Abstract
The membrane-spanning domain (MSD) of Human Immunodeficiency Virus Type I (HIV-1) envelope glycoprotein (Env) is critical for its biological activity. Initial studies have defined an almost invariant “core” structure in the MSD and demonstrated that it is crucial for anchoring Env in the membrane and virus entry. We show here that amino acid substitutions in the MSD “core” do not influence specific virus-cell attachment, nor CD4 receptor and CXCR4 coreceptor recognition by Env. However, substitutions within the MSD “core” delayed the kinetics and reduced the efficiency of cell-cell fusion mediated by Env. Although we observed no evidence that membrane fusion mediated by the MSD core mutants was arrested at a hemifusion stage, impaired Env fusogenicity was correlated with minor conformational changes in the V2, C1, and C5 regions in gp120 and the immunodominant loop in gp41. These changes could delay initiation of the conformational changes required in the fusion process.
Keywords: HIV-1, Env, gp120, gp41, membrane-spanning domain, GxxxG, conformational changes, membrane fusion, hemifusion
INTRODUCTION
Human immunodeficiency virus Type I (HIV-1) infection is initiated by fusion of the viral membrane with that of the target cell and is mediated by the viral envelope glycoprotein (Env). On the virus surface, a trimer, composed of three non-covalently linked dimers of gp120, the receptor-binding surface (SU) component, and gp41, the membrane-spanning, trans-membrane (TM) component, forms the functional Env complex (Eckert and Kim, 2001; Moore et al., 1990; Willey and Martin, 1993; Wyatt and Sodroski, 1998). The gp120 and gp41 glycoproteins are synthesized as a precursor gp160 glycoprotein, which is encoded by the env gene. The gp160 precursor is co-translationally glycosylated and, following transport to the trans-Golgi network, is cleaved into the mature products by a member of the furin family of endoproteases (Wyatt and Sodroski, 1998). Mature Env proteins are transported to the plasma membrane where they are rapidly endocytosed or incorporated into virions (Byland et al., 2007; Rowell, Stanhope, and Siliciano, 1995; Wilk et al., 1996).
The HIV-1 gp120 glycoprotein binds to the CD4 receptor and chemokine coreceptors and consists of 5 conserved domains (C1–C5) and 5 variable domains (V1–V5) (Fig. 1A). Several studies suggest that the C1 and C5 regions directly interact with the immunodominant loop in gp41 (Binley et al., 2000; Helseth et al., 1990; Ivey-Hoyle, Clark, and Rosenberg, 1991). The C2 region is sequestered in the oligomeric structure and involved in the oligomerization of gp120 (Lemasson et al., 1995). The binding of CD4 to HIV-1 Env requires multiple conserved regions in gp120, including C1 (Kropelin et al., 1998; Orloff et al., 1995), C3 (Howie et al., 1998; Howie et al., 1999), and C4 (Morrison, Kirchhoff, and Desrosiers, 1995). A comparison of the sequence and structural profiles of HIV-1 and SIV Env shows that the C2-V3-C3 region is involved in the contact with chemokine receptors (Chen et al., 2005; Shimizu and Gojobori, 2000). The V3 loop is a hyper variable disulfide-bonded structure and is the major determinant of the tropism of HIV-1 virions (Hartley et al., 2005). The V1V2 region also influences HIV-1 cellular tropism, probably via an interaction with regions of the V3 loop (Boyd et al., 1993; Koito et al., 1994). The V4 and V5 regions of gp120 may also be necessary for efficient utilization of CXCR4 (Cho et al., 1998; Labrosse et al., 2001). Moreover, the V1V2 and V3 regions have been demonstrated to play a more important role when HIV-1 uses, in addition to CCR5 or CXCR4, other chemokine coreceptors such as CCR2b, CCR3, STRL33, and APJ (Hoffman et al., 1998). In the dual tropic strain 89.6, the V3, V4, and V5 regions are involved in CCR5, CXCR4, and CCR3 utilization (Smyth et al., 1998).
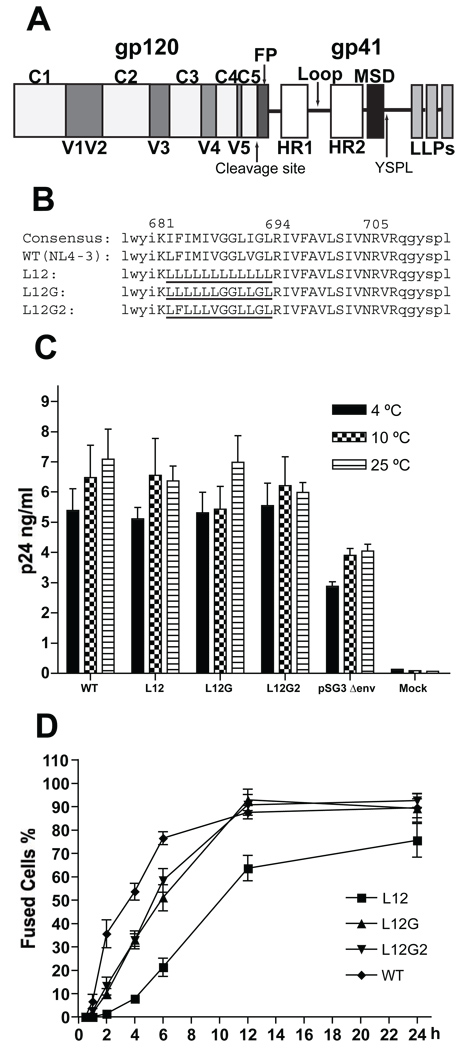
FIG. 1. Kinetics of cell-cell fusion mediated by the Env.
(A) A schematic structure of the HIV-1 Env sequence (FP: fusion peptide; HR: heptad repeat; Loop: immunodominant loop; MSD: membrane-spanning domain; LLP: Lentivirus lytic peptide). (B) Amino acid sequences of HIV-1 MSDs. The consensus sequence of the HIV-1 MSD was generated by the alignment of the Env MSD sequences of all M and N group HIV-1 isolates from the Los Alamos HIV Sequences Database. The amino acid residues of the putative MSD region are shown as upper case letters. The flanking sequences of the HIV-1 MSD are shown as lower case letters. The position of the MSD in the sequence of HIV-1 NL4-3 is marked above the positively charged amino acid residues. The mutated portions of the HIV-1 MSD are underlined. (C) Virus-cell attachment assay. Purified viruses were co-incubated with JC53BL cells at specific temperature for 2 h. Cells were then washed three times in PBS and lysed in PBS containing 0.5% Triton X-100. The amount of p24 in cell lysate was measured by p24 ELISA. The error bars represent the standard deviations of three independent experiments. (D) Cell-cell fusion kinetics. 293T cells were transfected with the proviral vector, pTN6-GFP, containing the MSD mutants. At 48 h posttransfection, the transfected 293T cells were cocultured with the CMAC-blue-loaded JC53BL indicator cells for specific times. Fused cells were observed and quantitated by fluorescent microscopy. For each mutant, at least 50 fusion events were counted at each time point.
