Abstract
DNA polymerase (pol) λ is homologous to pol β and has intrinsic polymerase and terminal transferase activities. However, nothing is known about the amino acid residues involved in these activites. In order to precisely define the nucleotide-binding site of human pol λ, we have mutagenised two amino acids, Tyr505 and the neighbouring Phe506, which were predicted by structural homology modelling to correspond to the Tyr271 and Phe272 residues of pol β, which are involved in nucleotide binding. Our analysis demonstrated that pol λ Phe506Arg/Gly mutants possess very low polymerase and terminal transferase activities as well as greatly reduced abilities for processive DNA synthesis and for carrying on translesion synthesis past an abasic site. The Tyr505Ala mutant, on the other hand, showed an altered nucleotide binding selectivity to perform the terminal transferase activity. Our results suggest the existence of a common nucleotide-binding site for the polymerase and terminal transferase activities of pol λ, as well as distinct roles of the amino acids Tyr505 and Phe506 in these two catalytic functions.
INTRODUCTION
DNA polymerase (pol) λ is a nuclear enzyme that has been assigned, based on sequence homology, to the family X polymerases, comprising pol β, pol µ and terminal transferase (TdT) (1). The gene encoding the novel pol λ was cloned and mapped to mouse chromosome 19 and to human chromosome 10. Furthermore, isolation of endogenous pol λ from calf thymus has been described (2). Pol λ has 32% amino acid identity to pol β and contains all four pol β structural subdomains, named fingers, palm, thumb and the 8 kDa 5′-deoxyribose phosphate lyase (dRP lyase) domain. How ever, the high homology of pol λ with pol β is exclusively confined to the C-terminal half of the protein (amino acids 244–575) containing the catalytic site. Pol λ contains an intrinsic dRP lyase activity and can substitute for pol β in in vitro base excision repair (3). Both the human and the calf thymus enzymes were able to synthesise DNA on a template containing abasic (AP) sites with the same efficiency as on undamaged DNA, thus suggesting a potential role of pol λ in translesion synthesis (2,4). Moreover, it has recently been demonstrated that human pol λ interacts functionally and physically with proliferating cell nuclear antigen (PCNA). This interaction stabilises the binding of pol λ to the primer template, thus increasing its affinity for the hydroxyl primer and its processivity in DNA synthesis, without affecting the nucleotide incorporation rate. PCNA also stimulated efficient synthesis by pol λ across an AP site leading to elongation past the lesion (4).
Beside pol β and pol λ, other members of polymerase family X are TdT and pol µ (5). Interestingly, whereas pol β does not possess terminal transferase activity, both pol λ and pol µ are able to add nucleotides to the 3′-end of a single-stranded (ss)DNA (6,7). These different properties of pol β and pol λ might reflect a different architecture of the active site. Sequence alignments of the members of the polymerase X family showed two long motifs of conserved residues, which have been shown to constitute the dNTP-binding site in the crystal structures of pol β and TdT (8–13). The first motif is LYFTGS in human pol β (amino acids 271–275) and human pol λ (amino acids 504–509) and LGWTGS (amino acids 447–452) in human TdT. Essential roles in dNTP binding and catalysis have been shown for the pol β residues Y271 and F272 (12,14,15). These residues correspond to Y505 and F506 of human pol λ and to G449 and W450 of human TdT. Additional important amino acids involved in nucleotide binding by pol β are R254 and R258 (8), the latter being adjacent to the F272 side chain in the crystal structure of pol β complexed with its substrates. Interestingly, homology modelling revealed that the putative nucleotide-binding pocket of pol λ has a more hydrophobic character with respect to that of pol β, since the pol β residue R258 is replaced in pol λ by the moderately hydrophobic residue I492, which lies close to F506.
Since it is not known whether the same residues of pol λ are responsible for both the template-dependent (i.e. DNA polymerase) and template-independent (i.e. terminal transferase) activities, we have generated three pol λ mutants: Y505A, F506G and F506R. While the first two (Y505A and F506G) allowed us to test the effects of the lack of the corresponding aromatic side chains, the F506R mutant was generated to test the effect, if any, of replacement of the Phe ring with a positively charged chain within the hydrophobic nucleotide-binding pocket of pol λ. We have analysed the effects of these mutations on both the DNA polymerisation and terminal transferase activities of pol λ. Our data suggest a common nucleotide-binding site for both the DNA polymerase and terminal transferase functions of pol λ and that the Y505A and F506G/R mutations affect these two activities differently.
MATERIALS AND METHODS
Chemicals
[3H]dTTP (30 Ci/mmol), [3H]dCTP (18 Ci/mmol), [3H]dUTP (19 Ci/mmol), [3H]dATP (73 Ci/mmol) and [γ-32P]ATP (3000 Ci/mmol) were from Amersham. Unlabelled dNTPs, poly(dA) and oligo (dT)12–18 were from Roche Molecular Biochemicals. The tetrahydrofuran (dSpacer) was from Glen Research. Activated calf thymus DNA was prepared as described (16). Whatman was the supplier of the GF/C filters. All other reagents were of analytical grade and purchased from Merck or Fluka.
Nucleic acid substrates
The d73mer, either undamaged or containing the synthetic (tetrahydrofuran) AP site and the corresponding primers were chemically synthesized and purified on denaturing polyacrylamide gels (4). The sequence of d73mer is 5′-GATCGGGA GGGTAGGAATATTGAG[X/G]AGTGGAGATAGTGGAGGGTAGTATGGTGGATA-3′. The sequences complementary to the 17mer and 18mer primers are singly and double underlined, respectively. The position of the lesion or the corresponding G residue in the undamaged template are indicated in bold in brackets (X, AP site). The sequence of the 66mer single-stranded oligonucleotide used as a substrate for the terminal transferase assays was 5′-AGGATGTATGTTTAGTAGGTACATAACTATCT ATTGATACAGACCTAAAACAAAAAATTTTCCGAG-3′.
