDnaB (Mut) did not show any ATPase activity. Release of radiolabeled inorganic phosphate (Pi) is also marked. (B) Analysis of DNA-dependent ATPase activity of HpDnaB. Helicase activity was measured in the presence of 0.9 pmol of HpDnaB and increasing amounts of 36mer oligonucleotides. Release of inorganic phosphate was quantitated and plotted against protein or oligo concentration. (C) Effect of divalent cations on helicase activity. Helicase assays were performed in the presence of various divalent cations (2 mM each) as indicated. Percent unwinding activity in the presence of nucleotides and divalent cations is also shown. 18mer oligos annealed to M13mp19 ssDNA were used as substrate.
Hydrolysis of NTPs also depends on divalent cations. We first titrated the Mg2+ ion concentration in helicase reactions and 2 mM Mg2+ was found to be optimal for the helicase activity (data not shown). Later, we used several divalent cations to check their effect on helicase activity. At 2 mM concentration of all the divalent cations used, Mg2+ was found to be the most effective for helicase activity (Fig. 7C). It has been shown earlier that DnaB helicase exists as a stable hexamer in a large protein concentration range and this form is specifically stabilized by magnesium (25). The order of divalent cation preference on helicase activity is as follows: Mg2+ > Mn2+ > Ca2+ > Co2+ > Zn2+. No activity was measured in the absence of divalent cations.
HpDnaB has 5′ to 3′ polarity
The direction of unwinding by a helicase is defined by the strand to which the enzyme binds and moves along that strand. All the DNA helicases characterized so far exhibited either 5′ to 3′ or 3′ to 5′ polarity. Escherichia coli DnaB protein unwinds DNA in a 5′ to 3′ direction (26). In order to test the polarity of HpDnaB helicase action, two radiolabeled (at 5′ or 3′) partial duplex substrates were linearized by digestion with SmaI restriction enzyme to generate a duplex DNA at both ends of a long linear ssDNA molecule (26). In the presence of HpDnaB, the displacement of a specific labeled fragment (5′ or 3′ end-labeled) was followed to determine directionality of action.
Experimental results as shown in Figure 8A, exhibited only displacement of the 3′-end-labeled ssDNA fragment, indicating that HpDnaB possesses a 5′ to 3′ polarity. The partial duplex substrates used in this set of experiments did not have any short 5′ or 3′ tail attached to them, however, in subsequent experiments substrates containing 5′ or 3′ tails were also tested. Under the same experimental conditions, oligos having a 5′ single-stranded tail were released more efficiently than oligos having no tail or having a 3′ single-stranded tail (Fig. 8B).
Figure 8.
Effect of various substrates on DnaB helicase activity. (A) Polarity of DnaB helicase. Helicase reaction was carried out either using, 5′-end-labeled 18mer synthetic oligo substrate or 3′-end-labeled 18mer synthetic oligo substrate (generated by SmaI digestion of 36mer-labeled DNA annealed to M13mp19 ssDNA, M13mp19). All the oligos are described in the Materials and Methods. Lane 1 contains no protein, lane 2 contains 0.9 pmol of HpDnaB and lane 3 contains heated substrate. (B) Influence of 5′ and 3′ tail on helicase activity. Substrates containing no tail (lanes 1–3), 5′ tail (lanes 4–6) or 3′ tail (lanes 7–9) were used for the helicase assay.
HpDnaB rescues the temperature-sensitive phenotype of E.coli dnaBts
The comparison of primary sequences of E.coli and H.pylori DnaB (Fig. 1A) shows ∼32% identity and 57% similarity between the two proteins. In vitro characterization of the HpDnaB also indicated that, like its E.coli counterpart, it is an ATP-dependent helicase, possesses ATPase activity and exists in solution as a hexamer. We were interested to test whether HpDnaB can rescue a temperature-sensitive DnaB function in E.coli. To carry out complementation analysis, a His-tagged dnaB gene was subcloned into pBR322 under the control of Bla-P2 promoter (see Materials and Methods) and E.coli strain DJ58 (dnaBts) was transformed with the recombinant plasmid. HpDnaB was found to complement the defective dnaB gene in E.coli at 40°C, whereas pBR322 and HpDnaB (Mut) failed to do so (Fig. 9A) at the same temperature. When the same E.coli strain was transformed with a plasmid containing wild-type E.coli dnaB (pdnaBC), it also complemented the defective function (Fig. 9A, sector 4) at 40°C. We further tested HpDnaB expression in E.coli strain DJ58 at 40°C (Fig. 9B) by western blotting using anti-His antibody. These results taken together suggest that, in fact, the ORF HP1362 encodes a functional homolog of replicative DnaB helicase.