The HIV-1 gp41 glycoprotein is the fusion machinery that mediates virus-cell membrane fusion. When activated, the ectodomain of gp41 carries out the fusion function, while the membrane-spanning domain and cytoplasmic domain are both important for its fusogenecity. The ectodomain consists of a fusion peptide, two heptad repeats (HR1 and HR2) with the immunodominant loop in between, and a membrane proximal tryptophan-rich domain (Fig. 1A). The cytoplasmic domain contains signals for intracellular trafficking of the Env, and three lipid lytic peptides (LLP1, LLP2, and LLP3) that play important roles in membrane fusion (Comardelle et al., 1997; Miller et al., 1993; Tencza et al., 1995).
After the sequential binding of gp120 to the CD4 receptor and chemokine coreceptors, changes in the conformation of gp120 activate the fusion competency of gp41. The N-terminal hydrophobic fusion peptide is released from a compact structure of gp41 and becomes associated with the outer monolayer of the target membrane. Meanwhile, the ectodomains of the gp41 trimer are rearranged into a 6-helix bundle structure with the three HR1 segments in the center, forming a coiled-coil structure, and the three HR2 segments on the outside, being tightly packed into the hydrophobic grooves of the coiled-coil. This process brings together the cell membrane associated fusion peptides with the viral membrane-binding MSDs, and results in a close proximity between the viral envelope and the cellular membrane, which is necessary for virus-cell membrane fusion (Eckert and Kim, 2001; White et al., 2008).
The membrane-spanning domain of Env is defined as a stretch of 25 predominantly hydrophobic amino acids that spans residues K681 to R705 (NL4-3). In the previous C-terminal truncation studies of HIV-1 Env, we have demonstrated that the entire 25 amino acid region is not required for the biological function of Env (Yue, Shang, and Hunter, 2009). The 17 amino acid residues (from K681 to A697) are sufficient for stably anchoring the truncated gp41 in the membrane and mediating cell-cell fusion at a WT level. Serial small deletions (3 amino acid residues) in the region between R694 and R705 showed normal cell-cell fusion, although larger deletions were detrimental, suggesting that, with respect to the biological functions of the Env glycoprotein, the length of this region is more important than its amino acid conservation (Owens et al., 1991). From these data, we have proposed a topology where the flanking K681 and R694 residues “snorkel” in the membrane, and 12 amino acid residues (from L682 to L693) form a hydrophobic core buried within the membrane. Arginines 705 and 707 are located at the interface of membrane and cytoplasm in this model (Yue, Shang, and Hunter, 2009).
The membrane-spanning domain, particularly within the “core” region, is one of the most conserved sequences in Env. This argues for a functional role of the MSD “core” in the biological activity of the HIV-1 Env complexes. Recent studies of the MSD “core” region have focused on the GXXXG motif in which the glycine residues are the most conserved among all MSD residues. Statistical studies and bacterial models suggested that the GXXXG motif probably constructs a framework for the association of transmembrane helixes (Russ and Engelman, 2000; Senes, Engel, and DeGrado, 2004; Senes, Gerstein, and Engelman, 2000). Previously, we substituted all of the residues for leucine within the HIV-1 MSD core, and then sequentially reintroduced the GGXXG motif, and then two other highly conserved residues, F683 with V687 (Shang, Yue, and Hunter, 2008). These studies showed that the specific amino acid residues in the helical core of HIV-1 MSD are critical for the fusogenicity of Env complexes and infectivity of HIV-1 virions. The loss of infectivity correlated with impaired virus-cell entry. However, these mutations in the MSD core did not influence the biogenesis, intracellular transport and incorporation of Env complexes.
In order to further understand the roles played by the more conserved HIV-1 MSD core residues in virus-cell entry, we attempted in the current study to define the mechanism by which the MSD core mutants impact Env-mediated membrane fusion. We show here that the amino acid composition of the MSD core does not influence viral attachment to the cell surface or CD4 and CXCR4 recognition by HIV-1 Env. However, the cell-cell fusion kinetics of the recovery-of-function mutants was delayed compared to WT, and minor conformational changes in localized regions of gp120 and gp41 were observed, suggesting that these changes might delay the triggering of the fusion process after receptor binding.
RESULTS
Previously, we used a recovery-of-function mutagenesis strategy to define the roles of the specific amino acid residues within the 12-amino acid MSD “core” in HIV-1 assembly and entry. The amino acid sequences and nomenclature of the MSD “core” mutants are shown in Fig. 1B. We have shown that the MSD “core” region of HIV-1 is critical for its viral infectivity. Replacement of the conserved amino acid residues with leucine residues blocked virus entry and subsequent viral infection. Truncation of the cytoplasmic domain of the Env was not able to rescue the impaired virus entry. In this study, we examined the competence of these MSD “core” mutants at each step of virus entry, in order to further understand the mechanistic basis for the defects observed in this process.
MSD mutants bind efficiently to target cells
Initially, we carried out a virus-cell attachment assay at low temperatures (4°C, 10° C, and 25°C for 2 h). Virions containing the MSD mutant Envs adsorbed to the surface of JC53BL cells with the same efficiency as those with WT Env (Fig. 1C), indicating that the amino acid composition of the MSD “core” does not affect the ability of HIV-1 Env to mediate a virus-specific binding to the cell surface. Additionally, using an Env-free virus, we demonstrated that nonspecific binding accounts for approximately 59% of virus-cell attachment measured in this assay.
MSD mutants showed delayed cell-cell fusion kinetics
In order to examine how the MSD “core” mutants influence the fusogenicity of the HIV-1 Env, we measured the kinetics of Env mediated cell-cell fusion, using a fluorescent marker transfer system. The WT Env and MSD mutants were cloned into the pTN6-GFP proviral vector, in which the nef gene is replaced by a GFP reporter gene, and then were transfected into 293T cells. In these cells, the Env MSD mutants were transported to the cell surface as efficiently as WT, since cells expressing WT and mutant Envs demonstrated equivalent binding of the anti-gp120 monoclonal antibody (MAb) 902 (data not shown). The transfected cells were mixed with JC53BL cells, which were pre-loaded with the fluorescent dye CMAC-blue, and cell mixtures were co-cultured for 0.5 h, 1 h, 2 h, 4 h, 6 h, 12 h, and 24 h. Fused cells fluorescing with both colors were then quantitated as a percent of total cells. As shown in the Fig. 1D, the cell-cell fusion kinetics mediated by the Env MSD mutants was slower than that of WT. After a lag period of approximately 1 h, WT Env showed the highest initial rate of mediating cell-cell fusion (28.87% cells per hour) between 1 h and 2 h and a decreased rate (35.3% of the initial rate) in the following 4 h. Cell-cell fusion rates of the mutants L12G and L12G2 were relatively constant during the first 12 h of incubation and were nearly 25% of the WT initial rate. The L12 mutant exhibited a prolonged lag period of between 1 h and 4 h prior to a linear increase in cell-cell fusion rate, which is equivalent to those of the other two mutants. The WT Env reached 50% fusion at 4 h after cell mixing, while the mutants L12G and L12G2 were at 6 h, and the L12 mutant was at 10 h. Moreover, at 12 h after cell mixing, cell-cell fusion mediated by the Env mutants L12G and L12G2 was equivalent to that of WT, while even at 24 h mutant L12 showed only 84% of WT cell-cell fusogenicity. These data indicated that the MSD “core” mutants impaired the rate of cell-cell fusion by Env.