Enzymes and proteins
Site-directed mutations were introduced into a human pol λ overexpression plasmid (pRSETb-hpol λ) by a PCR-based method with the following oligonucleotides: Y505A-forward, 5′-CGCCGCTAGCCTTCACCGGCTCTGCACAC (NheI); Y505A-reverse, 5′-CGCCGCTAGCAGGGCACAGGCAAA CTC (NheI); F506R-forward, 5′-CGCCCGTACGGGCTCTG CACACTTCAACCGC (BsiWI); F506R-reverse, 5′-CGCCC GTACGGTAGAGCAGGGCACAGGCAA (BsiWI); F506G-forward, 5′-CTGAGGTACCGGCTCTGCACACTTCAA (KpnI); F506G-reverse, 5′-CTGAGGTACCGTAGAGCAGGGCAC AGGCA (KpnI); wt pol λ, forward, 5′-CGCCGGATCCCA GGGGTATCTTGAAG (BamHI); wt pol λ, reverse, 5′-CCG GAATTCTCACCAGTCCCGCTCAGCAG (EcoRI). Restric tion sites used for further cloning are in italic and changed codons are in bold. PCR products were double digested with the appropriate enzymes, agarose gel purified and ligated into plasmid pRSETb (BamHI and EcoRI cloning sites). Pfu DNA polymerase used for PCR was from Promega. Sequencing of Y505A, F506G and F506R pol λ mutants was performed with a BigDye Terminator cycle sequencing kit (Perkin-Elmer) and analysed in an ABI 310 automated sequencer. Recombinant human wild-type (wt) pol λ and the Y505A, F506G and F506R pol λ mutants were expressed and purified as described (7). After purification, the proteins were >90% homogeneous, as judged by SDS–PAGE and Comassie staining, and had specific activities of 200 000 U/mg for pol λ wt, 80 000 U/mg for the Y505A pol λ mutant, 10 700 U/mg for the F506G pol λ mutant and 11 000 U/mg for the F506R pol λ mutant. One unit of polymerase activity corresponds to the incorporation of 1 pmol total dTMP into acid-precipitable material in 60 min at 37°C in a standard assay containing 0.5 µg (as nucleotides) of poly(dA)·oligo(dT)10:1 and 10 µM dTTP. Recombinant human PCNA was isolated as described (4).
Enzymatic assays
DNA polymerase assay. Pol λ activity on poly(dA)· oligo(dT)10:1 was assayed in a final volume of 25 µl containing 50 mM Tris–HCl (pH 7.0), 0.25 mg/ml bovine serum albumin (BSA), 1 mM dithiothreitol (DTT), 0.5 mM MnCl2, 0.2 µM poly(dA)·oligo(dT)10:1 (3′-OH ends), 50 nM pol λ and 5 µM [3H]dTTP (5 Ci/mmol), unless otherwise indicated in the figure legends. All reactions were incubated for 15 min at 37°C unless otherwise stated and the DNA precipitated with 10% trichloroacetic acid. Insoluble radioactive material was determined by scintillation counting as described (16). For denaturing gel analysis, the reaction mixture included 50 mM Tris–HCl (pH 7.0), 0.25 mg/ml BSA, 1 mM DTT, 0.5 mM MnCl2. Enzymes, unlabelled dNTPs and template were as indicated in the figure legends. For the processivity assay, the reactions were performed in the presence of 40-fold molar excess of poly(dA)·oligo(dT) as a trap (4) over the labelled template.
Terminal transferase assay. Pol λ terminal transferase activity on a single-stranded 66mer oligonucleotide was assayed in a final volume of 25 µl containing 50 mM Tris–HCl (pH 7.0), 0.25 mg/ml BSA, 1 mM DTT, 0.5 mM MnCl2, 0.2 µM of single-stranded 66mer (3′-OH ends). Pol λ and [3H]dNTPs (10 Ci/mmol) were added as indicated in the figure legends. All reactions were incubated for 15 min at 37°C unless otherwise stated and the DNA precipitated with 10% trichloroacetic acid. Insoluble radioactive material was determined by scintillation counting as described (16).
Steady-state kinetic data analysis
For kinetic analysis, enzymatic activity was measured in the presence of 25 nM pol λ wt and Y505A and 75 nM pol λ F506G and F506R and increasing concentrations of DNA or nucleotide substrate. For dTTP incorporation, concentrations tested were 0.1, 0.2, 0.75, 1.25, 5, 10 and 25 µM. For DNA substrate utilisation, poly(dA)·oligo(dT) concentrations used (as 3′-OH ends) were 10, 20, 40, 80 and 200 nM. Enzymatic activity was measured at the 16 min time point, which corresponded to the mid-point of the linear phase of the reaction, as determined by time-dependent incorporation experiments. The Km and Vmax values were calculated by plotting the initial velocity dependence on substrate concentration and fitting the data to the Michaelis–Menten equation in the form:
v = (kcatE0)/{1 + (Km/[S])}
where kcatE0 = Vmax.
For quantification of the reaction products after separation on sequencing gels, the products were calculated from the values of integrated gel band intensities I*T/IT – 1, where T is the target site, the template position of interest, and I*T is the sum of the integrated intensities at positions T, T + 1 … T + n
All the intensity values were normalised to the total intensity of the corresponding lane to correct for differences in gel loading. An empty lane was scanned and the corresponding value subtracted as background.
RESULTS
Substitutions of Y505 and F506 have different effects on the polymerase activity of DNA polymerase λ
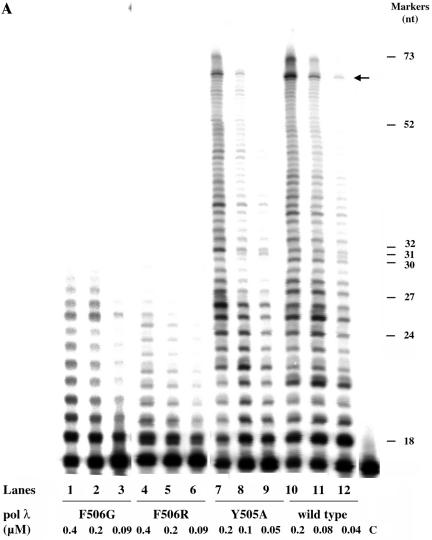
Human pol λ, either wild-type or the Y505A, F506G or F506R mutant, was titrated in DNA polymerase assays with a 5′-end-labelled 17:73 primer/template DNA oligonucleotide substrate. All the reactions were performed in the presence of Mn2+, which has been shown to be the optimal divalent cation for pol λ (21), and the products were resolved by sequencing gel analysis. As shown in Figure 1A, both the F506 pol λ mutants (lanes 1–6) displayed a strongly reduced activity compared to the wild-type (lanes 10–12). On the other hand, the pol λ Y505A mutant (lanes 7–9) showed only a moderate reduction in catalytic activity at low enzyme concentrations (compare lanes 8 and 9 to lanes 11 and 12), but nearly identical activity at high concentrations (compare lane 7 with lane 10). These results indicated a highly distributive mode of synthesis by pol λ, as already reported (4). Both the pol λ wt and the Y505A mutant showed a strong stop site around positions +42 to +45 (giving a product of ∼59–62 nt, indicated by the arrow), corresponding to a G-rich sequence. The kinetic parameters for nucleotide incorporation and DNA binding were determined for the four enzymes as described in Materials and Methods, and are summarised in Table 1. The pol λ F506R and F506G mutants showed a 50- to 100-fold decrease in the apparent kcat value with respect to the wild-type. Interestingly, the F506G mutant had nearly 30-fold lower Km values for both the nucleotide and the DNA substrates with respect to the wild-type enzyme, resulting in similar efficiencies of substrate utilisation (kcat/Km values). On the other hand, the F506R substitution did not have any significant effect on the apparent Km value for the nucleotide substrate with respect to pol λ wt, but showed a 27-fold decrease in the Km value for the 3′-OH primer end with respect to the wild-type enzyme. As a result, the F506R mutant displayed a 77-fold reduction in nucleotide utilisation efficiency (kcat/Km values), whereas the efficiency of DNA substrate utilisation was reduced only 4-fold. The pol λ Y505A mutant, on the other hand, showed only a 3-fold reduction in its apparent kcat value, with nucleotide and DNA substrate utilisation efficiencies nearly identical to pol λ wt.
