Abstract
Background
Eosinophilic esophagitis (EE) is an emerging disorder with poorly understood pathogenesis.
Objective
Whereas prior studies have primarily focused on the role of eosinophils in disease diagnosis and pathogenesis, this study investigates the involvement of mast cells.
Methods
Total and degranulated mast cell counts were correlated to microarray and RT-PCR data to generate transcriptome expression profiles related to mast cell number and degranulation in EE patients and normal controls.
Results
Esophageal mastocytosis and mast cell degranulation was readily apparent in EE patients compared with controls (p < 0.01) as assessed by staining for total mast cells and the presence of extracellular mast cell tryptase (p < 0.01). Microarray analysis revealed that mast cell levels correlated with the dysregulation of 0.8% (301 genes) of the genome which were partially distinct from the genes that correlated with tissue eosinophilia. The expression of transcripts for the mast cell proteases carboxypeptidase A3 (CPA3) and tryptase, but not chymase, correlated with mast cell levels and distinguished EE patients from controls. Suprabasilar mast cell counts (p < 0.01) and degranulation (p < 0.01) were proportional with KIT ligand mRNA expression. Treatment of EE patients with swallowed fluticasone propionate (FP) normalized levels of mast cells and the mast cell related transcriptome in responder patients.
Conclusion
Herein we have identified local mastocytosis and mast cell degranulation in the esophagus of EE patients; identified an esophageal mast cell associated transcriptome that is significantly divergent from the eosinophil-associated transcriptome with CPA3 mRNA levels serving as the best mast cell surrogate marker; and provide evidence for the involvement of KIT ligand in the pathogenesis of EE.
Keywords: Mast Cells/Basophils, Eosinophils, Human
Introduction
Eosinophilic esophagitis (EE) is an emerging worldwide disease, as documented by case series from all continents except Africa, which appears to be a growing health problem with an annual incidence of at least 1:10,000 children1–3. The primary symptoms of EE (chest and abdominal pain, dysphagia, heartburn, vomiting, and food impaction) are also observed in patients with chronic esophagitis (CE) including those patients with gastroesophageal reflux disease (GERD)4–6. However, in contrast to GERD, EE occurs more frequently in males (80%), appears to have a common familial form, has a high rate of associated atopic disease (70%), and is typically associated with a normal pH probe recording of the esophagus1;7–10. EE patients respond inadequately to anti-GERD therapy alone, but may respond to anti-inflammatory therapy and/or allergen elimination as determined by allergen testing, or empiric dietary elimination11–13.
Dissection of experimental EE models in mice has revealed that EE is triggered by both food and aeroallergens, whereas pollen exposure has been associated with cases of EE in humans, and is clearly associated with atopic disorders such as allergic rhinitis, asthma, and eczema in pediatric and adult patients12;14–17. However, nearly 25% of individuals with EE are non-atopic individuals and have no discernable evidence of allergic sensitization1;8;18;19. It is important to understand the relationship between the allergic and non-allergic variants of EE; whether allergic and non-allergic esophagitis involve similar effector pathways, such as localized mastocytosis and mast cell activation, which may have significant implications for therapeutic strategies.
Previously, whole genome wide expression analysis of esophageal tissue has uncovered a striking EE transcript signature that correlated with eosinophil levels and was similar across gender and patient age, but completely distinct from CE. Notably, the top induced transcript in EE was eotaxin-3 and levels of eotaxin-3, strongly correlated with disease severity; furthermore, a single nucleotide polymorphism in the eotaxin-3 gene was associated with disease susceptibility20;21. Additional analyses of the EE transcriptome with respect to IL-13, demonstrated a dose-dependent increase in eotaxin-3 mRNA and protein from primary esophageal epithelial cell cultures following stimulation with IL-1322 consistent with prior reports demonstrating elevated levels of antigen specific IL-5+ Th2 cells in the blood of EE patients23. Recently, a variant in TSLP was also found to be associated with EE, and may play a role in the male predominance found in this disorder, as its receptor is encoded within in a pseudoautosomal region for the X and Y chromosomes24.
The high level of eosinophils in the esophagus of EE patients, the identification of eotaxin-3, as a primary process in EE pathogenesis and the correlation of eosinophils with the degree of epithelial cell hyperplasia, all implicate the eosinophil as a primary effector cell in EE20;21. Indeed, murine models have established that eosinophils are required for induction of allergen-induced epithelial hyperplasia and remodeling in models of EE14. However, mast cells may be particularly important in disease pathogenesis as they produce an abundance of cytokines that activate eosinophils as well as molecules that directly promote tissue remodeling including fibrosis, a process that has been recently identified in EE, even in pediatric patients25;26. Recently, elevated levels of mast cells in the esophagus of EE patients have been identified; however their phenotype, regulation, and role in disease have not been explored in detail19;26–28. We now provide substantial evidence for the involvement of KIT ligand, esophageal mastocytosis, and mast cell activation in the pathogenesis of EE. In addition, we demonstrate that the esophageal genes associated with mast cell levels are distinct from those associated with eosinophil levels, at least in part, and that CPA3 mRNA serves as the best surrogate tissue marker for mast cells. Furthermore, we present molecular evidence that these processes of tissue mastocytosis and dysregulation of the mast cell transcriptomes are reversible with FP therapy and distinguishes EE from normal and CE patients.
Methods
Esophageal samples
Patients were selected without regard to age, race, atopic status or sex and their characteristics are described in Table 1. A total of 29 patients were selected for microarray analysis based on their diverse clinical features. Except when indicated, none of the patients were undergoing therapy with glucocorticoids or dietary modification. An additional 13 patients with active EE were evaluated for mast cell counts, degranulation, and the impact on mast cell transcriptomes following treatment with fluticasone propionate. These patients were treated with 880 mcg/day of swallowed fluticasone propionate, for 3 months and are described in detail in Konikoff et. al., 200627. Two biopsies from each patient were collected from the distal esophagus < 5 cm from the lower esophageal sphincter, with one sample immediately fixed in formalin prior to H&E staining and the second stored separately for microarray and RT-PCR analysis. A pathologic diagnosis from the H&E stained sample was determined based on the maximum eosinophil count per high power field (HPF) (400×, area of microscopic field = 0.22 mm2) and basal layer expansion according to established criteria and correlated with clinical findings4;8;20;29. Normal biopsies were obtained from patients that presented with symptoms consistent with GERD and EE, but whose endoscopic and histologic appearance were ultimately normal. Patients with CE were defined as having ≤ 12 eosinophils/HPF and/or mild expansion of the basal layer (<1/3 of epithelium) and not requiring treatment with either steroid or dietary therapies. EE patients were defined ≥ 24 eosinophils/HPF and extensive basal layer hyperplasia (expansion to ≥ 1/3 of epithelium) and clinical evidence of disease. The maximum eosinophil counts and thickness of basal layer were assessed following H&E staining. Mast cell counts were determined utilizing immunohistochemistry for tryptase. Biopsy samples were also assessed via staining for chymase utilizing immunohistochemistry and chloroacetate esterase activity30–32. This study was approved by the Institutional Review Board of the Cincinnati Children’s Hospital Medical Center.
Table 1. Characterization of patients used to generate mast cell transcriptomes, therapy, and distal esophageal biopsies.