CPZ treatment did not increase cell-cell fusion
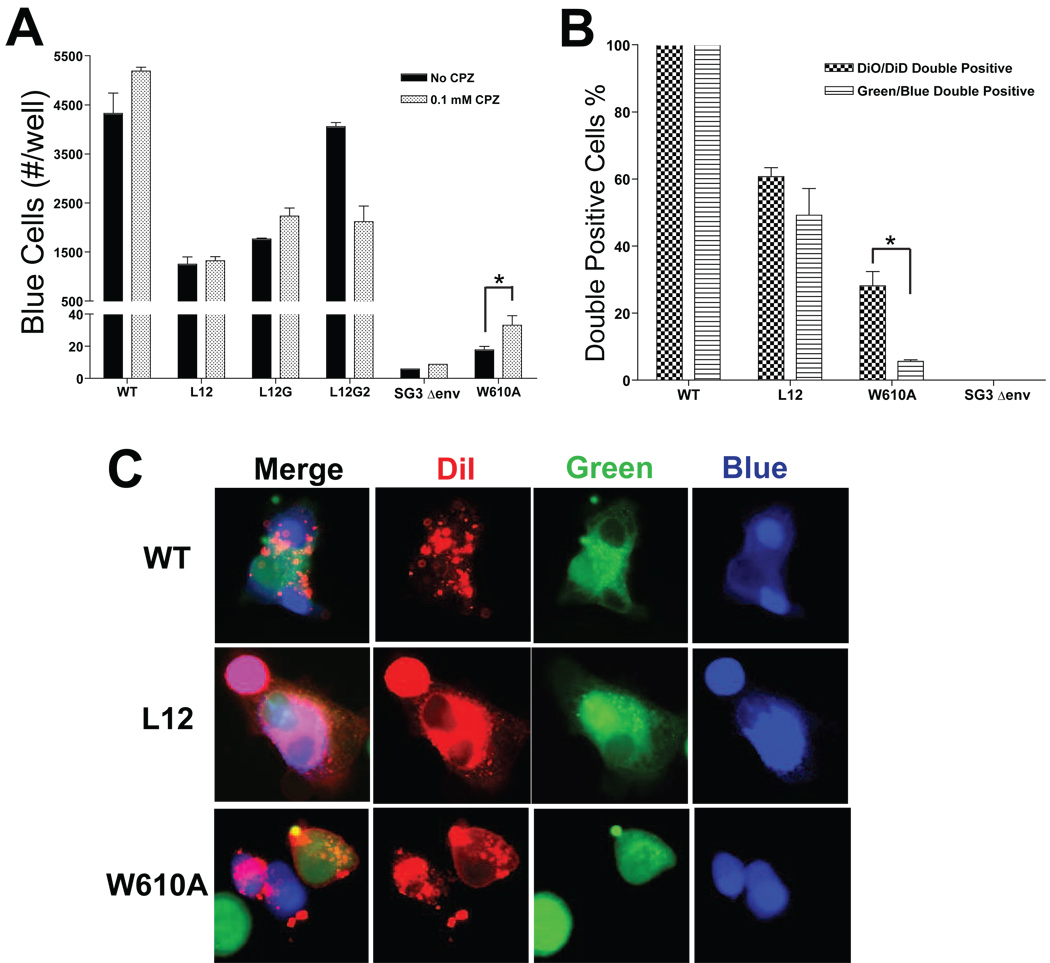
The delayed cell-cell fusion kinetics suggested that the MSD “core” mutants might have to overcome a higher energy barrier to mediate membrane fusion than WT. In order to analyze whether the higher energy barrier was created by a hemifusion intermediate, we treated cells with 0.1 mM CPZ (chlorpromazine) for 30 seconds during the cell-cell fusion mediated by the Env. CPZ specifically disrupts the lipid bilayer structure in a hemifusion intermediate, which is formed by the two inner layers of the membranes involved in the fusion, thereby enhancing membrane fusion. An Env mutant W610A (Bar and Alizon, 2004), which was previously reported to form hemifusion intermediates during Env-mediated cell-cell fusion, was used as a positive control for CPZ treatment. As shown in Fig. 2A, although cell-cell fusogenicity of the Env mutant W610A is much less than WT (240 fold of Env W610A) and Env MSD mutants, CPZ treatment significantly enhanced the Env-mediated cell-cell fusion by 2-fold, indicating a break of the lipid bilayer structure in hemifusion intermediates. However, CPZ-treatment was unable to promote a significant increase in fusion for WT or any of the MSD mutants (Fig. 2A). These results argue that the reduction in fusion observed with the MSD “core” mutants is not due to fusion-arrest at a hemifusion intermediate.
FIG. 2. Examination of hemifusion intermediates.
(A) Cell-cell fusion mediated by Env. HeLa cells, which were transfected with proviral DNAs, were cocultured with HeLa P4 indicator cells for 12 h, and then mixed cells were treated with 0.1mM CPZ for 30 seconds. After extensive wash, cells were incubated in complete DMEM for 10 h and then were subjected to a β-Gal staining. The cell-cell fusogenicity of Env mutants were measured by counting the number of cells involved into syncytia (blue cells) per well (at least two different wells were counted). (B) Flow cytometry analysis of cell-cell fusion. The proviral DNA-transfected HeLa cells were loaded with either membrane dye DiO or cytoplasmic dye Cell Tracker Green, and then were cocultured with DiD-loaded or Cell Tracker Blue-loaded HeLa P4 indicator cells, respectively, for 6 h. The cell mixtures then were subjected to flow cytometry analysis. The double-positive cell populations are shown as percentage to WT. The error bars represent standard deviations of three independent experiments. (“*” – statistically significant) (C) Three color microscopy of cell-cell fusion mediated by the Env. HeLa cells, which were transfected with proviral DNAs and loaded with CMFDA-Green, were cocultured with HeLa P4 indicator cells, which were double stained with DiI (red) and CMAC-blue, for 3 h. Fused cells were observed by fluorescent microscopy.
MSD “core” mutants can mediate full cell-cell fusion
In order to confirm the absence of hemifusion intermediates during membrane fusion mediated by the MSD mutants, we used dual-color flow cytometry analysis (Fig. 2B) and three-color microscopy (Fig. 2C) to examine cell-cell fusion mediated by the least fusogenic mutant, L12. To measure Env-mediated lipid exchange during cell-cell fusion, HeLa cells, which were transfected with NL4-3 proviral DNA and loaded with membrane dye DiO, were cocultured with DiD-loaded HeLa P4 indicator cells. Similarly, cytoplasmic dye CMFDA-green and CMAC-blue were used to stain HeLa cells and HeLa P4 cells, respectively, to measure the exchange of cytoplasm during cell-cell fusion. The DiO/DiD and Green/Blue double positive cells were examined by flow cytometry. As shown in Fig. 2B, the Env-mediated lipid exchange for the W610A mutant was 5-fold higher than the corresponding mixture of cytoplasmic contents, consistent with fusion being blocked at a hemifusion intermediate. In contrast, equivalent levels of lipid and cytoplasmic mixing were observed in the assays of Env mutant L12, suggesting that fusion proceeded to completion despite a reduction in fusion efficiency.