Figure 1.
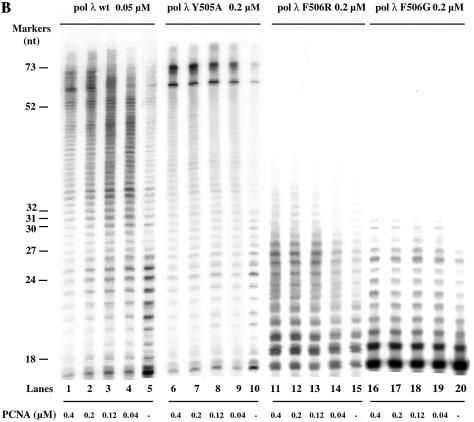
The F506G/R mutations reduce DNA polymerase λ DNA synthesis activity. (A) Titration of pol λ wt, Y505A, F506R and F506G on a 5′-end-labelled 17:73mer primer template oligonucleotide (see Materials and Methods for details of the sequence). Reactions were carried out under the conditions described in Materials and Methods, in the presence of 50 nM oligonucleotide substrate (3′-OH ends) and increasing concentrations of pol λ F506G (lanes 1–3), F506R, (lanes 4–6), Y505A (lanes 7–9) and wild-type (lanes 10–12). C, control reaction without added enzyme. The concentrations of the enzymes are indicated at the bottom of the panel. The size of the molecular weight markers (in nucleotides) is indicated on the right side. The arrow indicates the position of a strong pausing site on the template strand. (B) Stimulation of the DNA synthesis activity of pol λ wt and the Y505A, F506G and F506R mutants by PCNA. Reactions were carried out under the conditions described in Materials and Methods, in the presence of 50 nM 5′-end-labelled 17:73mer primer/template oligonucleotide (3′-OH ends) and 0.05 µM pol λ wild-type (lanes 1–5), 0.2 µM Y505A (lanes 6–10), 0.2 µM F506R (lanes 11–1 5) or 0.2 µM F506G (lanes 16–20). The DNA synthesis activity was measured in the absence (lanes 5, 10, 15 and 20) or presence of increasing concentrations (0.04, 0.12, 0.2 and 0.4 µM as trimer) of PCNA. Molecular weight marker sizes (in nucleotides) are indicated on the left.
Table 1. Kinetic parameters for the DNA polymerisation reaction catalysed by DNA polymerase λ wild-type and the Y505A, F506R and F506G mutants.
| Substrate | Wild-type | Y505A | F506R | F506G | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km | kcat (min–1) | kcat/Km (min–1 µM–1) | Km | kcat (min–1) | kcat/Km (min–1 µM–1) | Km | kcat (min–1) | kcat/Km (min–1 µM–1) | Km | kcat (min–1) | kcat/Km (min–1 µM–1) | |
| dTTP | 14.3 µM | 11 (± 1) | 0.8 (± 0.1) | 6.8 µM | 3.7 (± 0.5) | 0.5 (± 0.01) | 9.7 µM | 0.1 (± 0.2) | 0.01 (± 0.02) | 0.5 µM | 0.2 (± 0.02) | 0.4 (± 0.06) |
| dUTP | 14.6 µM | 9 (± 2) | 0.6 (± 0.1) | 2.5 µM | 2.6 (± 0.5) | 1 (± 0.1) | n.da | n.d. | n.d. | n.d | n.d. | n.d. |
| 3′-OH | 270 nM | 8.7 (± 0.7) | 32 (± 5) | 82 nM | 2.8 (± 6) | 34 (± 4) | 10 nM | 0.08 (± 0.01) | 8 (± 1) | 8.5 (nM) | 0.23 (± 0.02) | 27 (± 1) |
an.d., not determined.
The DNA polymerase λ F506G and F506R mutants show reduced processivity with respect to both the wild-type and the Y505A mutant
Human pol λ has been shown to interact with PCNA, which stimulates its processivity (4). In order to test the responsiveness of the mutant enzymes to PCNA, pol λ wt and the Y505A, F506G and F506R mutants were tested in the absence or presence of increasing concentrations of human PCNA, under distributive conditions. As shown in Figure 1B, PCNA stimulated synthesis by pol λ wt and the Y505A mutant in a very similar manner (compare lanes 1–5 with lanes 6–10). On the other hand, the pol λ F506G and F506R mutants were severly impaired in their polymerase activity compared to the wild-type, synthesising products of 10–15 nt in both the absence and presence of PCNA (compare lane 15 with lanes 11–14 and lane 20 with lanes 16–19). Due to the experimental conditions used, the products shown in Figure 1B were synthesised in distributive mode (i.e. with multiple primer turnover by the enzyme). In order to more precisely assess the impact of PCNA on processivity of the enzyme, we tested the four different pol λ (wt, Y505A, F506G and F506R) under processive (i.e. single primer turnover) conditions.