Age, race, sex, atopic status, and therapy are derived from self-reported questionnaires and medical histories. Patients with any history of asthma, allergic rhinitis, or eczema were considered atopic. Average counts per HPF (400×) are based upon the maximum count noted for eosinophils and mast cells per patient esophageal sample. P-values were determined with the Student’s T-test. +compared between Normal and EE.
| Table 1. Patient Characterization |
Normal (n=10) | CE (n=6) | EE (n=13) | p-value |
|---|---|---|---|---|
| Age at collection (Y): | 6.7 ±1.2 | 10.8 ± 1.9 | 14.6 ± 3.4 | N.S. |
| Age Range: | 1.5 to 12.9 | 4.6 to 17.8 | 1.8 to 45.2 | - |
| Race: | Caucasian: 7 Native Hawaiian/Pacific Islander: 1 Other: 2 |
Caucasian: 5 Other: 1 |
Caucasian: 12 Other: 1 |
N.S. |
| Sex: | Female: 2 Male: 8 |
Female: 3 Male: 3 |
Female: 3 Male: 10 |
N.S. |
| History of atopy (%): | 5/10 (50%) | 1/6 (17%) | 10/13 (77%) | N.S. |
|
Treatments one month prior to esophageal biopsy: |
||||
| Systemic steroids: | 0 | 0 | 0 | N.S. |
| Swallowed steroids: | 0 | 0 | 0 | N.S. |
| Inhaled steroids: | 4 | 0 | 4 | N.S. |
| Leukotriene inhibitors: | 0 | 1 | 3 | N.S. |
| Acid suppressive therapy: | 4 | 0 | 6 | N.S. |
| Elimination diet: | 0 | 0 | 3 | N.S. |
| Elemental diet: | 0 | 0 | 0 | N.S. |
|
Average eosinophil count per HPF: |
0 | 5.0 ± 2.0 | 84.7 ± 19.8 | <0.01 |
|
Range of eosinophil counts per HPF: |
0 | 0 to 12 | 25 to 248 | - |
|
Average total mast cell count per HPF (Tryptase): |
0.5 ± 0.3 | 1.8 ± 0.7 | 6.9 ± 1.4 | <0.01 |
|
Range of total mast cell counts per HPF (Tryptase): |
0 to 3 | 0 to 4 | 0 to 17 | - |
|
Average degranulated mast cell count per HPF (Tryptase): |
0.2 ± 0.2 | 0.8 ± 0.5 | 3.9 ± 0.9 | <0.01 |
|
Range of degranulated mast cell counts per HPF (Tryptase): |
0 to 2 | 0 to 3 | 0 to 13 | - |
DNA microarray analysis
For each patient, a distal esophageal mucosal biopsy sample was subjected to DNA microarray as previously reported33. The genome wide Human Genome U133 Plus 2.0 Array genechip that encompasses 38,572 genes with 54,120 probesets (http://www.affymetrix.com, technical note, Part No. 701483 Rev. 2) was used, and gene transcript levels were determined using algorithms in the Microarray Analysis Suite and GeneSpring software (Agilent Technologies). A base set of probesets in transcriptome analyses was generated by requiring a minimum raw expression on the microarray of 250 (average difference) in at least 3 patient samples identifying 16,425 probesets for use in all subsequent analyses.
Ontology assessment
We subjected the list of differentially expressed transcripts to gene ontology analysis using DAVID (database for annotation, visualization and integrated discovery) and EASE (expression analysis systematic explorer), a web-based (http://david.abcc.ncifcrf.gov) application that provides access to a relational database of functional annotations34;35.
Real-time polymerase chain reaction (PCR) analysis
The RNA samples (500 ng) were subjected to reverse transcription analysis using Bioscript reverse transcriptase (Biorad) according to the manufacturer’s instructions. Human tryptase, chymase, CPA3, CXCL6, and HPRT were quantified by real-time PCR using the Roche LightCycler 480 instrument and Roche LightCycler 480 SYBR Green I master mix as a ready-to-use reaction mix. Results were then normalized to HPRT amplified from the same cDNA mix and expressed as fold induction compared to the controls. cDNA were amplified using following primers: hTryptase (70 bp): gcgatgtggacaatgatgag and tccattatggggaccttcac; hChymase (64 bp): acggaactttgtgctgacg and ggctccaagggtgactgtta; hCPA3 (94 bp) cccagatgctattgtttcccta and agaacatcagtgccaatctttg; hCXCL6 (131 bp): agccccttttctaaagaaagtca and tccagggatctccagaaaac; and hHPRT (120 bp): cagactgaagagctattgtaatg and ccagtgtcaattatatcttccac.
Immunohistochemistry
Esophageal sections were individually immunostained with anti-tryptase (Cell Marque, CMA890), as previously reported36. Immunoreactive cells were counted (400×) and are expressed as maximum mast cell number/HPF.
Mast cell number and extent of degranulation were assessed in the suprabasilar epithelium from normal, CE, and EE patients in a blinded fashion. While blinded, each esophageal sample was assessed for overall quality, and those deemed to be of inadequate quality were excluded from our final analyses with 3 or fewer high powered fields that could be evaluated. The remaining samples had between 7 to 10 high powered fields counted per subject and were used in generating the mast cell transcriptomes were quantified for the total number of mast cells and the number of degranulated mast cells per 400× HPF in the epithelium exclusive of the basal cell layer. Categorizing mast cells as intact or degranulated was dependent upon the following definitions: mast cells were counted as intact if they had discrete, focal, and well-circumscribed immunostaining for tryptase. Degranulated mast cells had discrete and relatively focal staining for tryptase but did not have well-circumscribed immunostaining (Figure 1). Up to a maximum of 10 HPF were evaluated for each patient’s sample and the maximum counts for mast cell number and degranulation were used in the subsequent analyses. For samples evaluated following therapy with fluticasone propionate, the maximum mast cell count for superficial epithelium was also counted and used for further analyses.
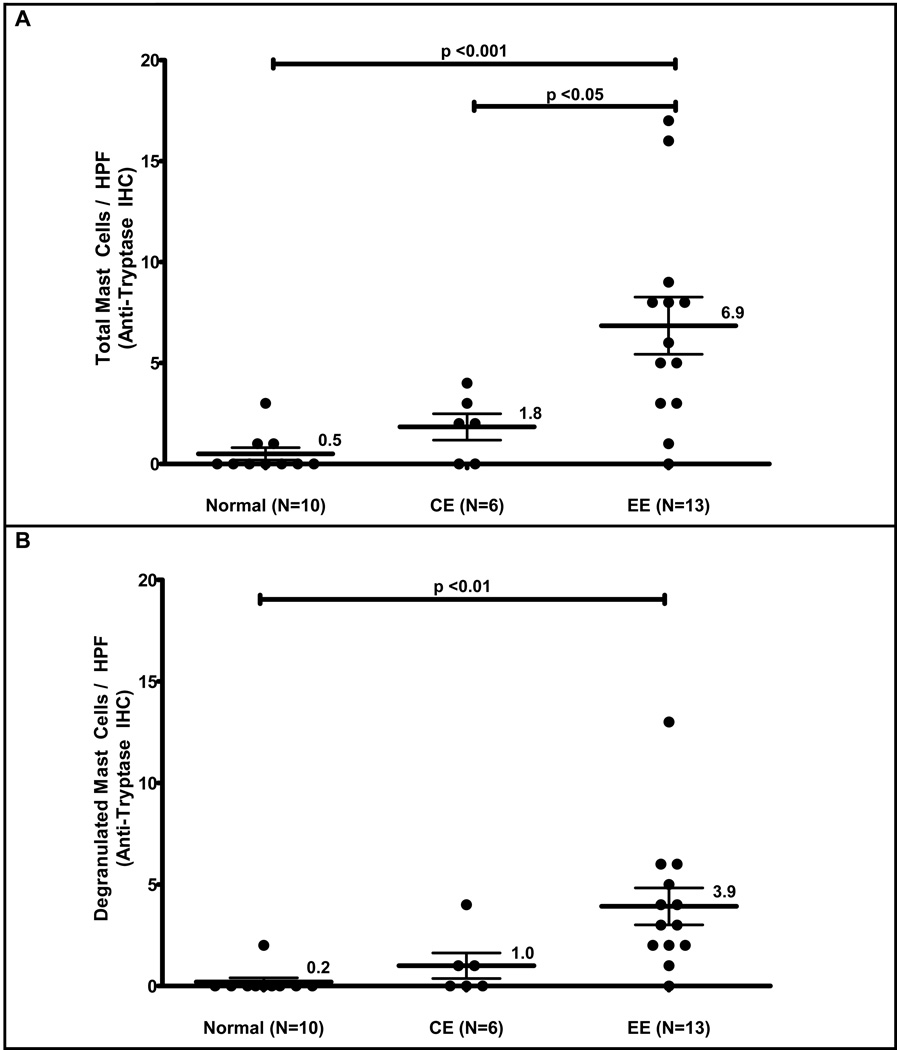
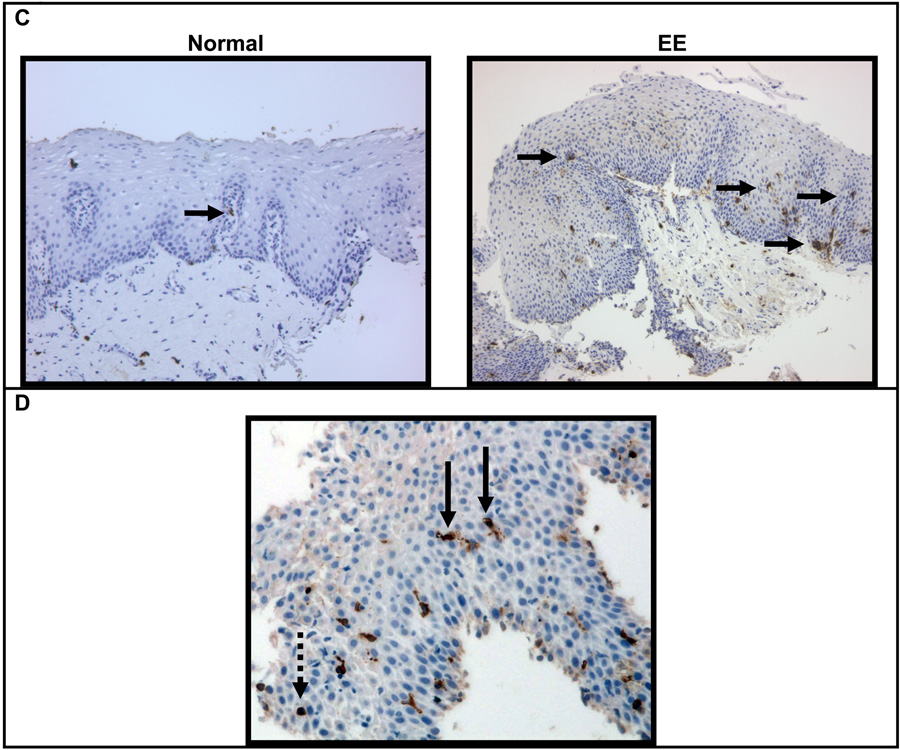
Figure 1. Mast cell counts in EE patient samples versus normal or chronic esophagitis samples.