In the three-color microscopy experiment, Env-expressing HeLa cells, which were loaded with CMFDA-green, were co-cultured with DiI (red) and CMAC-blue double-stained HeLa P4 indicator cells. Even though cell-cell fusion induced by mutant L12 was only 11.1% of WT after 3 h of co-incubation (Fig. 1D), only complete cell-cell fusion events, in which both the lipid dye and cytoplasmic dyes are mixed, were observed (Fig. 2C). In contrast, hemifusion intermediates, in which the lipid dye but not the cytoplasmic dyes are mixed, were observed for mutant W610A (Fig. 2C). Similar studies, in which the L12 mutant was expressed from the proviral vector pTN6-GFP, demonstrated that those fusion pores which did form, were expanded to a size that allowed at least 30 KD molecules (GFP) to pass through (data not shown).
High levels of CD4 receptors or CXCR4 coreceptors on the surface of target cells can not rescue the defects of the MSD “core” mutants in viral infectivity
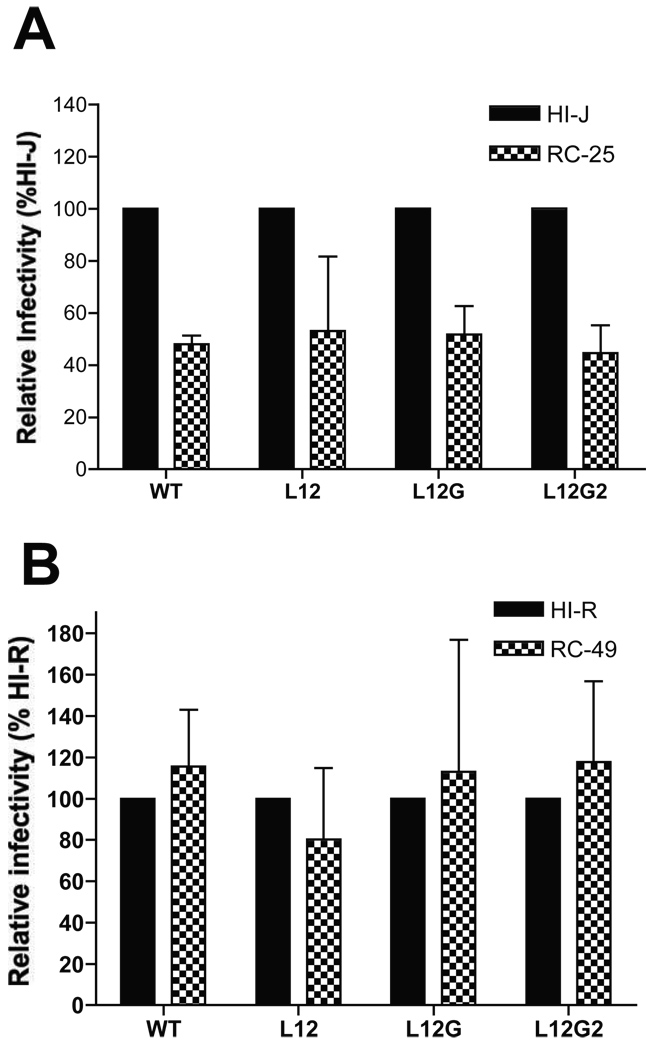
In order to determine whether the MSD core mutants exhibited differential utilization of the CD4 receptor and CXCR4 co-receptor, cell lines expressing different levels of these molecules were employed. In the first series of experiments, the HI-J, and RC-25 cell lines (Kabat), which express similar levels of CXCR4 but differ in their CD4 expression by 15-fold (HI-J>>RC-25) were utilized. The WT and Env mutants were cloned into the pTN7 proviral vector, in which the nef coding sequence is replaced by a renilla luciferase reporter gene. Viruses produced by transfecting 293T cells with these proviral DNAs were used to infect the two cell lines. At 48 h postinfection, cells were subjected to a luciferase activity assay. For each mutant, the viral infectivity in the HI-J cell lines was measured as 100% and compared with that in the RC-25 cell line. As shown in Fig. 3A, the higher level of surface CD4 enhanced virus infectivity of both the WT and MSD mutants, with both the MSD mutants and the WT exhibiting an approximately 2-fold increase in infectivity in the HI-J cells.
FIG. 3. Sensitivity of MSD core mutants to the levels of CD4 and CXCR4 on cell surface.
The Env mutants were cloned into a proviral vector, pTN7, in which the nef coding sequence was replaced by a renilla luciferase reporter gene. Viruses produced by transfecting 293T cells with pTN7 constructs were used to infect HI- and RC- cell lines (Kabat). Luciferase activities were measured 48 h postinfection. (A) Cell lines HI-J and RC-25 have the same surface level of CXCR4, but the CD4 level of HI-J is 15-fold higher than that of RC-25. (B) The cell surface levels of CD4 are similar between HI-R and RC-49 cells, while the surface level of CXCR4 on RC-49 is 6-fold lower than that of HI-R cells. The standard deviations of three independent experiments are shown in the error bars.
To investigate sensitivity to changes in CXCR4, the HI-R and RC-49 cells were utilized. These cells have equivalent levels of surface CD4, but a 6-fold difference in the surface level of CXCR4 (HI-R>>RC-49). In contrast to changes in CD4, changes in the surface level of CXCR4 had no effect on the infectivity of either the WT or MSD “core” mutants (Fig. 3B). These results suggest that the amino acid substitution in the MSD “core” does not affect CD4 or CXCR4 recognition by Env during viral entry.