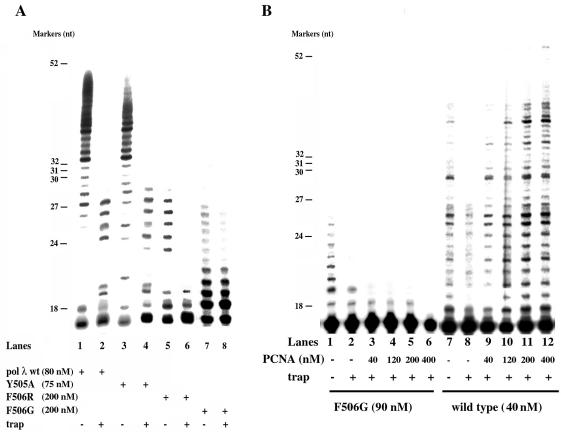
The approach used consisted of measuring DNA synthesis on a 5′-end-labelled synthetic 18:73mer primer/template DNA oligonucleotide substrate, in the absence or presence of an excess of unlabelled poly(dA)·oligo(dT) trapping substrate. When challenged with the trapping DNA, all the polymerase molecules that dissociated from the oligonucleotide substrate should be sequestered by the cold poly(dA)·oligo(dT). Thus, the number of nucleotides incorporated in the presence of the trap should be equal to the processivity of the enzyme, defined as the number of incorporation events for a single DNA binding event. The effectiveness of this method has been proven previously with pol λ wt (4). The results of this experiment are shown in Figure 2A. In the absence of the trapping agent, pol λ wt (lane 1) and the pol λ Y505A mutant (lane 3) were able to synthesise full-length products, whereas the pol λ F506R and F506G mutants (lanes 5 and 7, respectively) stopped after ∼10–11 incorporation events. In the presence of the trap, pol λ wt and the pol λ Y505A mutant again showed comparable processivity of ∼10 nt (compare lane 2 with lane 4). On the other hand, the pol λ F506R mutant showed a processivity of 1–2 nt (lane 6) and the F506G of 3–4 nt only (lane 8). We next tested the effect of PCNA on the processivity of pol λ wt and the F506G mutant only. As shown in Figure 2B, processivity of the F506G mutant could not be stimulated by increasing amounts of PCNA (compare lane 2 with lanes 3–6), whereas stimulation was observed with the wild-type enzyme (compare lane 8 with lanes 9–11). Since PCNA has been shown to increase the stability of the enzyme:primer complex, without affecting its rate of nucleotide incorporation, these results suggest that the main defect induced by the F506G mutation was a reduction of the catalytic rate.
Figure 2.
Effect of the Y505A, F506G and F506R mutations on the processivity of DNA polymerase λ. (A) The F506G/R mutations reduce the processivity of pol λ. Reactions were carried out under the conditions described in Materials and Methods, in the presence of 50 nM 5′-end-labelled 18/73mer primer-template oligonucleotide (see Materials and Methods for details on the sequence) and 0.08 µM pol λ wt (lanes 1 and 2), 0.075 µM Y505A (lanes 3 and 4), 0.2 µM F506R (lanes 5 and 6) or 0.2 µM F506G (lanes 7 and 8). DNA synthesis was measured in the absence (lanes 1, 3, 5 and 7) or presence (lanes 2, 4, 6 and 8) of 2 µM (3′-OH ends) unlabelled poly(dA)·oligo(dT) as a trapping agent. Molecular weight marker sizes (in nucleotides) are indicated on the left. (B) PCNA does not rescue the defect in the processivity of pol λ F506G. Reactions were carried out under the conditions described in Materials and Methods, in the presence of 50 nM 5′-end-labelled 18/73mer primer-template oligonucleotide (3′-OH ends) and 0.09 µM F506G mutant (lanes 1–6) or 0.04 µM pol λ wild-type (lanes 7–12). DNA synthesis was measured in the absence (lanes 1 and 7) or presence (lanes 2–6 and 8–12) of 2 µM (3′-OH ends) unlabelled poly(dA)·oligo(dT) as a trapping agent and in the absence (lanes 1, 2, 7 and 8) or presence (lanes 3–6 and 9–12) of increasing amounts of PCNA, as indicated at the bottom of the panel.
The DNA polymerase λ F506G mutant has a reduced ability to bypass an abasic site
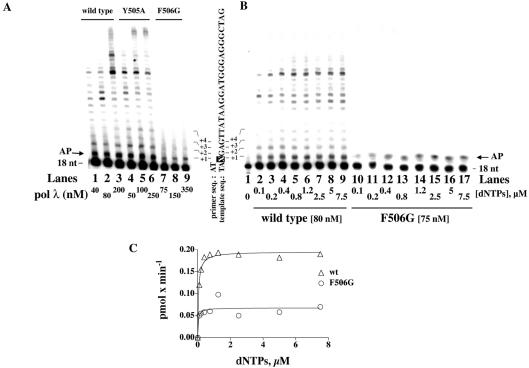
Pol λ has been shown to be able to synthesise DNA across an AP site lesion on the template strand (2,4). Pol λ wt and the Y505A and F506G mutants were titrated under standing start conditions, using a 18/73mer primer/template DNA oligonucleotide substrate. The 18mer primer was annealed to nucleotides 26–44 of the 73mer template, thus leaving a single-stranded 5′-tail of 25 nt available for synthesis (see Materials and Methods for details). The 73mer oligonucleotide contained a single synthetic AP site (tetrahydrofuran moiety) in place of the G at position 25. Thus, upon annealing of the 18mer primer, an abasic site resulted at position +1 of the template. As shown in Figure 3A, in the presence of all four dNTPs pol λ wt (lanes 1–3) and the Y505A mutant (lanes 4–6) were able to synthesise a full-length product starting from the lesion. On the other hand, the F506G mutant aborted DNA synthesis after incorporation of one nucleotide (lanes 7–9), corresponding to the position of the AP site. These results suggested that the F506G mutant, albeit able to incorporate one nucleotide in front of the lesion, was severly impaired in its ability to elongate the resulting primer end. Next, pol λ wt and the F506G mutant were tested on the same 18/73mer substrate with the lesion, in the presence of increasing concentrations of all four dNTPs. As shown in Figure 3B, while the wild-type enzyme was able to catalyse both incorporation in front of the lesion and subsequent elongation (lanes 2–9) independent of the dNTP concentration, the F506G mutant could only add one nucleotide at the position corresponding to the AP site and was unable to further elongate the nascent DNA chain, even at high dNTPs concentrations (lanes 10–17). When total DNA synthesis (calculated from the intensities of the bands) was plotted as a function of the nucleotide substrate concentration (Fig. 3C), pol λ wt showed an apparent maximal rate of elongation of 0.32 min–1, whereas the F506G mutant exhibited a rate of 0.05 min–1.