The maximum number of tryptase positive mast cells per high power field in EE samples is increased (mean ± SEM) relative to NL, and CE patients (A). The maximum number of degranulated mast cells per high field is increased in EE relative to NL patients (B). Immunohistochemistry with anti-tryptase antibody in normal and EE patients (40×) (C). High power field (400×) from an EE patient demonstrating an intact mast cell with a dashed arrow and degranulated mast cells with solid arrows (D).
Statistical analysis
Gene lists on microarray were generated by studying differences in gene expression levels between two groups using Welch T Test with Benjamini and Hochberg false discovery rate correction21. The mast cell-related transcriptome was generated via positive and negative correlations of gene expression with epithelial mast cell counts as determined by tryptase immunohistochemistry in normal, CE, and EE patients. A p-value < 0.05 for Spearman correlations between gene expression and epithelial mast cell counts was used as a cutoff and this gene list was then filtered based upon Spearman r coefficient with fold changes in expression noted in the supplementary tables.
Results
Patient and sample characterization for generation of the mast cell transcriptomes
No significant differences were noted for patient age, race and sex between normal, CE and EE patients; however, atopy was common in patients with EE (Table 1), consistent with previous studies12;37;38. At the baseline visit, none of the patients were undergoing treatment with either systemic or swallowed steroids; however, several patients were undergoing therapy with leukotriene inhibitors, intranasal or inhaled steroids, and PPIs at the time of biopsy. Elimination diets were ongoing in three of the EE patients and none of the normal control or CE patients. None of the patients in either control group or patients with EE were undergoing therapy with an elemental diet at the time of this evaluation. The average peak eosinophil count in EE patients was 84.7 ± 19.8 and ranged between 24–248 eosinophils per HPF.
Mast cell distribution, number, and degranulation in EE
In all patients, mast cells could readily be found in peripapillary regions. In EE patients relative to normal patients, mast cells were also often found within the epithelium outside of the basal layer, both in the interpapillary locations and in the superficial epithelium outside of the papillae. The average peak mast cell count per HPF ± SEM based upon tryptase immunohistochemical staining was increased approximately 13-fold higher in patients with EE (6.9 ± 1.3 mast cell/HPF, n=13) relative to normal patients (0.5 ± 0.3 mast cell/HPF, n=10) (Figure 1A, p < 0.01). However, while peak mast cell counts in the suprabasilar epithelium were elevated in EE patients, there was overlap between normal and EE patients based on tryptase staining alone, with a range of 0 to 3 mast cells/HPF in normal patients and 0 to 17 mast cells/HPF in EE patients. Patients with CE had an intermediate level relative to both normal and EE patients with a range 0 to 4 mast cells/HPF, and an average of 1.8 ± 0.7 mast cells/HPF, n=6 (Figure 1A). The CE patients could be differentiated from EE patients averaging approximately 3-fold fewer mast cells compared with EE patients (p < 0.05), while normal patients could not be differentiated from CE patients on the basis of mast cell counts alone.
Mast cell degranulation was also assessed via tryptase staining, with nearly all EE patient samples (92%, 12/13) demonstrated evidence of degranulation. The average number of degranulated mast cells ± SEM/HPF was 0.2 ± 0.2 in normal patients, 1.0 ± 0.6 in CE patients, and 3.9 ± 0.9 in EE patients. These findings represent an approximately 20-fold increase in mast cell degranulation in EE patients versus normal patients (Figure 1B, p < 0.01), while no differences could be detected in comparisons of CE patients to either normal or EE patients. Immunohistochemistry was also performed for chymase, which could not be detected (data not shown). Furthermore, when using chloroacetate esterase staining for mast cells, which is dependent upon chymase activity, we were unable to demonstrate histochemical staining for mast cells in either EE or normal patients using standard methodologies (data not shown).
Mast cell-associated and EE-associated transcriptomes
Prior studies have demonstrated several dysregulated mast cell-related genes within the EE transcriptome20. In the current study, a mast cell related transcriptome was generated based on correlations of transcript levels from normal, CE, and EE patients with total mast cell counts as determined by tryptase immunohistochemistry (IHC). A minimum p-value < 0.05 for the Spearman correlation coefficient identified a total of 1632 genes that correlated with mast cell counts. These genes were then selected for a minimum 2-fold difference in EE patients relative to normal patients (301 genes) and represent approximately 0.8% of the entire tested genome. We define these 301 genes as the mast cell-related transcriptome (Supplementary Table 1) of which approximately 50% (148/301 genes) overlapped with of the original EE transcriptome that was based on eosinophil counts. Several genes historically related to mast cells were readily identified within this mast cell-related transcriptome including KIT ligand (KITLG), tryptase (TPSB2), cathespin C (CTSC/DPPI), carboxypeptidase A3 (CPA3), interleukin-8 receptor (IL8RB/CXCR2), phospholipase A2 (PLA2G4A), and protein phosphatase 2 (PPP2R2C)39;39–41;41–43;43;44;44;45;45;46. Consistent with the absent findings for chymase IHC and chloroacetate esterase staining, the expression of chymase (CMA1)47;48, as detected by microarray analysis, was not different for EE, CE, and normal patients and was not found within the mast cell-related transcriptome. Additionally the mast cell restricted protease, tryptase gamma 1 (TPSG1)49, was below the limits of detection in all studied groups.
We aimed to determine which genes correlated with mast cell degranulation. Accordingly, we generated a mast cell degranulation-related transcriptome with a minimum cutoff p-value < 0.05 for the Spearman correlation coefficient. A total of 1298 genes were identified that correlated with mast cell degranulation, with a 116 of those genes with a minimum 2-fold difference between EE and normal patients (Supplementary Table 2). Two mast cell-related genes demonstrated altered steady state transcript expression on microarray (KITLG and PPP2R2C), that were highly correlated with mast cell degranulation with Spearman coefficients r > ±0.85, KITLG (+0.86) and PPP2R2C (−0.91). The products of these genes have previously been directly associated with mast cell degranulation in vivo and in vitro45;50–55. A comparison of the mast cell transcriptome (Supplemental Table 1) and the mast cell degranulation transcriptome (Supplemental Table 2) revealed significant overlap, with 112 genes in common between both transcriptomes, 184 genes unique to the mast cell transcriptome, and 5 genes unique to the degranulation transcriptome.