Figure 9.
Complementation analysis. (A) Complementation of E.coli DJ58ts with plasmids expressing wild-type or mutated HpDnaB. Escherichia coli DJ58ts cells were transformed with either pBR322 (1), pBR322+HpDnaB (Wt) (2), pBR322+HpDnaB (Mut) or pdnaBC (E.coli) (4). Cells were plated on LB agar plates and incubated either at the permissive temperature (30°C) or at the non-permissive temperature (40°C). (B) Detection of HpDnaB expression in E.coli DJ58ts by immuno-blotting using anti-His antibody. Lane 1, purified HpDnaB; lanes 2 and 3, extract from pBR322 and HpDnaB transformed cells at the permissive temperature (30°C); lanes 4 and 5, extract from pBR322 and HpDnaB transformed cells at the non-permissive temperature (40°C). (C) Gel filtration analysis of His-EcDnaC, His-HpDnaB or a mixture of both. The above proteins were subjected to gel filtration chromatography using a Bio-Sil SEC 250-5 column. Fractions of 0.5 ml were collected in each case. The peak fraction in each case was subjected to SDS–PAGE followed by western blot analysis using anti-His antibodies. Fraction numbers are shown on the top. Input lane shows the presence of both the proteins used for the gel filtration analysis.
In E.coli, DnaC is essential to load DnaB on oriC. The complementation of E.coli dnaBts with HpDnaB raises the issue as to whether HpDnaB interacts with DnaC to facilitate the loading of HpDnaB. In order to address this issue, we performed gel filtration analysis using either purified HpDnaB or E.coli DnaC (EcDnaC) alone or the mixture of HpDnaB and EcDnaC, followed by western blot analysis of the peak fractions of gel filtration eluates using anti-His antisera in each case. Purified EcDnaC runs as a monomer when compared with the molecular mass marker (data not shown). We have already shown that HpDnaB runs as a multimer following gel filtration (Fig. 6A). When HpDnaB and EcDnaC were mixed together and subjected to gel filtration chromatography, both proteins were co-eluted in the same fraction, suggesting that these proteins may physically interact with each other (Fig. 9C). Therefore, the loading of HpDnaB on oriC could be mediated through EcDnaC.
DISCUSSION
In this work, we report the identification and characterization of an important H.pylori DNA replication protein, DnaB helicase. Unlike E.coli, H.pylori lacks an obvious helicase loader DnaC homolog from the genome sequence. Therefore, the characterization of HpDnaB homolog might help in understanding the basic mechanism of helicase loading in this organism.
This report shows that HpDnaB is a helicase and it is active in the presence of a number of hydrolysable NTPs. ATP, GTP or UTP are equally active as co-factors whereas CTP is somewhat less active. In contrast, E.coli DnaB was shown to be inactive in the presence of UTP (26). Among the four dNTPs, only dATP supported helicase activity of HpDnaB, whereas both dATP and dCTP were shown to support the helicase activity of E.coli DnaB. Like E.coli, HpDnaB requires Mg2+ for the optimum activity (26). Mn2+ and Ca2+ are also well tolerated by HpDnaB for the helicase activity, although it has been reported in the literature that for several helicases, Ca2+ could not be a substitute for Mg2+ (27,28). Like other helicases, HpDnaB showed the optimal helicase activity in the presence of 2 mM ATP. Monovalent cations (K+) inhibited the helicase activity at a concentration higher than 100 mM. This phenomenon is also common for many other helicases.