Figure 3.
The DNA polymerase λ F506G mutant shows a reduced ability to bypass an abasic site. (A) AP site bypass under standing start conditions. Reactions were carried out under the conditions described in Materials and Methods, in the presence of 50 nM 5′-end-labelled 18/73mer primer-template oligonucleotide (see Materials and Methods for details of the sequence) containing a single synthetic AP site at position +1. Pol λ wt (lanes 1–3), Y505A (lanes 4–6) or F506G (lanes 7–9) were titrated in the presence of 5 µM dNTPs. The concentrations of the enzymes are indicated at the bottom of the panel. The arrow on the left indicates the position of the product resulting from incorporation of a nucleotide in front of the lesion. The sequence of the template is shown on the right side, with the X corresponding to the AP site. (B) Dependence on nucleotide substrate concentration of AP site bypass by pol λ wt and the F506G mutant, under standing start conditions. Reactions were carried out under the conditions described in Materials and Methods, in the presence of 50 nM 5′-end-labelled 18/73mer primer-template oligonucleotide (see Materials and Methods for details of the sequence) containing a single synthetic AP site at position +1. Pol λ wild-type (80 nM, lanes 2–9) or F506G (75 nM, lanes 10–17) was tested in the presence of increasing concentrations of nucleotides, as indicated at the bottom of the panel. The sequence of the template strand is shown on the left of the panel, with the X corresponding to the AP site. The arrow on the right side indicates the position of the product resulting from incorporation of a nucleotide in front of the lesion. (C) The bands of the gel shown in (B) were quantified by scanning densitometry. Since the concentration of the input substrate was known, the relative intensities, normalised for the total intensity of each lane, were converted into apparent velocities (pmols min–1), which were then plotted against the nucleotide substrate concentrations. Curves were fitted to the Michelis–Menten equation as described in Materials and Methods.
The terminal transferase activity of DNA polymerase λ is reduced in the mutant Y505A and abolished in the mutants F506G/R
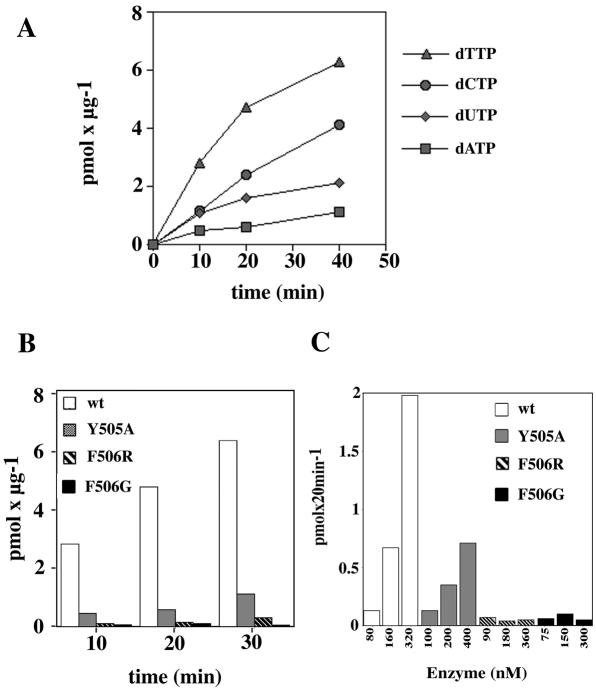
We have recently shown that pol λ has intrinsic terminal transferase activity, with a preference for pyrimidine nucleotides (7). This is also evident from the time–course experiments shown in Figure 4A, where the terminal transferase activity of pol λ wt was tested with a 66mer ssDNA oligonucleotide as substrate, in the presence of different radioactively labelled deoxynucleotides. Pol λ incorporated all three pyrimidine nucleotides (dTTP, dCTP and dUTP) with higher efficiency than the purine dATP, in accordance with previous data (7). Next, the pol λ mutants Y505A, F506G and F506R were compared under the same conditions to pol λ wt in the presence of labelled dTTP. As can be seen from Figure 4B and C, the Y505A mutant showed a reduced ability to incorporate dTTP, compared to the wild-type enzyme, whereas the mutants F506G and F506R showed almost no detectable activity. These results indicated that the two pol λ residues Y505 and F506 are important for polymerisation in both the DNA polymerase and terminal transferase activities.
Figure 4.
The DNA polymerase λ Y505A and F506G/R mutants have reduced terminal transferase activity. (A) Time course of the terminal transferase activity of pol λ wt (0.2 µM) on a single-stranded 66mer oligonucleotide substrate. Terminal transferase activity was measured under the conditions described in Materials and Methods, in the presence of 10 µM labelled dTTP (triangles), dCTP (circles), dUTP (rhombi) or dATP (squares). (B) Time course of the terminal transferase activity of pol λ wt (0.2 µM, white boxes), Y505A (0.2 µM, grey boxes), F506R (0.4 µM, striped boxes) or F506G (0.4 µM, black boxes) on a single-stranded 66mer oligonucleotide substrate. Terminal transferase activity was measured under the conditions described in Materials and Methods, in the presence of 10 µM labelled dTTP. (C) Titration of pol λ wt (white boxes), Y505A (grey boxes) F506R (striped boxes) and F506G (black boxes) on a single-stranded 66mer oligonucleotide substrate. Terminal transferase activity was measured under the conditions described in Materials and Methods, in the presence of 10 µM labelled dTTP.
The DNA polymerase λ Y505A mutant specifically reduces pyrimidine nucleotide utilisation by the terminal transferase activity
Pol λ wt and pol λ Y505A were compared in a terminal transferase assay for their ability to incorporate dTTP or dATP in the presence of either Mn2+ or Mg2+ as the metal activators. Both metals were tested since it is known that the nature of the divalent ion can influence the nucleotide specificity of polymerases. In particular, pol λ has recently been shown to utilise both Mg2+ and Mn2+, but with very different efficiencies (18). As shown in Figure 5A and B, the terminal transferase activity of pol λ, either wild-type or the mutant, showed a clear preference for Mn2+ as the activating metal. However, in the presence of both Mn2+ and Mg2+, the pol λ Y505A mutant showed reduced dTTP incorporation with respect to pol λ wt, whereas the incorporation of dATP was comparable for the two enzymes. In order to better understand the molecular basis for this effect, a detailed kinetic and thermodynamic analysis was performed for the incorporation of pyrimidine (dTTP and dUTP) and purine (dATP) nucleotides by the terminal transferase activity of pol λ wt and the Y505A mutant. The results are summarised in Table 2. The apparent rate of incorporation (kcat) of pol λ wt was 4- and 3-fold higher for dTTP and dUTP, respectively, than for dATP, thus explaining the observed preference for pyrimidine nucleotides as substrates for the terminal transferase activity. Pol λ Y505A, on the other hand, showed comparable efficiency of dATP incorporation (kcat/Km values) with respect to the wild-type, but a 3- to 4-fold reduction in the incorporation efficiencies of dTTP and dUTP. This effect was mainly due to differences in the Km values for the pyrimidine nucleotides of the pol λ Y505A mutant with respect to pol λ wt. Interestingly, comparison of the kinetic parameters for dTTP and dUTP incorporation for the DNA polymerisation activity reported in Table 1 with the corresponding values for the terminal transferase activity reported in Table 2 revealed that the difference between pol λ wt and the Y505A mutant for pyrimidine utilisation was specific for the terminal transferase activity.