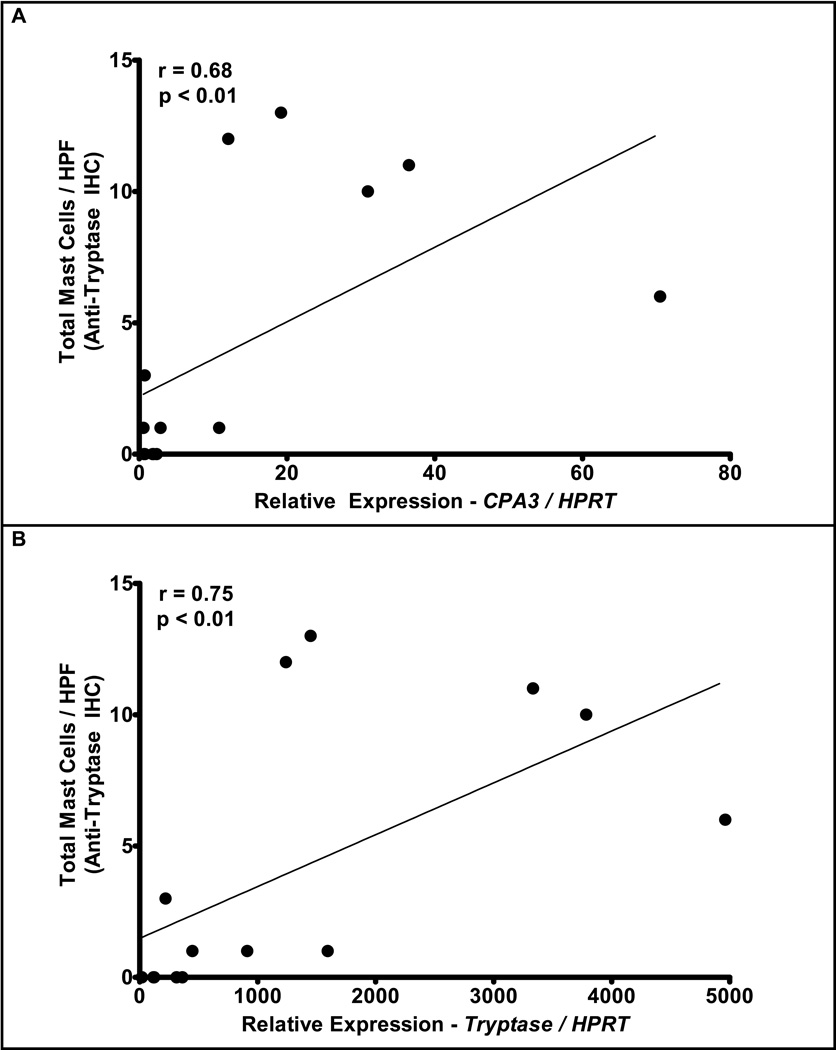
When assessing the genes found in either the mast cell-related or the degranulation-related transcriptomes, the mast cell-specific proteases CPA3 and tryptase underwent the largest fold-change in the expression of their respective transcripts in normal versus EE patients. Validation studies demonstrated a 25-fold induction in the expression of CPA3 mRNA in EE patients relative to normal control patients as determined by real-time PCR analysis. CPA3 expression directly correlated with mast cell counts as determined by tryptase positive cells (r = 0.68, p < 0.01, Figure 2A). Tryptase mRNA (TPSB2) expression had a 10-fold increase in EE patients relative to normal controls and correlated with tryptase IHC (r = 0.75, p < 0.01, Figure 2B). No increase in chymase mRNA expression was noted by real-time PCR in EE versus normal patients, and there was no correlation between tryptase-derived mast cell counts and chymase expression (data not shown). Along with the increase in the primary mast cell proteases, CPA3 and tryptase, on microarray analysis demonstrated a 5-fold increase in the dipeptidyl peptidase I (DPPI, cathespin C or CTSC) an enzyme that activates mast cell chymase41;56.
Figure 2. Relationship between mast cell counts and relative expression of CPA3 and tryptase.
Mast cell counts were determined by anti-tryptase IHC and correlated against the relative expression CPA3 (r = 0.68, p < 0.01)(A) and tryptase (r = 0.75, p < 0.01)(B) as determined by real-time PCR.
Mast cell related genes
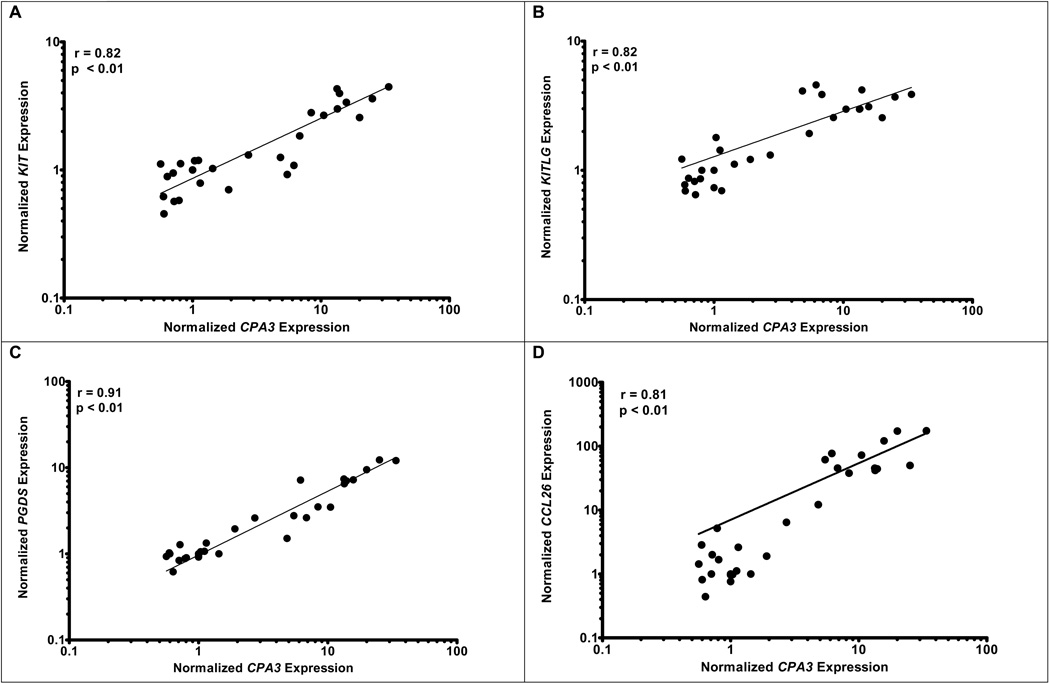
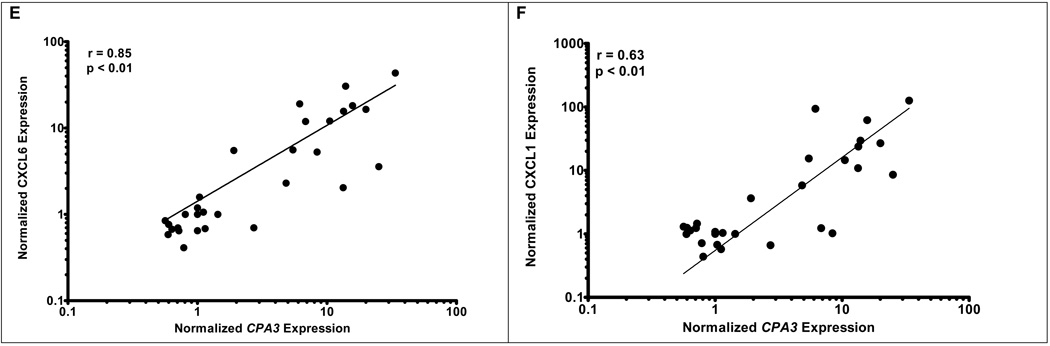
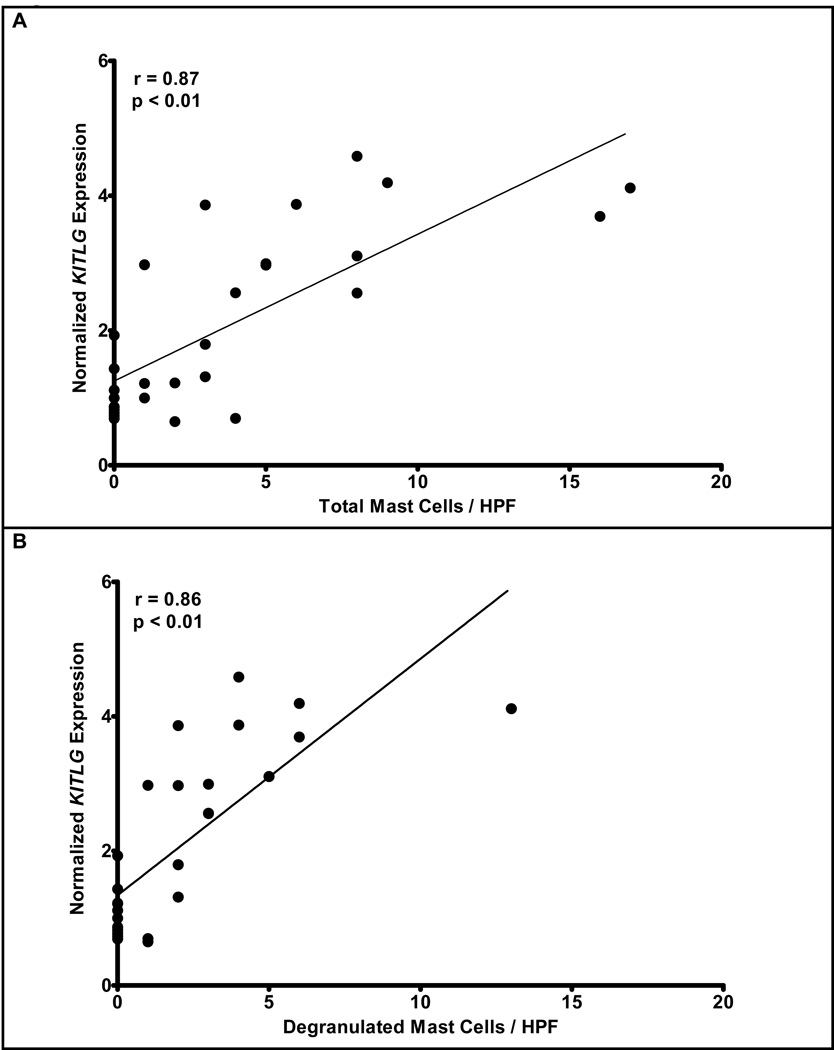
Identifying CPA3 as a grossly dysregulated surrogate genetic marker for esophageal mast cells in EE provided a valuable opportunity to dissect molecular processes associated with human mastocytosis. We therefore correlated global gene expression with the expression of CPA3 mRNA. The ontology of genes that most correlated with CPA3 are broadly based and associated with combined innate and adaptive immune responses including complement activation, natural killer cell cytoxicity, mast cell activation, and programmed cell death (Supplementary Table 3). Strong correlations between CPA3 and c-Kit, KITLG, and prostaglandin D2 synthase (PGDS) expression were apparent with r = 0.82, 0.82, and 0.91 respectively (Figure 3A, 3B, & 3C). When CPA3 was compared to the chemokines overexpressed in EE (CCL26, CXCL6, and CXCL1), strong correlations were readily apparent for CCL26 (r = 0.81) and