MSD core mutants change the accessibility of epitopes to monoclonal antibodies in the gp120 glycoprotein
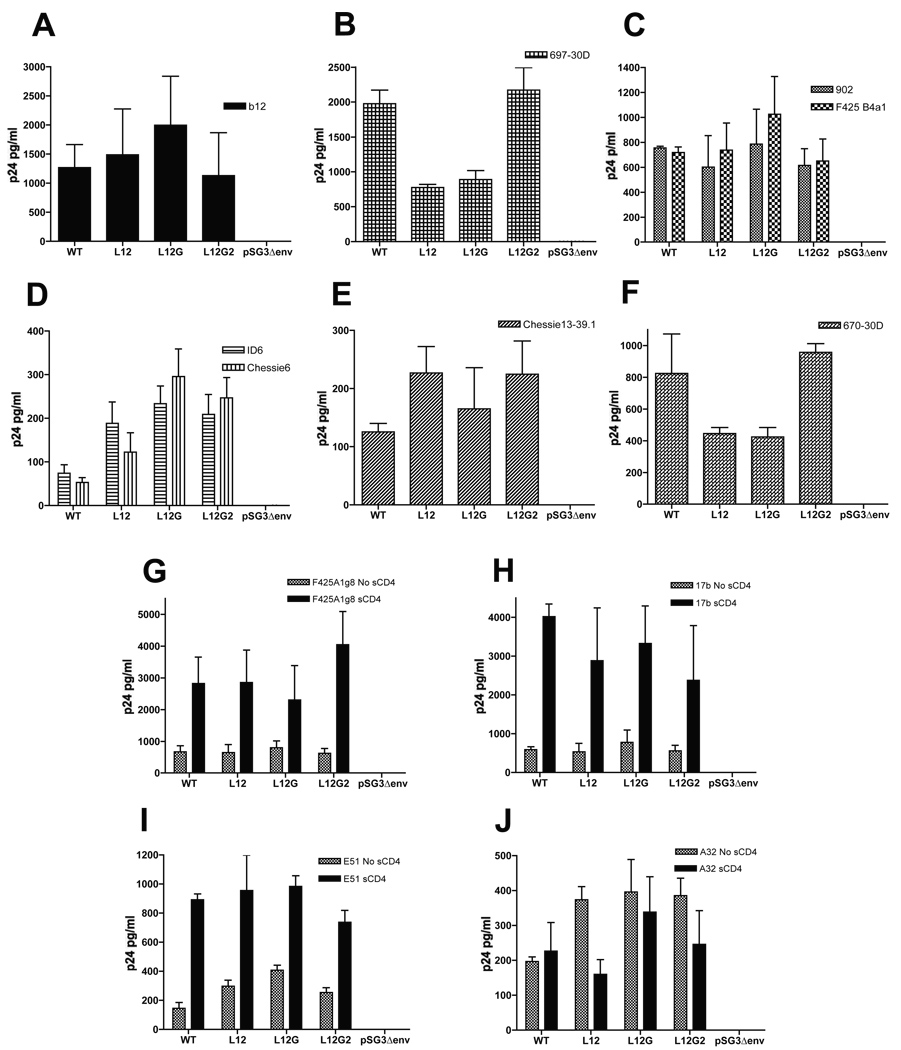
In order to determine whether the MSD mutants alter the conformational integrity of gp120, we measured the binding of anti-gp120 MAb to the Env glycoprotein on the viral surface by a virus capture assay. Viruses purified by ultracentrifugation through 25% sucrose were incubated in 96-well plates precoated with the specific anti-gp120 MAbs. The bound viruses were lysed after extensive washing and then subjected to a p24 ELISA assay. Two groups of MAbs were used to capture viruses, including (i) the CD4-independent MAbs: b12 (Fig. 4A), which binds to the CD4-binding site; 697-30D (Fig. 4B) - the V2 region; F425-B4a1 and 902 (Fig. 4C) - the V3 loop; ID6 and Chessie6 (Fig. 4D) - the C1 region; Chessie13–39.1 (Fig. 4E) - the C2 region; 670-30D (Fig. 4F) - the C5 region, and (ii) the CD4-induced MAbs: F425 A1g8 (Fig. 4G), 17b (Fig. 4H) and E51 (Fig. 4I) which bind the coreceptor-binding region; and A32 (Fig. 4J) - the C1/C4 region. The MSD mutants L12 and L12G showed an approximately 2-fold decrease in the binding of antibodies to epitopes in the V2 and C5 region (MAbs 697-30D and 670-30D), while the mutant L12G2 showed a WT binding ability to the MAbs in these regions. Moreover, the MSD “core” mutants resulted in a 2- to 4-fold increases in the binding of MAbs (ID6 and Chessie6) to epitopes in the C1 region. Consistent with the conformational changes in the C1 region, the binding of A32 to its conformational epitope in the C1 and C4 regions also increased approximately 2-fold. In spite of the conformational changes observed in the V2, C1, and C5 region, binding of antibodies to the V3 and C2 regions was not affected by the MSD “core” mutants. In addition, the accessibility of the MAb b12 to the CD4-binding region and CD4i MAbs F425a1g8, 17b and E51 to coreceptor-binding regions were not affected either, which is consistent with the infectivity data based on the Kabat cell lines, indicating an intact CD4 binding and co-receptor recognition of the Env mutants.
FIG. 4. gp120 virus capture assay.
Purified viruses were incubated at 37°C in 96-well plates which have been pre-coated with anti-gp120 monoclonal antibodies. The coating antibodies target to different epitopes of the gp120 glycoprotein: b12 (A) binds to the CD4-binding site, 697-30D (B) to the V2 region, F425 B4a1 and 902 (C) to the V3 loop, ID6 and Chessie6 (D) to the C1 region, Chessie13–39.1 (E) to the C2 region, 670-30D (F) to the C5 region, F425 A1g8 (G), 17b (H) and E51 (I) to the coreceptor-binding region, and A32 (J) to the C1/C4 region. The bound viruses were lysed in PBS containing 0.5% Triton X-100 and then subjected to a p24 ELISA assay. The experiment was carried out in triplicates for each antibody and the standard deviation is shown in the error bar.
MSD “core” mutants change the conformation of the gp41 ectodomain
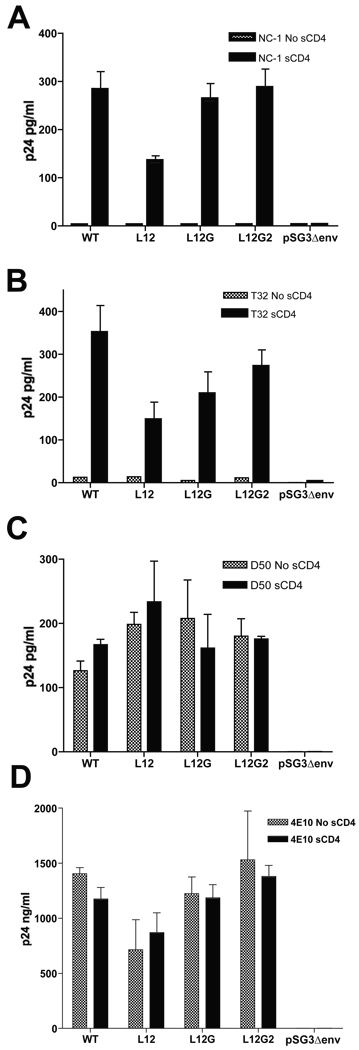
We used the same virus capture assay to examine potential conformational changes in the gp41 ectodomain. Four anti-gp41 MAbs were used: NC-1 (Fig. 5A) specifically binds to the 6-helix bundle, T32 (Fig. 5B) to the immunodominant loop in between the HR1 and HR2 regions, D50 (Fig. 5C) to the HR2 region, and 4E10 to the MPER region (Fig. 5D). Binding of antibody to the 6-helix bundle in the mutant L12 decreased to 50% of WT following induction by sCD4, while the mutants L12G and L12G2 were not significantly affected. Moreover, the exposure of the immunodominant loop was also reduced in the MSD mutants: L12 43% of WT, L12G 57%, and L12G2 81%, which suggested a stoichiometric relationship between the conformational changes in the gp41 ectodomain and the number of substitutions in the MSD “core” region. While the HR2 region of the MSD “core” mutants was fully accessible to the MAb D50 (Fig. 5C), exposure of the downstream MPER region, as assayed by MAb 4E10 binding, was decreased in the L12 mutant. These results were confirmed by immunoprecipitating viral gp41 with the same MAbs in the presence or absence of sCD4, followed by a western blotting using the anti-gp41 MAb Chessie8 (data not shown).
FIG. 5. gp41 virus capture assay.
Purified viruses were incubated at 37°C in 96-well plates which were pre-coated with anti-g41 monoclonal antibodies. The coating antibodies target to different epitopes of the gp41 glycoprotein: NC-1 (A) to the 6-helix bundles, T32 (B) to the immunodominant loop of gp41 ectodomain, D50 (C) to the HR2 region, and 4E10 (D) to the MPER region. Bound viruses were lysed in PBS containing 0.5% Triton X-100 and then subjected to a p24 ELISA assay. The experiment was carried out in triplicates for each antibody and the standard deviation is shown in the error bar.