Figure 5.
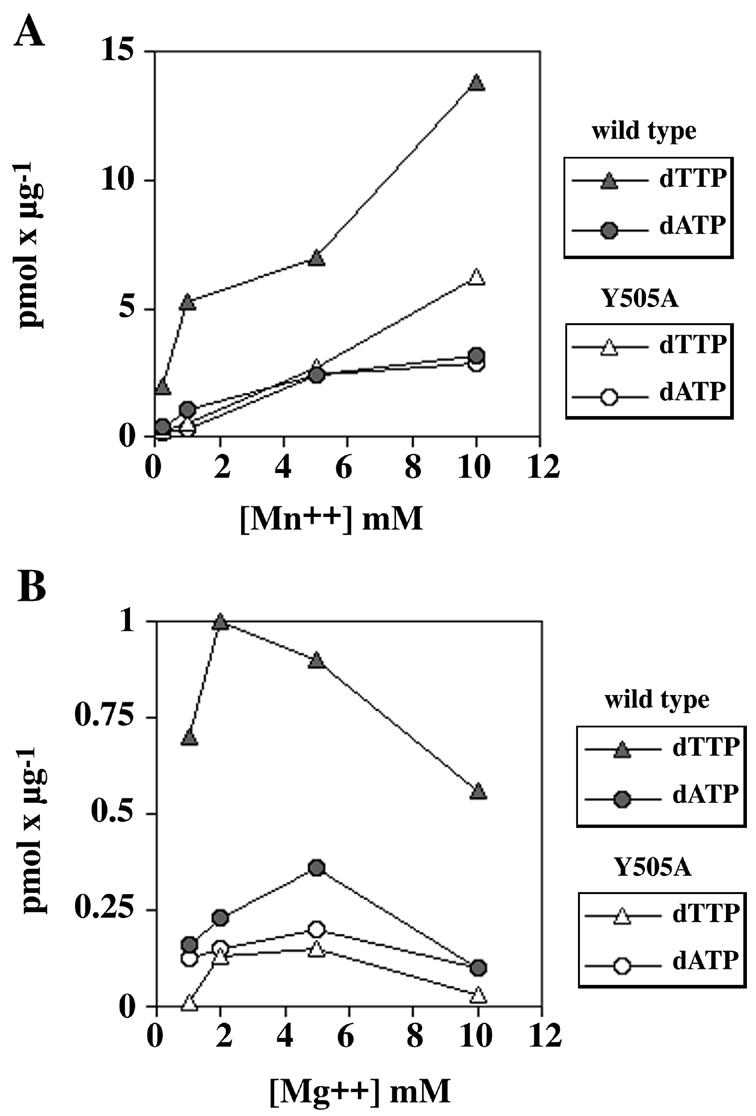
The DNA polymerase λ Y505A mutation specifically reduces the efficiency of pyrimidine utilisation by its terminal transferase activity. (A) Dependence of the terminal transferase activity of pol λ wt (0.2 µM, filled symbols) or Y505A (0.2 µM, open symbols) on Mn2+ as the activating metal ion. Terminal transferase activity was measured under the conditions described in Materials and Methods, in the presence of 10 µM labelled dTTP (triangles) or dATP (circles). (B) Dependence of the terminal transferase activity of pol λ wt (0.2 µM, filled symbols) or Y505A (0.2 µM, open symbols) on Mg2+ as the activating metal ion. Terminal transferase activity was measured under the conditions described in Materials and Methods, in the presence of 10 µM labelled dTTP (triangles) or dATP (circles).
Table 2. Kinetic parameters for the incorporation of pyrimidine and purine deoxynucleotides by the terminal transferase activity of DNA polymerase λ wild-type and the Y505A mutant.
| dATP | dTTP | dUTP | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Km (µM) | kcat (min–1) | kcat/Km (min–1 µM–1) | Km (µM) | kcat (min–1) | kcat/Km (min–1 µM–1) | Km (µM) | kcat (min–1) | kcat/Km (min–1 µM–1) | |
| wt | 25(± 2) | 0.12(± 0.01) | 0.005(± 0.0006) | 14(± 3) | 0.08(± 0.01) | 0.0057(± 0.0004) | 37(± 3) | 0.06(± 0.01) | 0.0016(± 0.0002) |
| Y505A | 32(± 3) | 0.09(± 0.007) | 0.003(± 0.0002) | 60(± 4) | 0.09(± 0.01) | 0.0015(± 0.0003) | 96(± 4) | 0.06(± 0.01) | 0.0006(± 0.0001) |
DISCUSSION
In mammalian cells, four members of the pol X family have been identified so far: pol β, TdT, pol λ and pol µ (5). Pol β performs template-directed incorporation of nucleotides, whereas TdT adds nucleotides to the 3′-end of a DNA strand, in a template-independent manner. Strikingly, pol λ and pol µ possess both template-directed polymerisation and terminal transferase activities (7,17). Crystal structures are available for pol β and TdT (8–13). Based on these structures and on mutagenesis studies, the residues important for nucleotide binding and catalysis have been identified. Sequence alignments with the other members of the polymerase X family showed that these residues belong to highly conserved domains. One of the conserved dNTP-binding domains is shown in Figure 6. Its sequence is LYFTGS in human pol β (amino acids 270–275) and human pol λ (amino acids 504–509) and LGWTGS (amino acids 447–452) in human TdT. In pol β, the Y271 residue has been shown to constitute the floor of the dNTP-binding pocket and was proposed to contribute to stabilisation of the enzyme–primer complex by engaging a hydrogen bond with the primer base (12). It is interesting to note that Y271 is conserved in pol λ (corresponding to Y505), but not in TdT, which bears a glycine at this position (G447). This suggests that the function of this residue is different for template-directed polymerases and for TdT. The adjacent amino acid (F272 in pol β) plays an essential role in catalysis. When pol β adopts its ‘closed’ conformation prior to catalysis, the phenyl ring of F272 disrupts the salt bridge between D192 and R258, freeing D192 to coordinate the nucleotide binding metal ion (12). Interestingly, the pol β residue R258 is not conserved in pol λ, whose corresponding amino acid is I492. However, in a structural model of the pol λ structure, based on the known structures of pol β and TdT, the F506 side chain has been predicted to be in close contact with the I492 residue, similarly to the F272 and R258 side chains in pol β. A F272L mutation has been described, which greatly increased the error rate of pol β (15). The F272 residue of pol β corresponds to W449 in human TdT. In the crystal structure of murine TdT, the aromatic ring of residue W450 was shown to be parallel and stacked with the base ring of the incoming nucleotide and also to make contacts with its sugar moiety (13). These observations suggest an important role of an aromatic residue at this position for both template-dependent and template-independent DNA polymerases.