CXCL6 (r = 0.85), while a weaker correlation was noted for CXCL1 (r = 0.63) (Figures 3D–3F). Clear correlation was noted between KITLG and total (r = 0.87) or degranulated (r = 0.86) mast cell counts (Figure 4). No statistically significant correlation was noted between peak eosinophil counts and the expression of KITLG (data not shown), consistent with the prominent role of KITLG in mast cell rather than eosinophil development.
Figure 3. Correlation of CPA3 expression with mast cell gene expression.
Correlations between microarray expression of CPA3 and mast cell related genes including: KIT (r = 0.82, p < 0.01), KITLG (r = 0.82, p < 0.01), PGDS (r = 0.91, p < 0.01), CCL26 (r = 0.81, p < 0.01), CXCL6 (r = 0.85, p < 0.01), CXCL1 (r = 0.63, p < 0.01)(A–F).
Figure 4. Correlation of KITLG expression with intact and degranulated mast cell.
The normalized expression of KITLG is plotted against the maximum number of intact mast cells (r = 0.87, p < 0.01)(A) or degranulated mast cells (r = 0.86, p < 0.01) (B).
Mast cell counts, gene expression and treatment with fluticasone propionate
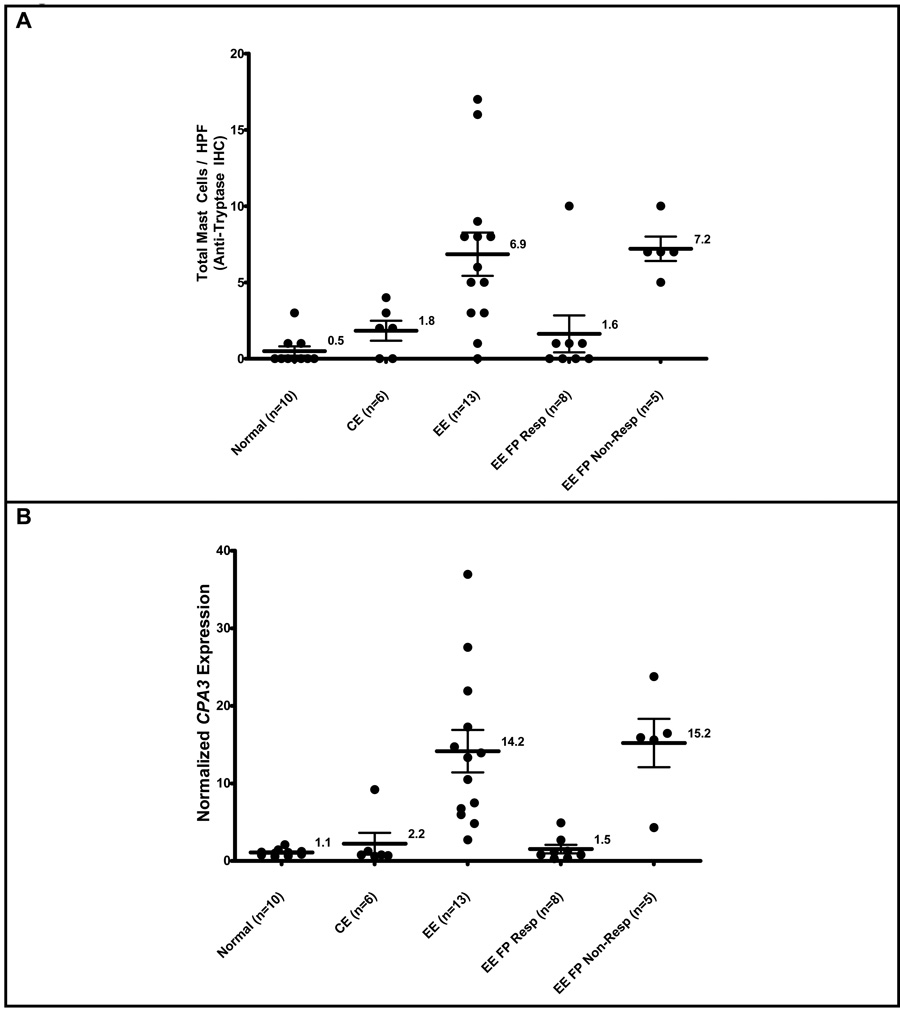
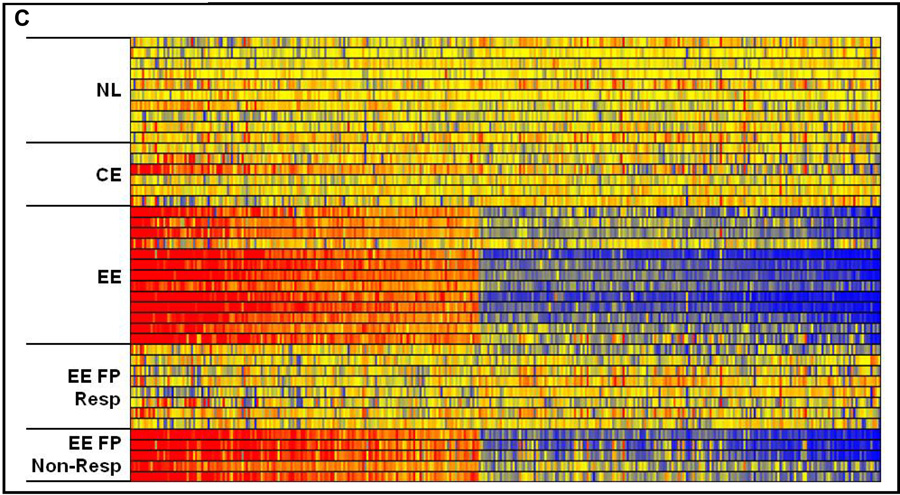
In those patients with evidence of response to FP treatment based upon the normalization of esophageal eosinophil counts, mast cell counts as determined by tryptase IHC (EE FP Resp, mast cell/HPF ± SEM, 1.5 ± 0.6) were equivalent to those seen in normal patients (NL, mast cells/HPF ± SEM, 0.5 ± 0.3 mast cell / HPF), and were reduced compared to EE patients with active disease (EE, mast cells/HPF ± SEM, 14,2 ± 2.7)(p < 0.05). For those patients who did not respond to swallowed FP therapy, the mast cell counts were elevated to the same extent as EE patients with active disease in comparison to normal patients (Figure 5A). The expression of CPA3 in patients whom responded to FP (EE FP Resp, CPA3 ± SEM, 1.5 ± 0.6) was equivalent to that seen in normal patients (NL, CPA3 ± SEM, 1.1 ± 0.1), and was reduced as compared to patients with active disease (EE, CPA3 ± SEM, 14.2 ± 2.7)(p < 0.05) (Figure 5B). Indeed, the expression of the entire mast cell transcriptome in EE patients who have responded to FP treatment cannot be readily differentiated from that seen in normal patients, while dysregulation of the mast cell transcriptome is readily apparent in the EE patient non-responders to FP therapy (Figure 5C). Similarly in CE patients, mast cell counts, CPA3 expression, and the mast cell transcriptome are essentially equivalent in comparison to untreated normal patients.