Helicases are known to bind to the single-stranded region and then proceed either in a 5′ to 3′ or 3′ to 5′ direction depending on their polarity and thereby unwinding the duplex using NTPs as an energy source (23). A similar type of polarity has also been observed for a number of proteins of the DnaB family (26,29–32). It has been shown earlier that the 3′ tail of a forked duplex DNA substrate stimulates unwinding in a length-dependent manner in the presence of the DnaB family of helicases (33). According to a recent model proposed by Kaplan (34), a forked structure containing a short 3′ tail allows DnaB to pass through the central channel without showing any helicase activity. The length of the 3′ tail determines whether one or two DNA strands will pass through the central channel of hexameric DnaB. In contrast, we observed unwinding activity in all the partial duplexes tested with or without a short 3′ tail. This discrepancy could be due to the differences in the substrate structure used. The possibility of non-specific activity of HpDnaB can be ruled out since the mutant protein with a mutation in the NTP binding motif fails to show helicase activity. Interestingly, we observed stimulation in helicase activity in the presence of a short 5′ tail. HpDnaB can be loaded simultaneously on 5′ tail and ssDNA in the partial duplex substrate followed by subsequent movement in the 5′ to 3′ direction.
HpDnaB is 32% identical and 57% similar to E.coli DnaB. The C-terminal half of the protein shares more homology with E.coli DnaB than the N-terminal half. In general, the C-terminal half of hexameric helicases contains all the motifs required for the helicase activity (35). Therefore, the similarity found in in vitro biochemical functions of HpDnaB with its E.coli counterpart could be due to the similarity in the C-terminal half. Escherichia coli DnaB has been dissected into three domains (35) and the first 150 amino acids have been assigned to energy transduction and protein–protein interaction. Thus, the less conserved N-terminal domain of H.pylori DnaB may functionally play some unique role in the initiation of DNA replication. Further dissection of the N-terminal half of the HpDnaB will be required to address these issues.
Unlike some enterobacteria such as Salmonella typhimurium (36) and Shigella flexneri (37), H.pylori lacks the E.coli dnaC homolog. The absence of a dnaC-like gene in many other bacteria whose genome sequences are available now, raises the question as to whether DnaC is essential for replication as a helicase loader. However, there are many examples of proteins both in prokaryotes and in eukaryotes that are used as helicase loaders without sharing much homology with DnaC. The best examples are DnaI in Bacillus (38–40) and Cdc6 in mammals which load Mcm helicases (41).
It is still not clear whether DnaB can be loaded at origin in the absence of DnaC.
Recently, it has been shown that P.putida and P.aeruginosa DnaB helicases could be loaded on plasmid RK2 oriV in the absence of DnaC-like ATPases (16). Whether the independent loading of DnaB is possible at the chromosomal origin of Pseudomonas is still not known.
The temperature-sensitive phenotype of the E.coli mutant DnaB252 containing a mutation in the amino acid residue 299 can be rescued by over-expression of DnaC, suggesting that this region could be important for DnaB–DnaC interaction (42). Interestingly, the residue 299 and surrounding amino acids are highly divergent between E.coli and H.pylori DnaB counterparts. Therefore, apparently HpDnaB may not interact with E.coli DnaC. However, we have shown clearly that HpDnaB can functionally complement the E.coli DnaB temperature-sensitive mutant. Since the loading of E.coli DnaB is dependent on DnaC, it is possible that HpDnaB also interacts with EcDnaC to facilitate the loading of HpDnaB. By gel filtration analysis we showed that HpDnaB and EcDnaC co-eluted in the same fraction. This clearly suggests that HpDnaB has an affinity towards EcDnaC. These findings strengthen the hypothesis that there could be a functional homolog of DnaC in H.pylori which helps loading of HpDnaB on an oriC equivalent of H.pylori. However, it is also not unlikely that HpDnaB can be loaded on oriC by itself, independent of DnaC. Further careful studies are required to address these issues.