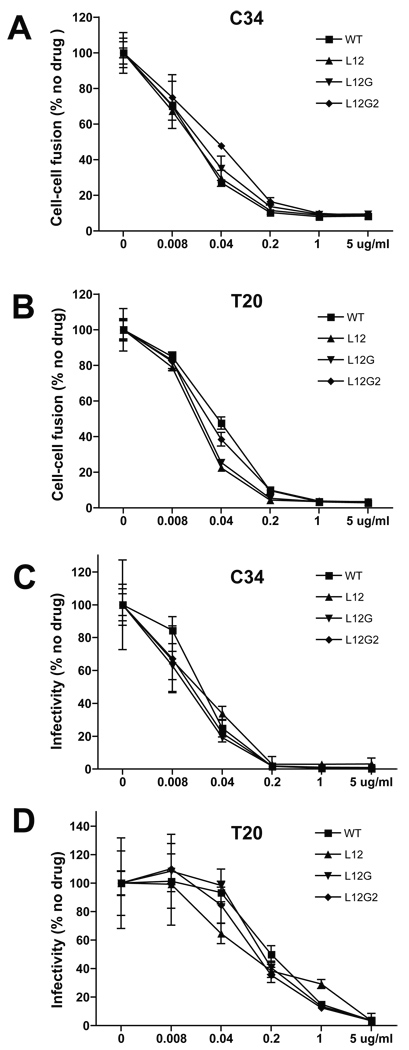
MSD “core” mutants exhibit the same sensitivity to the fusion inhibitors, T20 and C34, as WT
In order to understand the degree to which the conformation of the gp41 ectodomain has been changed by the MSD “core” mutants, we compared sensitivity of the Env mutants to the fusion inhibitors, C34 and T20, to that of WT. The inhibition kinetics of Env-mediated cell-cell fusion was measured by treating cells with the fusion inhibitors in a gradient of concentrations (Fig. 6A and 6B). The MSD mutants were as sensitive to either T20 or C34 as WT. Similar results were observed in the experiment measuring the T20 and C34 inhibition kinetics of viral infectivity (Fig. 6C and 6D). These data indicated that alterations in the gp41 ectodomain induced by the MSD “core” mutants were unlikely to slow formation of the 6-helix bundle once conformational changes were initiated and that access to the HR1 coiled-coil was retained in the MSD mutants, allowing the binding of the inhibitor peptides.
FIG. 6. C34 and T20 inhibition of cell-cell fusion mediated by the Env and viral infection.
293T cells transfected with the Env expression vector, pCDNA3.1, were cocultured with JC53BL indicator cells. Cell mixtures were incubated for 24 h in the presence of fusion inhibitors, C34 (A) and T20 (B), at specific concentrations, and then were subjected to a luciferase activity assay. JC53BL indicator cells were infected with the p24-normalized viruses in DMEM containing 1% fetal bovine serum, 80µg/ml DEAE-Dextran and fusion inhibitors, C34 (C) and T20 (D), at specific concentrations. Complete DMEM was added following 2 h incubation and cells were analyzed for luciferase activity 48 h post infection. Error bars represent standard deviations of at least three independent experiments.
DISCUSSION
Previous studies have demonstrated that, in addition to anchoring the Env in membrane, the membrane-spanning domain of HIV-1 gp41 plays a role in Env incorporation and virus-cell entry during the viral life cycle (Shang, Yue, and Hunter, 2008; Yue, Shang, and Hunter, 2009). The MSD, and in particular its N-terminal “core” region, is a very highly conserved region in the Env sequence. We have demonstrated that the most conserved residue F683 and the GGxxG motif in the MSD “core” are critical for the virus-cell entry and subsequent viral infectivity by using a “recovery-of-function” mutagenesis approach (Shang, Yue, and Hunter, 2008). Truncation of the cytoplasmic domain of the MSD “core” mutants was not able to rescue the defect in virus-cell entry. In the current study, we investigated the molecular basis for the defects in viral entry observed with the MSD “core” mutants through an examination of conformational changes in the extracellular domains of Env, as well as changes in the efficiency of virus-cell attachment, receptor/coreceptor recognition, and membrane fusion.
Virions carrying the MSD “core” mutants all exhibited WT binding to the surface of target cells at different temperatures, indicating that substitutions in the MSD “core” regions did not influence the specific virus-cell binding mediated by the interaction of HIV-1 Env with CD4 receptors and chemokine coreceptors on cell surface. Consistent with this result, we demonstrated that changes in the surface levels of CD4 receptors and CXCR4 coreceptors on target cells gave rise to similar variations in viral infectivity for the WT and for the MSD mutants. This result suggested that substitutions in the MSD “core” region did not affect the utilization of CD4 receptors and CXCR4 coreceptors on cell surface by HIV-1 Env. Thus, in spite of the altered MSD “core” region in the gp41 glycoprotein, the general structure of the Env glycoprotein of the MSD mutants was retained, and the conformation of the CD4-binding site and chemokine receptor-binding site remained intact. However, the MSD “core” mutants showed significant differences in the kinetics of cell-cell fusion mediated by HIV-1 Env. This was most pronounced for L12, which exhibited a clear lag phase prior to initiation of fusion and a slower rate of increase in the extent of fusion. The mutants L12G and L12G2 demonstrated intermediate kinetics between L12 and WT in both the initiation and expansion period of cell-cell fusion. Therefore, the defect in virus-cell entry of the MSD mutants is most likely due to the impaired kinetics of membrane fusion mediated by the Env mutants, in particular at the initiation stage.
The utilization of a panel of MAb in a solid state virus binding assay allowed us to indirectly probe for changes in the conformation of gp120 and gp41 by determining whether exposure of a variety of epitopes had been altered by the MSD mutants. Consistent with the equivalent entry of all three mutants into cells expressing different levels of CD4, each mutant bound to the CD4-binding site antibody b12 similarly to WT. In addition, the regions involved in coreceptor recognition bound to the CD4 inducible MAbs (F425A1g8, 17b and E51) at WT levels. However, minor conformational changes, which were restricted to specific regions of gp120, were observed in the MSD mutants. Among all the constant and variable regions of gp120, only the V2, C1, and C5 regions exhibited altered binding to their cognate MAbs (697-30D, ID6, Chessie 6, 670-30D), while the remaining regions in gp120 showed no differences in antibody binding. Since C1 and C5 have been implicated in the non-covalent interactions between gp120 and gp41, the altered accessibility of these regions to antibodies could reflect a perturbation in the gp120-gp41 interface.
While CD4 induced epitopes in the bridging sheet were presented equally by the mutants and WT, minor localized conformational changes were also observed in the gp41 ectodomain of the MSD mutants by the reductions in the formation of the 6-helix bundle (MAb NC-1), and exposure of the immunodominant loop to MAb T32. This suggested that even though the open configuration of gp120, with an exposed bridging sheet, could be stabilized by the binding of CD4 in the MSD mutants, it was less efficient at inducing downstream conformational changes in gp41. Moreover, this could result in the lag phase and slower kinetics of fusion observed for the MSD “core” mutants. It is difficult to define quantitatively how the conformational changes observed above might correlate with the decrease in fusogenicity. However, one possible interpretation of these results is that changes in the MSD core alter in a subtle manner the interaction of gp120 and gp41 in such a way that there is a greater energy barrier to conformational changes in gp41 induced by CD4 and CXCR4. This would result in a reduction in overall efficiency of the fusion process. It seems likely, however, that once the conformation change is initiated it progresses with equivalent kinetics to WT, since all of the mutants showed similar sensitivity to the fusion inhibitors T20 and C34, which bind to the trimeric coiled-coil of HR1 and prevent 6-helix bundle formation. Previous studies have shown that reductions in the kinetics of gp41 conformation changes result in enhanced inhibition by these fusion inhibitors, and this was not observed with the MSD “core” mutants. The lack of any evidence for hemifusion intermediates or defects in fusion pore expansion in these studies, suggests that the defect in the MSD core mutants is in the initiation of conformational changes required for fusion rather than at these later stages of the fusion process. This is consistent with the studies of Miyauchi et al. (2005), who characterized HIV Env mutants with MSD replacements in this region (Miyauchi et al., 2005).