Figure 5. Mast cell counts, gene expression, and mast cell transcriptome in patients treated with swallowed FP.
Maximum mast cell counts from normal, CE, EE, and compared to EE patients whom responded to fluticasone propionate (EE FP Resp), or who did not respond to fluticasone propionate therapy (EE FP Non-Resp). Response to treatment with swallowed FP was defined as ≤ 1 eosinophil per HPF (A). CPA3 expression on microarray (B). The mast cell related-transcriptome under varying conditions listed above (C).
Discussion
Evidence is emerging that the immunopathogenesis of EE involves the complex interplay of a number of different cell types including epithelial cells, lymphocytes, and mast cells. Recent research indicates that mast cells accumulate in the esophagus and their activation may be regulated by locally generated IgE as well as other innate triggers including mast cell secretagogues released from eosinophils36;57;58. Notably, a detailed description of mast cell phenotype, activation, and the mechanisms of their recruitment and activation have not been delineated. We now confirm that substantial mastocytosis occurs in esophageal biopsies obtained from EE patients as compared with normal individuals and individuals with chronic (non-eosinophilic) esophagitis. In particular, mast cell accumulation in the epithelial layer distinguishes EE from the two control groups. Mastocytosis occurred independent of patient age, gender, atopic status, and PPI therapy indicating that it is a general phenomenon despite the distinct patient phenotypes. Furthermore, we report that mast cells undergo substantial degranulation in EE compared with both control groups. Using a combination of approaches (gene chip analysis, real-time PCR, and IHC), we report that a unique mast cell phenotype is predominantly associated with tryptase and CPA3, rather than chymase or tryptase gamma 1. This emphasizes the unique mast cell phenotype(s) in EE as CPA3 has typically been associated with connective tissue mast cells expressing both tryptase and chymase rather than the T-cell dependent mucosal mast cell phenotype that is associated with the expression of tryptase alone59. T cell derived products such as IL-5 have been shown to be involved in EE pathogenesis relative to patients with IgE-mediated food allergy; presumably this localized alteration in Th2 sub-populations could be influencing mast cell protease expression and general phenotype23. Of the known mast cell-specific genes, the transcript for CPA3 was strongly associated with mast cells and represents the most severely dysregulated mast cell gene within the mast cell related transcriptome. This finding has implications for other diseases with associated mastocytosis (e.g. asthma) as this gene provides a molecular “fingerprint” for mast cells. Using both tryptase IHC and CPA3 gene expression to quantitate mast cells, we identified a mast-cell-associated transcriptome in EE patients. Notably, only 148 out of 301 genes (~50%) composing the mast cell-related transcriptome overlapped with the eosinophil-associated EE transcriptome (490 genes) indicating that there are both shared and unique pathways involved in regulating these two cells types and their downstream pathways. In addition, the inflammatory consequences initiated by each of these two cells types are likely to be unique considering their distinct transcriptomes.
The mast cell-associated EE transcriptome provided a valuable opportunity to dissect mechanisms of mast cell accumulation and activation; indeed, we report that KITLG expression strongly correlated with both mast cell accumulation and activation. Several studies have found that the injection of KIT ligand into skin results in mast cell degranulation, histamine release, localized swelling, and fibrin deposition52–55. We report that the mast cell-associated transcriptome is completely reversible following successful therapy with swallowed FP, suggesting that the mast cell changes are likely secondary rather than constitutive to the elements of the esophagus that are assessed by the biopsy-associated transcriptome. It is notable that mast cell responses in asthma are not dramatically inhibited by glucocorticoids60;61, highlighting that this response is relatively unique and encouraging in terms of therapy and prognosis. In contrast treatment with mepolizumab does not clearly appear to dramatically impact mast cell numbers in at least two separate studies62;63. It is unknown to date if dietary therapies would alter these disease parameters as well.
In summary, we report that esophageal mastocytosis and degranulation occur in EE patients and have identified a mast cell-associated esophageal transcriptome that distinguishes EE patients from control individuals. This transcriptome only partially overlaps with the eosinophil-associated transcriptome and identifies unique upstream (KIT ligand) and downstream pathways (PGD2) involved in regulating mast cells and the inflammatory response characteristic of EE. Herein, we have identified that tissue levels of CPA3 serve as a strong surrogate marker for human mast cells. This finding is likely to have applications for other mast cell associated diseases where better mast cell assessments are needed (e.g. asthma). As such, we have identified mast cell-associated responses that may have diagnostic value and further significance in terms of pathogenesis and therapeutic strategies.
Key Messages
Mast cells are increased and degranulated in patients with eosinophilic esophagitis and are correlated with an atypical and modified phenotype associated with active disease.
Esophageal transcriptomes differentiates mast cell-associated and eosinophil-associated processes with CPA3 as the most dysregulated mast cell gene in EE.
Evidence is provided for the selective regulation of mast cells by KIT ligand.
Supplementary Material
Acknowledgments
We gratefully thank Cassie Kirby for her diligent work in support of this effort, and Shawna Hottinger for her final edits of this manuscript.
Abbreviations used in this paper
- CE
chronic esophagitis
- GERD
gastroesophageal reflux disease
- FP
fluticasone propionate
- PPI
proton pump inhibitor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was supported by NIH grants 12HD028827, AI057991 to J.P.A, AI070235, AI45898, P30 DK0789392 and DK076893, the Food Allergy Project, the Campaign Urging Research for Eosinophilic Disorders (CURED), and the Buckeye Foundations to M.E.R.