Recently, using a high-throughput yeast two-hybrid assay, 261 H.pylori proteins were screened against a highly complex library of genome-encoded polypeptides (43). According to this study, two ORFs (HP0897 and HP0340) with unknown functions were reported to be interacting partners of HpDnaB. It will be interesting to see whether these proteins physically interact with HpDnaB and thereby facilitate its loading on the chromosomal origin of replication. Considering the high prevalence of drug resistance in H.pylori, identification of such candidates will be useful for screening novel drug targets.
Acknowledgments
ACKNOWLEDGEMENTS
The authors acknowledge Dr Dhruba K. Chattoraj for providing the E.coli strain dnaBts, Dr Subhasis Biswas for E.coli anti-DnaB antisera and Dr Betty P. Guo for H.pylori extract. The authors also acknowledge Dr Sudha Bhattacharya, Professor Rajendra Prasad, Professor Kasturi Datta and Dr Balaji Prakash for their support. This work is supported by research grants from the University Excellence Programme (University Grant Commission, India), and Council of Scientific and Industrial Research (CSIR) to S.K.D. and G.M.. R.K.S. and P.M. acknowledge CSIR for fellowships.
REFERENCES
- 1.Peterson W.L. (1991) Helicobacter pylori and peptic ulcer disease. N. Engl. J. Med., 324, 1043–1048. [DOI] [PubMed] [Google Scholar]
- 2.Peterson W.L. (2002) Helicobacter pylori and gastric adenocarcinoma. Aliment. Pharmacol. Ther., 16 (Suppl. 1), 40–46. [DOI] [PubMed] [Google Scholar]
- 3.Parsonnet J., Friedman,G.D., Vandersteen,D.P., Chang,Y., Vogelman,J.H., Orentreich,N. and Sibley,R.K. (1991) Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med., 325, 1127–1131. [DOI] [PubMed] [Google Scholar]
- 4.Hansson L.E., Engstrand,L., Nyren,O., Evans,D.J.,Jr, Lindgren,A., Bergstrom,R., Andersson,B., Athlin,L., Bendtsen,O. and Tracz,P. (1993) Helicobacter pylori infection: independent risk indicator of gastric adenocarcinoma. Gastroenterology, 105, 1098–1103. [DOI] [PubMed] [Google Scholar]
- 5.Marshall B.J. and Warren,J.R. (1984) Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet, 1, 1311–1315. [DOI] [PubMed] [Google Scholar]
- 6.Warren J.R. (1984) Spiral bacteria of the gastric antrum. Med. J. Aust., 141, 477–478. [PubMed] [Google Scholar]
- 7.Wahle E., Lasken,R.S. and Kornberg,A. (1989) The dnaB–dnaC replication protein complex of Escherichia coli. I. Formation and properties. J. Biol. Chem., 264, 2463–2468. [PubMed] [Google Scholar]
- 8.Kornberg A. and Baker,T.A. (1992) DNA Replication. W.H. Freeman and Co., New York, NY. [Google Scholar]
- 9.Tomb J.F., White,O., Kerlavage,A.R., Clayton,R.A., Sutton,G.G., Fleischmann,R.D., Ketchum,K.A., Klenk,H.P., Gill,S., Dougherty,B.A. et al. (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature, 388, 539–547. [DOI] [PubMed] [Google Scholar]
- 10.Alm R.A., Ling,L.S., Moir,D.T., King,B.L., Brown,E.D., Doig,P.C., Smith,D.R., Noonan,B., Guild,B.C., deJonge,B.L., Carmel,G., Tummino,P.J., Caruso,A., Uria-Nickelsen. M.,Mills,D.M., Ives,C., Gibson,R., Merberg,D., Mills,S.D., Jiang,Q., Taylor,D.E., Vovis,G.F. and Trust,T.J. (1999) Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature, 397, 176–180. [DOI] [PubMed] [Google Scholar]