Figure 6.
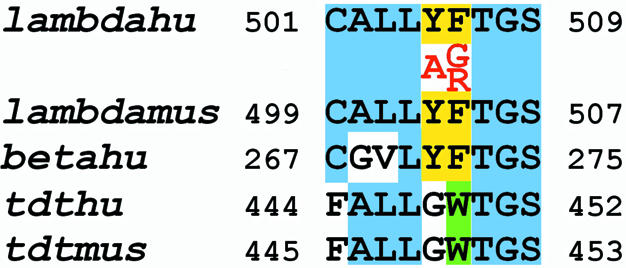
A highly conserved nucleotide-binding domain in X family DNA polymerases. The sequence of human pol λ (lambdahu) has been aligned to murine pol λ (lambdamus), human pol β (betahu), human TdT (tdthu) and murine TdT (tdtmus). The two residues considered in this study have been highlighted in yellow and the respective substitutions made by site-directed mutagenesis are indicated in red, below the human pol λ sequence. The Trp residue in the TdT sequence, which is functionally homologous to the Phe of pol λ and pol β, is highlighted in green. Light blue shading was used to box all the other amino acids conserved in at least three different enzymes. The identity within these nine amino acids is 100% between human and mouse pol λ, 100% between human and mouse TdT, 77.8% between pol λ and pol β, 66.7% between TdT and pol λ and 44.4% between TdT and pol β.
According to this prediction, the pol λ F506G/R mutants analysed in the present work showed a 50- to 100-fold reduction in the apparent catalytic rate (kcat) for template-dependent nucleotide incorporation (Table 1) and virtually undetectable terminal transferase activity (Fig. 4B and C) with respect to pol λ wt. Interestingly, we found a strong decrease in the processivity of the the pol λ F506G/R mutants, which dissociated after every two to four incorporation events (Fig. 2A), together with a concomitant increase in the apparent affinity of the mutated enzymes for the primer-template (Table 1). Since processivity is defined as the number (n) of incorporation events per dissociation event, n = kpol/koff, where kpol is the true polymerisation rate and koff is the rate of dissociation of the enzyme from the template, one possibility is that the F506G/R mutations lower both the dissociation rate (koff) and the catalytic rate (kpol), so that kpol ≈ 2–4 koff, hence n ≈ 2–4. The alternative possibility is that these mutations increased the koff rate of the enzyme from the DNA template. However, this would be in contrast to the observed increase in the affinity for the DNA template (Table 1). Moreover, in such a case we would have expected to see at least a partial rescue of the processivity upon addition of PCNA, since it has been shown that PCNA lowers the koff rate of pol λ for dissociation from the template. The experiments shown in Figures 1B and 2B, however, clearly indicated no effect of PCNA on the processivity of the F506G/R mutants.
The ability of a particular polymerase to bypass a non-instructional lesion like an AP site depends on its ability to adopt a less constrained conformation at the nucleotide-binding site (19), which usually preferentially accommodates the templating base and the complementary nucleotide. We have compared the AP translesion capacity of pol λ wt and the Y505A and F506G mutants under standing start conditions. The F506G mutant showed an impaired capacity to catalyse elongation past the lesion under these conditions (Fig. 3A). The apparent maximal rate of translesion synthesis was calculated as 0.32 min–1 for the wild-type enzyme and 0.05 min–1 for the F506G mutant. Comparison with the kcat values listed in Table 1 for normal DNA synthesis showed that pol λ wt had an ∼34-fold reduction in its catalytic rate, due to the presence of an abasic site at position +1. On the other hand, the F506G mutant showed only a 4-fold reduction in reaction rate. The mutant enzyme can catalyse only the first step of the AP site bypass process, namely incorporation in front of the lesion, but not elongation, whereas pol λ wt can do both. This suggests that, rather than incorporation in front of the lesion, the subsequent elongation is the major rate-limiting step for pol λ. Based on the increased distributivity of the F506G mutant (Fig. 2), we propose that during bypass of an abasic site by pol λ the enzyme can incorporate one nucleotide in front of the lesion without dissociating. Subsequent elongation of the resulting primer end, on the other hand, causes a major pausing of the enzyme. Due to its severely impaired catalytic efficiency and reduced processivity, the F506G mutant likely dissociates from the template during this pausing phase before having the chance to incorporate one additional nucleotide, resulting in virtually no elongation beyond the AP site. It is interesting here to note that the Y505A mutation had no detectable effect on the bypass ability of pol λ, however, as summarised in Table 1, both the apparent Km and kcat for nucleotide incorporation of the pol λ Y505A mutant were decreased with respect to the wild-type enzyme. This phenotype was very similar to that observed for the corresponding Y271A mutant of pol β (14), and might suggest a perturbation of the nucleotide-binding pocket.
Clear evidence for a role of the pol λ Y505 residue in nucleotide binding specificity comes from the results on the effects of this mutation on the terminal transferase activity of pol λ. As shown in Figure 4 and Table 2, the pol λ Y505A mutant had a reduced ability to catalyse the terminal addition of pyrimidine deoxynucleotides to single-stranded DNA, but showed similar efficiency to the wild-type enzyme when purine deoxynucleotides were used as substrates. The apparent catalytic rates (kcat values) for the wild-type pol λ terminal transferase activity (Table 2) were much lower than the corresponding rates for template-dependent nucleotide incorporation (Table 1). This was likely due to the very high distributivity of the terminal transferase reaction, which usually adds only a few nucleotides for each DNA binding event. Thus, at steady-state, the limiting rate of the reaction is indeed dissociation of the enzyme from the DNA substrate, hence kcat ≈ koff.