REFERENCES
- 1.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004;351:940–941. doi: 10.1056/NEJM200408263510924. [DOI] [PubMed] [Google Scholar]
- 2.Gill R, Durst P, Rewalt M, Elitsur Y. Eosinophilic esophagitis disease in children from West Virginia: a review of the last decade (1995–2004) Am J Gastroenterol. 2007;102:2281–2285. doi: 10.1111/j.1572-0241.2007.01352.x. [DOI] [PubMed] [Google Scholar]
- 3.Straumann A, Simon HU. Eosinophilic esophagitis: escalating epidemiology? J Allergy Clin Immunol. 2005;115:418–419. doi: 10.1016/j.jaci.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Kelly KJ, Lazenby AJ, Rowe PC, Yardley JH, Perman JA, Sampson HA. Eosinophilic esophagitis attributed to gastroesophageal reflux: improvement with an amino acid-based formula. Gastroenterology. 1995;109:1503–1512. doi: 10.1016/0016-5085(95)90637-1. [DOI] [PubMed] [Google Scholar]
- 5.Walsh SV, Antonioli DA, Goldman H, Fox VL, Bousvaros A, Leichtner AM, et al. Allergic esophagitis in children: a clinicopathological entity. Am J Surg Pathol. 1999;23:390–396. doi: 10.1097/00000478-199904000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Rothenberg ME. Eosinophilic gastrointestinal disorders (EGID) J Allergy Clin Immunol. 2004;113:11–28. doi: 10.1016/j.jaci.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 7.Collins MH, Blanchard C, Abonia JP, Kirby C, Akers R, Wang N, et al. Clinical, pathologic, and molecular characterization of familial eosinophilic esophagitis compared with sporadic cases. Clin Gastroenterol Hepatol. 2008;6:621–629. doi: 10.1016/j.cgh.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sant'Anna AM, Rolland S, Fournet JC, Yazbeck S, Drouin E. Eosinophilic esophagitis in children: Symptoms, histology and pH probe results. J Pediatr Gastroenterol Nutr. 2004;39:373–377. doi: 10.1097/00005176-200410000-00013. [DOI] [PubMed] [Google Scholar]
- 9.Lim JR, Gupta SK, Croffie JM, Pfefferkorn MD, Molleston JP, Corkins MR, et al. White specks in the esophageal mucosa: An endoscopic manifestation of non-reflux eosinophilic esophagitis in children. Gastrointest Endosc. 2004;59:835–838. doi: 10.1016/s0016-5107(04)00364-5. [DOI] [PubMed] [Google Scholar]
- 10.Jyonouchi S, Brown-Whitehorn TA, Spergel JM. Association of eosinophilic gastrointestinal disorders with other atopic disorders. Immunol Allergy Clin North Am. 2009;29:85–97. doi: 10.1016/j.iac.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Spergel JM, Brown-Whitehorn T, Beausoleil JL, Shuker M, Liacouras CA. Predictive values for skin prick test and atopy patch test for eosinophilic esophagitis. J Allergy Clin Immunol. 2007;119:509–511. doi: 10.1016/j.jaci.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 12.Assa'ad AH, Putnam PE, Collins MH, Akers RM, Jameson SC, Kirby CL, et al. Pediatric patients with eosinophilic esophagitis: an 8-year follow-up. J Allergy Clin Immunol. 2007;119:731–738. doi: 10.1016/j.jaci.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 13.Kagalwalla AF, Sentongo TA, Ritz S, Hess T, Nelson SP, Emerick KM, et al. Effect of Six-Food Elimination Diet on Clinical and Histologic Outcomes in Eosinophilic Esophagitis. Clin Gastroenterol Hepatol. 2006;4:1097–1102. doi: 10.1016/j.cgh.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 14.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001;107:83–90. doi: 10.1172/JCI10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. IL-5 promotes eosinophil trafficking to the esophagus. J Immunol. 2002;168:2464–2469. doi: 10.4049/jimmunol.168.5.2464. [DOI] [PubMed] [Google Scholar]
- 16.Fogg MI, Ruchelli E, Spergel JM. Pollen and eosinophilic esophagitis. J Allergy Clin Immunol. 2003;112:796–797. doi: 10.1016/s0091-6749(03)01715-9. [DOI] [PubMed] [Google Scholar]
- 17.Simon D, Marti H, Heer P, Simon HU, Braathen LR, Straumann A. Eosinophilic esophagitis is frequently associated with IgE-mediated allergic airway diseases. J Allergy Clin Immunol. 2005;115:1090–1092. doi: 10.1016/j.jaci.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Straumann A, Spichtin HP, Grize L, Bucher KA, Beglinger C, Simon HU. Natural history of primary eosinophilic esophagitis: a follow-up of 30 adult patients for up to 11.5 years. Gastroenterology. 2003;125:1660–1669. doi: 10.1053/j.gastro.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 19.Straumann A, Bauer M, Fischer B, Blaser K, Simon HU. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J Allergy Clin Immunol. 2001;108:954–961. doi: 10.1067/mai.2001.119917. [DOI] [PubMed] [Google Scholar]
- 20.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanchard C, Wang N, Rothenberg ME. Eosinophilic esophagitis: pathogenesis, genetics, and therapy. J Allergy Clin Immunol. 2006;118:1054–1059. doi: 10.1016/j.jaci.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 22.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 23.Prussin C, Lee J, Foster B. Eosinophilic gastrointestinal disease and peanut allergy are alternatively associated with IL-5+ and IL-5(−) T(H)2 responses. J Allergy Clin Immunol. 2009;124:1326–1332. doi: 10.1016/j.jaci.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010 doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aceves SS, Newbury RO, Dohil R, Bastian JF, Broide DH. Esophageal remodeling in pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2007;119:206–212. doi: 10.1016/j.jaci.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 26.Chehade M, Sampson HA, Morotti RA, Magid MS. Esophageal subepithelial fibrosis in children with eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2007;45:319–328. doi: 10.1097/MPG.0b013e31806ab384. [DOI] [PubMed] [Google Scholar]
- 27.Konikoff MR, Noel RJ, Blanchard C, Kirby C, Jameson SC, Buckmeier BK, et al. A randomized, double-blind, placebo-controlled trial of fluticasone propionate for pediatric eosinophilic esophagitis. Gastroenterology. 2006;131:1381–1391. doi: 10.1053/j.gastro.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 28.Kirsch R, Bokhary R, Marcon MA, Cutz E. Activated mucosal mast cells differentiate eosinophilic (allergic) esophagitis from gastroesophageal reflux disease. J Pediatr Gastroenterol Nutr. 2007;44:20–26. doi: 10.1097/MPG.0b013e31802c0d06. [DOI] [PubMed] [Google Scholar]
- 29.Ruchelli E, Wenner W, Voytek T, Brown K, Liacouras C. Severity of esophageal eosinophilia predicts response to conventional gastroesophageal reflux therapy. Pediatr Dev Pathol. 1999;2:15–18. doi: 10.1007/s100249900084. [DOI] [PubMed] [Google Scholar]
- 30.Leder LD. The chloroacetate esterase reaction. A useful means of histological diagnosis of hematological disorders from paraffin sections of skin. Am J Dermatopathol. 1979;1:39–42. [PubMed] [Google Scholar]
- 31.Beckstead JH, Halverson PS, Ries CA, Bainton DF. Enzyme histochemistry and immunohistochemistry on biopsy specimens of pathologic human bone marrow. Blood. 1981;57:1088–1098. [PubMed] [Google Scholar]
- 32.Friend DS, Gurish MF, Austen KF, Hunt J, Stevens RL. Senescent jejunal mast cells and eosinophils in the mouse preferentially translocate to the spleen and draining lymph node, respectively, during the recovery phase of helminth infection. J Immunol. 2000;165:344–352. doi: 10.4049/jimmunol.165.1.344. [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann N, King NE, Laporte J, Yang M, Mishra A, Pope SM, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest. 2003;111:1863–1874. doi: 10.1172/JCI17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 35.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vicario M, Blanchard C, Stringer KF, Collins MH, Mingler MK, Ahrens A, et al. Local B cells and IgE production in the oesophageal mucosa in eosinophilic oesophagitis. Gut. 2010;59:12–20. doi: 10.1136/gut.2009.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aceves SS, Newbury RO, Dohil R, Schwimmer J, Bastian JF. Distinguishing eosinophilic esophagitis in pediatric patients: clinical, endoscopic, and histologic features of an emerging disorder. J Clin Gastroenterol. 2007;41:252–256. doi: 10.1097/01.mcg.0000212639.52359.f1. [DOI] [PubMed] [Google Scholar]