- 11.Zawilak A., Cebrat,S., Mackiewicz,P., Krol-Hulewicz,A., Jakimowicz,D., Messer,W., Gosciniak,G. and Zakrzewska-Czerwinska,J. (2001) Identification of a putative chromosomal replication origin from Helicobacter pylori and its interaction with the initiator protein DnaA. Nucleic Acids Res., 29, 2251–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Y.B., Ratnakar,P.V., Mohanty,B.K. and Bastia,D. (1996) Direct physical interaction between DnaG primase and DnaB helicase of Escherichia coli is necessary for optimal synthesis of primer RNA. Proc. Natl Acad. Sci. USA, 93, 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S., Dallmann,H.G., McHenry,C.S. and Marians,K.J. (1996) Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell, 84, 643–650. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan D.L. and Steitz,T.A. (1999) DnaB from Thermus aquaticus unwinds forked duplex DNA with an asymmetric tail length dependence. J. Biol. Chem., 274, 6889–6897. [DOI] [PubMed] [Google Scholar]
- 15.Bird L.E. and Wigley,D.B. (1999) The Bacillus stearothermophilus replicative helicase: cloning, overexpression and activity. Biochim. Biophys. Acta, 1444, 424–428. [DOI] [PubMed] [Google Scholar]
- 16.Caspi R., Pacek,M., Consiglieri,G., Helinski,D.R., Toukdarian,A. and Konieczny,I. (2001) A broad host range replicon with different requirements for replication initiation in three bacterial species. EMBO J., 20, 3262–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolivar F., Rodriguez,R.L., Greene,P.J., Betlach,M.C., Heyneker,H.L. and Boyer,H.W. (1977) Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene, 2, 95–113. [PubMed] [Google Scholar]
- 18.Sambrook J.F., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 19.Tuteja N., Phan,T.N. and Tewari,K.K. (1996) Purification and characterization of a DNA helicase from pea chloroplast that translocates in the 3′-to-5′ direction. Eur. J. Biochem., 238, 54–63. [DOI] [PubMed] [Google Scholar]
- 20.Lohman T.M. and Bjornson,K.P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem., 65, 169–214. [DOI] [PubMed] [Google Scholar]
- 21.Patel S.S. and Picha,K.M. (2000) Structure and function of hexameric helicases. Annu. Rev. Biochem., 69, 651–697. [DOI] [PubMed] [Google Scholar]
- 22.Kaguni J.M. and Kornberg,A. (1984) Replication initiated at the origin (oriC) of the E. coli chromosome reconstituted with purified enzymes. Cell, 38, 183–190. [DOI] [PubMed] [Google Scholar]
- 23.Soultanas P. and Wigley,D.B. (2001) Unwinding the ‘Gordian knot’ of helicase action. Trends Biochem. Sci., 26, 47–54. [DOI] [PubMed] [Google Scholar]
- 24.Konieczny I. (2003) Strategies for helicase recruitment and loading in bacteria. EMBO Rep., 4, 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bujalowski W., Klonowska,M.M. and Jezewska,M.J. (1994) Oligomeric structure of Escherichia coli primary replicative helicase DnaB protein. J. Biol. Chem., 269, 31350–31358. [PubMed] [Google Scholar]
- 26.Le Bowitz J.H. and McMacken,R. (1986) The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem., 261, 4738–4748. [PubMed] [Google Scholar]
- 27.Tuteja N., Rahman,K., Tuteja,R., Ochem,A., Skopac,D. and Falaschi,A. (1992) DNA helicase III from HeLa cells: an enzyme that acts preferentially on partially unwound DNA duplexes. Nucleic Acids Res., 20, 5329–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuteja N., Phan,T.N. and Tewari,K.K. (1996) Purification and characterization of a DNA helicase from pea chloroplast that translocates in the 3′-to-5′ direction. Eur. J. Biochem., 238, 54–63. [DOI] [PubMed] [Google Scholar]