How might changes in the amino acid composition of the MSD “core” induce conformational changes and inhibit processes within the ectodomain of Env? The highly conserved GGXXG motif in the HIV-1 MSD “core” region has been postulated to facilitate the intramembrane helix-helix association by providing a relatively flat platform for the contact of the three helixes (Russ and Engelman, 2000; Senes, Gerstein, and Engelman, 2000). Moreover, residues F683 and V687 are also suggested to contribute to such an interaction via a (FXX)(V)GGXXG motif (Unterreitmeier et al., 2007). It is possible that these interactions are optimized to allow some flexibility within the trimer in order to facilitate conformational changes in gp41. By substituting a stretch of hydrophobic leucine residues into this region, it is likely that affinity of both protein-protein and protein-lipid interactions will be increased, or that the orientation of helixes in the membrane would be altered. If this resulted in a more rigid trimeric interaction, it could explain altered interactions with gp120 and the imposition of a greater energy barrier to gp41 conformational changes, with subsequent impact on the formation of 6-helix bundles and membrane fusion.
In summary, the experimental approaches described here have demonstrated that consistent with its high sequence conservation, the native MSD “core” amino acid sequence of HIV Env is critical for efficient membrane fusion during viral infection. Substitutions in the MSD “core” region reduced the fusogenicity of the Env mainly by inducing structural changes in the gp41 ectodomain, thereby, influencing gp120-gp41 interactions and consequently impairing initiation of the fusion process.
MATERIALS AND METHODS
Cells and antibodies
COS-1, 293T, JC53BL cells and KABAT cell lines were maintained in complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and penicillin-streptomycin (all from Gibco-BRL, Rockville, MD). Cells were grown under conditions of 37°C and 5% CO2 in humidified incubators. The anti-gp120 monoclonal antibodies b12 (Burton et al., 1994), 697-30D (Zolla-Pazner et al., 1995), 902 (Chesebro and Wehrly, 1988), F425b4a1, ID6 (Gomez-Roman et al., 2002), Chessie6 (Abacioglu et al., 1994), Chessie13–39.1 (Abacioglu et al., 1994), 670-30D (Zolla-Pazner et al., 1995), F425a1g8, 17b (Kwong et al., 1998), E51 (Huang et al., 2004), A32 (Wyatt et al., 1995), and anti-gp41 monoclonal antibodies NC-1 (Jiang, Lin, and Lu, 1998), T32 (Earl et al., 1997), 4E10 (Stiegler et al., 2001), and D50 (Earl et al., 1997) along with HIV-1 patient Ig, fusion inhibitors (T20 and C34) were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The pooled HIV-1 patient sera were provided through the Emory CFAR Clinical Core.
Virus-cell attachment assay
Viruses were harvested from proviral DNA (NL4-3)-transfected 293T cells and purified through a 25% sucrose cushion by ultracentrifugation. JC53BL indicator cells, which express the CD4 receptor and coreceptors (CXCR4 and CCR5), were co-incubated with the same amount of viruses (normalized by the amount of p24) at 4°C, 10°C, and 25°C for 2 h. After extensive wash in PBS to remove unbounded viruses, cells and bounded viruses were lysed in PBS containing 0.5% Triton X-100. The amount of viruses in cell lysate was measured by p24 ELISA.
Cell-cell fusion kinetics
MSD mutants were cloned into a proviral vector pTN6-GFP, in which the nef gene has been replaced by a GFP reporter gene. 293T cells, which were transfected with these proviral vectors, were resuspended 48 h post-transfection and were mixed with JC53BL indicator cells, which had been loaded with fluorescent dye CMAC-blue (Invitrogen, Carlsbad, CA). The cell mixtures were incubated at 37°C and 5% CO2 for 0.5 h, 1 h, 2 h, 4 h, 6 h, 12 h, and 24 h. Cell-cell fusion was observed and quantitated by fluorescent microscopy. The rate of cell-cell fusion was calculated as: (# of fused cells at t2 - # of fused cells at t1)/(total # of cells)(t2 - t1).
CPZ treatment of cell-cell fusion
HeLa cells were transfected with proviral DNAs (NL4-3). At 48 h after transfection, cells were co-cultured with HeLa P4 indicator cells, which express the CD4 receptor and CXCR4 coreceptor and also carry a β-galactosidase reporter gene, at a 1:1 ratio. Cell mixtures were incubated for 12 h, and then were treated with 0.1mM chlorpromazine (CPZ, in PBS) for 30 seconds. After extensive wash in PBS, cells were incubated in complete DMEM for 12 and then subjected to β-Gal staining. The numbers of blue cells (per well), which were involved into syncytia, were counted to measure the cell-cell fusogenicity of Env mutants.
Flow cytometry analysis of cell-cell fusion
HeLa cells were transfected with pNL4-3 proviral DNAs (NL4-3). At 48 h after transfection, cells were stained in serum-free DMEM containing either 5ul/ml DiO loading solution (Invitrogen, Carlsbad, CA) or Cell Tracker Green CMFDA (Invitrogen, Carlsbad, CA) for 45 min at 37 °C. HeLa P4 cells were stained in serum-free DMEM containing either 5ul/ml DiD loading solution (Invitrogen, Carlsbad, CA) or Cell Tracker Blue CMAC (Invitrogen, Carlsbad, CA) for 45 min at 37 °C. All cells were washed three times in PBS after incubation and resuspended in PBS containing 1mM EDTA. To measure the potential membrane exchange mediated by HIV-1 Env, HeLa (DiO) cells were mixed with HeLa P4 (DiD) cells; and to observe Env-mediated cell-cell fusion, HeLa (Green) cells were mixed with HeLa P4 (Blue) cells. After 6 h co-incubation at 37 °C, cells were resuspended by trypsinization, fixed in 4% paraformaldehyde, and then subjected to flow cytometry analysis.
Three color microscopy of cell-cell fusion
HeLa cells (2.5×105 cells per well in 6-well plates) were transfected with pNL4-3 proviral DNAs. At 48 h after transfection, cells were stained in serum-free DMEM containing 10mM cytoplasmic dye CMFDA-green (Invitrogen, Carlsbad, CA) for 45 min at 37 °C, followed by three washes in serum-free DMEM. HeLa P4 cells (2.5×105 cells per well in 6-well plates) were initially stained with 5 ul/ml DiI loading solution (Invitrogen, Carlsbad, CA) in serum-free DMEM. Cells were washed three times in serum-free DMEM after 45 min incubation at 37 °C and then were incubated with 10mM CMAC-blue (Invitrogen, Carlsbad, CA) in serum-free DMEM for 45 min, followed by wash in serum-free DMEM three times. The transfected HeLa and the double-stained HeLa P4 cells were both resuspended in PBS containing 1mM EDTA and then were mixed (1:1) in complete DMEM. The cell mixtures were cocultured at 37 °C for 3 h. Cell-cell fusion events were observed by fluorescent microscopy.