In summary, the results presented here suggest essential functional roles in both the template-dependent (i.e. DNA polymerase) and template-independent (i.e. terminal transferase) activities of pol λ for two highly conserved residues in the nucleotide-binding pocket of the X family polymerases. The F506 residue appears to have a critical role in catalysis, whereas the Y505 residue seems to be critical for the specificity of nucleotide incorporation by the pol λ terminal transferase activity.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the Swiss National Science Foundation (grant 3100.061361.00) to K.R., the EU (project no. QLK3-CT-202-02071 REBIOTECH) to U.H. and G.M. and the BBW (02.2286) to I.S. and grant 4373 from the Association pour la Recherche sur le Cancer to G.V.
REFERENCES
- 1.Garcia-Diaz M., Dominguez,O., Lopez-Fernandez,L.A., de Lera,L.T., Saniger,M.L., Ruiz,J.F., Parraga,M., Garcia-Ortiz,M.J., Kirchhoff,T., del Mazo,J. et al. (2000) DNA polymerase λ (Pol λ), a novel eukaryotic DNA polymerase with a potential role in meiosis. J. Mol. Biol., 301, 851–867. [DOI] [PubMed] [Google Scholar]
- 2.Ramadan K., Shevelev,I.V., Maga,G. and Hubscher,U. (2002) DNA polymerase λ from calf thymus preferentially replicates damaged DNA. J. Biol. Chem., 277, 18454–18458. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Diaz M., Bebenek,K., Sabariegos,R., Dominguez,O., Rodriguez,J., Kirchhoff,T., Garcia-Palomero,E., Picher,A.J., Juarez,R., Ruiz,J.F. et al. (2002) DNA polymerase lambda, a novel DNA repair enzyme in human cells. J. Biol. Chem., 277, 13184–13191. [DOI] [PubMed] [Google Scholar]
- 4.Maga G., Villani,G., Ramadan,K., Shevelev,I., Le Gac,N.T., Blanco,L., Blanca,G., Spadari,S. and Hubscher,U. (2002) Human DNA polymerase λ functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J. Biol. Chem., 277, 48434–48440. [DOI] [PubMed] [Google Scholar]
- 5.Hubscher U., Maga,G. and Spadari,S. (2002) Eukaryotic DNA polymerases. Annu. Rev. Biochem., 71, 133–163. [DOI] [PubMed] [Google Scholar]
- 6.Dominguez O., Ruiz,J.F., de Lera,L.T., Garcia-Diaz,M., Gonzalez,M.A., Kirchhoff,T., Martinez,A.C., Bernad,A. and Blanco,L. (2000) DNA polymerase µ (Pol µ), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J., 19, 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramadan K., Maga,G., Shevelev,I.V., Villani,G., Blanco,L. and Hubscher,U. (2003) Human DNA polymerase λ possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. J. Mol. Biol., 328, 63–72. [DOI] [PubMed] [Google Scholar]
- 8.Pelletier H., Sawaya,M.R., Kumar,A., Wilson,S.H. and Kraut,J. (1994) Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer and ddCTP. Science, 264, 1891–1903. [PubMed] [Google Scholar]
- 9.Sawaya M.R., Pelletier,H., Kumar,A., Wilson,S.H. and Kraut,J. (1994) Crystal structure of rat DNA polymerase β: evidence for a common polymerase mechanism. Science, 264, 1930–1935. [DOI] [PubMed] [Google Scholar]
- 10.Pelletier H., Sawaya,M.R., Wolfle,W., Wilson,S.H. and Kraut,J. (1996) A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase β. Biochemistry, 35, 12762–12777. [DOI] [PubMed] [Google Scholar]
- 11.Pelletier H., Sawaya,M.R., Wolfle,W., Wilson,S.H. and Kraut,J. (1996) Crystal structures of human DNA polymerase β complexed with DNA: implications for catalytic mechanism, processivity and fidelity. Biochemistry, 35, 12742–12761. [DOI] [PubMed] [Google Scholar]
- 12.Sawaya M.R., Prasad,R., Wilson,S.H., Kraut,J. and Pelletier,H. (1997) Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry, 36, 11205–11215. [DOI] [PubMed] [Google Scholar]
- 13.Delarue M., Boule,J.B., Lescar,J., Expert-Bezancon,N., Jourdan,N., Sukumar,N., Rougeon,F. and Papanicolaou,C. (2002) Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J., 21, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kraynov V.S., Werneburg,B.G., Zhong,X., Lee,H., Ahn,J. and Tsai,M.D. (1997) DNA polymerase β: analysis of the contributions of tyrosine-271 and asparagine-279 to substrate specificity and fidelity of DNA replication by pre-steady-state kinetics. Biochem. J., 323, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S.X., Vaccaro,J.A. and Sweasy,J.B. (1999) Involvement of phenylalanine 272 of DNA polymerase β in discriminating between correct and incorrect deoxynucleoside triphosphates. Biochemistry, 38, 4800–4808. [DOI] [PubMed] [Google Scholar]
- 16.Hubscher U. and Kornberg,A. (1979) The δ subunit of Escherichia coli DNA polymerase III holoenzyme is the dnaX gene product. Proc. Natl Acad. Sci. USA, 76, 6284–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz J.F., Dominguez,O., de Lera,L.T., Garcia-Diaz,M., Bernad,A. and Blanco,L. (2001) DNA polymerase µ, a candidate hypermutase? Philos. Trans. R. Soc. Lond. B Biol. Sci., 356, 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanca G., Shevelev,I., Ramadan,K., Villani,G., Spadari,S., Hübscher,U. and Maga,G. (2003) Human DNA polymerase λ diverged in evolution from DNA polymerase β toward specific Mn+1 dependence: a kinetic and thermodynamic study. Biochemistry, 42, 7467–7476. [DOI] [PubMed] [Google Scholar]
- 19.Taylor J.S. (2002) New structural and mechanistic insight into the A-rule and the instructional and non-instructional behavior of DNA photoproducts and other lesions. Mutat. Res., 510, 55–70. [DOI] [PubMed] [Google Scholar]