- 29.Lee E.H., Kornberg,A., Hidaka,M., Kobayashi,T. and Horiuchi,T. (1989) Escherichia coli replication termination protein impedes the action of helicases. Proc. Natl Acad. Sci. USA, 86, 9104–9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manna A.C., Pai,K.S., Bussiere,D.E., Davies,C., White,S.W. and Bastia,D. (1996) Helicase–contrahelicase interaction and the mechanism of termination of DNA replication. Cell, 87, 881–891. [DOI] [PubMed] [Google Scholar]
- 31.Richardson R.W. and Nossal,N.G. (1989) Characterization of the bacteriophage T4 gene 41 DNA helicase. J. Biol. Chem., 264, 4725–4731. [PubMed] [Google Scholar]
- 32.Venkatesan M., Silver,L.L. and Nossal,N.G. (1982) Bacteriophage T4 gene 41 protein, required for the synthesis of RNA primers, is also a DNA helicase. J. Biol. Chem., 257, 12426–12434. [PubMed] [Google Scholar]
- 33.Ahnert P. and Patel,S.S. (1997) Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J. Biol. Chem., 272, 32267–32273. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan D.L. (2000) The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol., 301, 285–299. [DOI] [PubMed] [Google Scholar]
- 35.Biswas E.E. and Biswas,S.B. (1999) Mechanism of DnaB helicase of Escherichia coli: structural domains involved in ATP hydrolysis, DNA binding and oligomerization. Biochemistry, 38, 10919–10928. [DOI] [PubMed] [Google Scholar]
- 36.Parkhill J., Dougan,G., James,K.D., Thomson,N.R., Pickard,D., Wain,J., Churcher,C., Mungall,K.L., Bentley,S.D., Holden,M.T., Sebaihia,M., Baker,S., Basham,D., Brooks,K., Chillingworth,T., Connerton,P., Cronin,A., Davis,P., Davies,R.M., Dowd,L., White,N., Farrar,J., Feltwell,T., Hamlin,N., Haque,A., Hien,T.T., Holroyd,S., Jagels,K., Krogh,A., Larsen,T.S., Leather,S., Moule,S., O’Gaora,P., Parry,C., Quail,M., Rutherford,K., Simmonds,M., Skelton,J., Stevens,K., Whitehead,S. and Barrell,B.G. (2001) Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature, 413, 848–852. [DOI] [PubMed] [Google Scholar]
- 37.Wei J., Goldberg,M.B., Burland,V., Venkatesan,M.M., Deng,W., Fournier,G., Mayhew,G.F., Plunkett,G.,III, Rose,D.J., Darling,A., Mau,B., Perna,N.T., Payne,S.M., Runyen-Janecky,L.J., Zhou,S., Schwartz,D.C. and Blattner,F.R. (2003) Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun., 71, 2775–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Velten M., McGovern,S., Marsin,S., Ehrlich,S.D., Noirot,P. and Polard,P. (2003) A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol. Cell, 11, 1009–1020. [DOI] [PubMed] [Google Scholar]
- 39.Polard P., Marsin,S., McGovern,S., Velten,M., Wigley,D.B., Ehrlich,S.D. and Bruand,C. (2002) Restart of DNA replication in Gram-positive bacteria: functional characterisation of the Bacillus subtilis PriA initiator. Nucleic Acids Res., 30, 1593–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soultanas P. (2002) A functional interaction between the putative primosomal protein DnaI and the main replicative DNA helicase DnaB in Bacillus. Nucleic Acids Res., 30, 966–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bell S.P. and Dutta,A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem., 71, 333–374. [DOI] [PubMed] [Google Scholar]
- 42.Saluja D. and Godson,G.N. (1995) Biochemical characterization of Escherichia coli temperature-sensitive dnaB mutants dnaB8, dnaB252, dnaB70, dnaB43 and dnaB454. J. Bacteriol., 177, 1104–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rain J.C., Selig,L., DeReuse,H., Battaglia,V., Reverdy,C., Simon,S., Lenzen,G., Petel,F., Wojcik,J., Schachter,V., Chemama,Y., Labigne,A. and Legrain,P. (2001) The protein–protein interaction map of Helicobacter pylori. Nature, 409, 211–215. [DOI] [PubMed] [Google Scholar]