- 38.Spergel JM, Andrews T, Brown-Whitehorn TF, Beausoleil JL, Liacouras CA. Treatment of eosinophilic esophagitis with specific food elimination diet directed by a combination of skin prick and patch tests. Ann Allergy Asthma Immunol. 2005;95:336–343. doi: 10.1016/S1081-1206(10)61151-9. [DOI] [PubMed] [Google Scholar]
- 39.Geissler EN, Liao M, Brook JD, Martin FH, Zsebo KM, Housman DE, et al. Stem cell factor (SCF), a novel hematopoietic growth factor and ligand for c-kit tyrosine kinase receptor, maps on human chromosome 12 between 12q14.3 and 12qter. Somatic Cell & Molecular Genetics. 1991;17:207–214. doi: 10.1007/BF01232978. [DOI] [PubMed] [Google Scholar]
- 40.Miller JS, Moxley G, Schwartz LB. Cloning and characterization of a second complementary DNA for human tryptase. J Clin Invest. 1990;86:864–870. doi: 10.1172/JCI114786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheth PD, Pedersen J, Walls AF, McEuen AR. Inhibition of dipeptidyl peptidase I in the human mast cell line HMC-1: blocked activation of tryptase, but not of the predominant chymotryptic activity. Biochem Pharmacol. 2003;66:2251–2262. doi: 10.1016/j.bcp.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds DS, Gurley DS, Stevens RL, Sugarbaker DJ, Austen KF, Serafin WE. Cloning of cDNAs that encode human mast cell carboxypeptidase A, and comparison of the protein with mouse mast cell carboxypeptidase A and rat pancreatic carboxypeptidases. Proc Natl Acad Sci U S A. 1989;86:9480–9484. doi: 10.1073/pnas.86.23.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abonia JP, Austen KF, Rollins BJ, Joshi SK, Flavell RA, Kuziel WA, et al. Constitutive homing of mast cell progenitors to the intestine depends on autologous expression of the chemokine receptor CXCR2. Blood. 2005;105:4308–4313. doi: 10.1182/blood-2004-09-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murakami M, Kudo I, Inoue K. Characteristics and possible functions of mast cell phospholipases A2. Adv Exp Med Biol. 1992;318:27–34. doi: 10.1007/978-1-4615-3426-6_3. [DOI] [PubMed] [Google Scholar]
- 45.Ludowyke RI, Holst J, Mudge LM, Sim AT. Transient translocation and activation of protein phosphatase 2A during mast cell secretion. J Biol Chem. 2000;275:6144–6152. doi: 10.1074/jbc.275.9.6144. [DOI] [PubMed] [Google Scholar]
- 46.Caughey GH, Raymond WW, Blount JL, Hau LW, Pallaoro M, Wolters PJ, et al. Characterization of human gamma-tryptases, novel members of the chromosome 16p mast cell tryptase and prostasin gene families. J Immunol. 2000;164:6566–6575. doi: 10.4049/jimmunol.164.12.6566. [DOI] [PubMed] [Google Scholar]
- 47.Urata H, Kinoshita A, Perez DM, Misono KS, Bumpus FM, Graham RM, et al. Cloning of the gene and cDNA for human heart chymase. J Biol Chem. 1991;266:17173–17179. [PubMed] [Google Scholar]
- 48.Caughey GH, Zerweck EH, Vanderslice P. Structure, chromosomal assignment, and deduced amino acid sequence of a human gene for mast cell chymase. J Biol Chem. 1991;266:12956–12963. [PubMed] [Google Scholar]
- 49.Wong GW, Tang Y, Feyfant E, Sali A, Li L, Li Y, et al. Identification of a new member of the tryptase family of mouse and human mast cell proteases which possesses a novel COOH-terminal hydrophobic extension. J Biol Chem. 1999;274:30784–30793. doi: 10.1074/jbc.274.43.30784. [DOI] [PubMed] [Google Scholar]
- 50.Costa JJ, Demetri GD, Harrist TJ, Dvorak AM, Hayes DF, Merica EA, et al. Recombinant human stem cell factor (kit ligand) promotes human mast cell and melanocyte hyperplasia and functional activation in vivo. J Exp Med. 1996;183:2681–2686. doi: 10.1084/jem.183.6.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bischoff SC, Dahinden CA. c-kit ligand: a unique potentiator of mediator release by human lung mast cells. J Exp Med. 1992;175:237–244. doi: 10.1084/jem.175.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Columbo M, Horowitz EM, Botana LM, MacGlashan DW, Jr, Bochner BS, Gillis S, et al. The human recombinant c-kit receptor ligand, rhSCF, induces mediator release from human cutaneous mast cells and enhances IgE-dependent mediator release from both skin mast cells and peripheral blood basophils. J Immunol. 1992;149:599–608. [PubMed] [Google Scholar]
- 53.Wershil BK, Tsai M, Geissler EN, Zsebo KM, Galli SJ. The rat c-kit ligand, stem cell factor, induces c-kit receptor-dependent mouse mast cell activation in vivo. Evidence that signaling through the c-kit receptor can induce expression of cellular function. J Exp Med. 1992;175:245–255. doi: 10.1084/jem.175.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammerberg B, Olivry T, Orton SM. Skin mast cell histamine release following stem cell factor and high-affinity immunoglobulin E receptor cross-linking in dogs with atopic dermatitis. Vet Dermatol. 2001;12:339–346. doi: 10.1046/j.0959-4493.2001.00273.x. [DOI] [PubMed] [Google Scholar]
- 55.Feldweg AM, Friend DS, Zhou JS, Kanaoka Y, Daheshia M, Li L, et al. gp49B1 suppresses stem cell factor-induced mast cell activation-secretion and attendant inflammation in vivo. Eur J Immunol. 2003;33:2262–2268. doi: 10.1002/eji.200323978. [DOI] [PubMed] [Google Scholar]
- 56.Sakai K, Ren S, Schwartz LB. A novel heparin-dependent processing pathway for human tryptase. Autocatalysis followed by activation with dipeptidyl peptidase I. J Clin Invest. 1996;97:988–995. doi: 10.1172/JCI118523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lucendo AJ, Navarro M, Comas C, Pascual JM, Burgos E, Santamaria L, et al. Immunopenotypic characterization and quantification of the epithelial inflammatory infiltrate in eosinophilic esophagitis through stereology: an analysis of the cellular mechanisms of the disease and the immunologic capacity of the esophagus. Am J Surg Pathol. 2007;31:598–606. doi: 10.1097/01.pas.0000213392.49698.8c. [DOI] [PubMed] [Google Scholar]
- 58.Lucendo AJ, Bellon T, Lucendo B. The role of mast cells in eosinophilic esophagitis. Pediatr Allergy Immunol. 2009;20:512–518. doi: 10.1111/j.1399-3038.2008.00798.x. [DOI] [PubMed] [Google Scholar]
- 59.Pejler G, Knight SD, Henningsson F, Wernersson S. Novel insights into the biological function of mast cell carboxypeptidase A. Trends Immunol. 2009;30:401–408. doi: 10.1016/j.it.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 60.Wallin A, Sandstrom T, Soderberg M, Howarth P, Lundback B, la-Cioppa G, et al. The effects of regular inhaled formoterol, budesonide, and placebo on mucosal inflammation and clinical indices in mild asthma. Am J Respir Crit Care Med. 1999;159:79–86. doi: 10.1164/ajrccm.159.1.9801007. [DOI] [PubMed] [Google Scholar]
- 61.Wallin A, Sue-Chu M, Bjermer L, Ward J, Sandstrom T, Lindberg A, et al. Effect of inhaled fluticasone with and without salmeterol on airway inflammation in asthma. J Allergy Clin Immunol. 2003;112:72–78. doi: 10.1067/mai.2003.1518. [DOI] [PubMed] [Google Scholar]
- 62.Stein ML, Collins MH, Villanueva JM, Kushner JP, Putnam PE, Buckmeier BK, et al. Anti-IL-5 (mepolizumab) therapy for eosinophilic esophagitis. J Allergy Clin Immunol. 2006;118:1312–1319. doi: 10.1016/j.jaci.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 63.Straumann A, Conus S, Grzonka P, Kita H, Kephart G, Bussmann C, et al. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: a randomised, placebo-controlled, double-blind trial. Gut. 2010;59:21–30. doi: 10.1136/gut.2009.178558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.