Virus capture assay
Viruses were harvested from the supernatant of proviral vector transfected 293T cells by ultracentrifugation through 25% sucrose cushion. Pelleted virus (in PBS) was added into 96-well plates (10ng p24 per well), which had been coated with anti-gp120 and anti-gp41 monoclonal antibodies in 50mM carbonate/bicarbonate buffer pH9.6, and then incubated at 37 °C for 2 h. For studies involving CD4-induced exposure of epitopes, 1ug/ml sCD4 was mixed with the virus immediately prior to addition to the wells. Unbound viral particles were removed by extensive washes in PBS. Bound viruses were lysed in PBS containing 0.5% Triton X-100 and then subjected to a p24 ELISA assay.
C34 and T20 inhibition of cell-cell fusion and viral infection
293T cells were transfected with pCDNA3.1 expression vectors, which also express HIV-1 Tat protein. At 36–48h after transfection, cells resuspended by trypsinization were combined with JC53BL indicator cells at a 5:1 ratio. The JC53BL indicator cells carry a HIV LTR promoter-driven luciferase reporter gene, which is activated by Tat. Cell mixtures were incubated for 24 h in the presence of T20 or C34 in specific concentrations (0, 0.008, 0.04, 0.2, 1, and 5 ug/ml) before the analysis of luciferase activity.
Medium from 293T cells transfected with proviral vectors was harvested 72 h posttransfection, filtered through a 0.45µm membrane to remove cellular debris, and total virions were quantified using a p24 ELISA assay. The p24-normalized virus-containing supernatant was added to JC53BL indicator cells cultured in DMEM containing 1% fetal bovine serum, 80µg/ml DEAE-Dextran and T20 or C34 in specific concentrations. Complete DMEM was added following 2 h incubation and cells were analyzed for luciferase activity at 48 h postinfection.
ACKNOWLEDGEMENTS
We thank Drs. Lara Pereira and Malinda Schaefer for critical reading of the manuscript. The pooled HIV-1 patient sera were kindly provided by Jeffery Lennox through the Clinical Core, and flow cytometry was performed in the Immunology Core of the Emory Center for AIDS Research (P30 AI050409). The antibodies, immunoglobulin, and fusion inhibitors used in this study were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: D50 and T32 from Dr. Patricia Earl; NC-1 from Dr. Shibo Jiang; b12 from Dr. Dennis Burton and Carlos Barbas; F425 A1g8 and F425 B4a1 from Dr. Marshall Posner and Dr. Lisa Cavacini; 17b, E51, and A32 from Dr. James E. Robinson; 902 from Dr. Bruce Chesebro; 697-30D and 670-30D from Dr. Susan Zolla-Pazner; ID6 from Dr Kenneth Ugen and Dr. David Weiner; Chessie 6 and Chessie 13–39.1 from Dr. George K. Lewis, and 4E10 from Dr. Hermann Katinger. This work was supported by grant R01 AI33319 (E.H.) from the National Institute of Allergy and Infectious Diseases at the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- Abacioglu YH, Fouts TR, Laman JD, Claassen E, Pincus SH, Moore JP, Roby CA, Kamin-Lewis R, Lewis GK. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retroviruses. 1994;10(4):371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- Bar S, Alizon M. Role of the ectodomain of the gp41 transmembrane envelope protein of human immunodeficiency virus type 1 in late steps of the membrane fusion process. J Virol. 2004;78(2):811–820. doi: 10.1128/JVI.78.2.811-820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd MT, Simpson GR, Cann AJ, Johnson MA, Weiss RA. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67(6):3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266(5187):1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Byland R, Vance PJ, Hoxie JA, Marsh M. A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol Biol Cell. 2007;18(2):414–425. doi: 10.1091/mbc.E06-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Determining the structure of an unliganded and fully glycosylated SIV gp120 envelope glycoprotein. Structure. 2005;13(2):197–211. doi: 10.1016/j.str.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Chesebro B, Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J Virol. 1988;62(10):3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MW, Lee MK, Carney MC, Berson JF, Doms RW, Martin MA. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72(3):2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comardelle AM, Norris CH, Plymale DR, Gatti PJ, Choi B, Fermin CD, Haislip AM, Tencza SB, Mietzner TA, Montelaro RC, Garry RF. A synthetic peptide corresponding to the carboxy terminus of human immunodeficiency virus type 1 transmembrane glycoprotein induces alterations in the ionic permeability of Xenopus laevis oocytes. AIDS Res Hum Retroviruses. 1997;13(17):1525–1532. doi: 10.1089/aid.1997.13.1525. [DOI] [PubMed] [Google Scholar]
- Earl PL, Broder CC, Doms RW, Moss B. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J Virol. 1997;71(4):2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- Gomez-Roman VR, Cao C, Bai Y, Santamaria H, Acero G, Manoutcharian K, Weiner DB, Ugen KE, Gevorkian G. Phage-displayed mimotopes recognizing a biologically active anti-HIV-1 gp120 murine monoclonal antibody. J Acquir Immune Defic Syndr. 2002;31(2):147–153. doi: 10.1097/00126334-200210010-00004. [DOI] [PubMed] [Google Scholar]
- Hartley O, Klasse PJ, Sattentau QJ, Moore JP. V3: HIV's switch-hitter. AIDS Res Hum Retroviruses. 2005;21(2):171–189. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W, Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J Virol. 1990;64(12):6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman TL, Stephens EB, Narayan O, Doms RW. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc Natl Acad Sci U S A. 1998;95(19):11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie SE, Cotton GJ, Heslop I, Martin NJ, Harrison DJ, Ramage R. Synthetic peptides representing discontinuous CD4 binding epitopes of HIV-1 gp120 that induce T cell apoptosis and block cell death induced by gp120. FASEB J. 1998;12(11):991–998. doi: 10.1096/fasebj.12.11.991. [DOI] [PubMed] [Google Scholar]
- Howie SE, Fernandes ML, Heslop I, Hewson TJ, Cotton GJ, Moore MJ, Innes D, Ramage R, Harrison DJ. A functional, discontinuous HIV-1 gp120 C3/C4 domain-derived, branched, synthetic peptide that binds to CD4 and inhibits MIP-1alpha chemokine binding. FASEB J. 1999;13(3):503–511. doi: 10.1096/fasebj.13.3.503. [DOI] [PubMed] [Google Scholar]
- Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A. 2004;101(9):2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey-Hoyle M, Clark RK, Rosenberg M. The N-terminal 31 amino acids of human immunodeficiency virus type 1 envelope protein gp120 contain a potential gp41 contact site. J Virol. 1991;65(5):2682–2685. doi: 10.1128/jvi.65.5.2682-2685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Lin K, Lu M. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1998;72(12):10213–10217. doi: 10.1128/jvi.72.12.10213-10217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koito A, Harrowe G, Levy JA, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68(4):2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropelin M, Susal C, Daniel V, Opelz G. Inhibition of HIV-1 rgp120 binding to CD4+ T cells by monoclonal antibodies directed against the gp120 C1 or C4 region. Immunol Lett. 1998;63(1):19–25. doi: 10.1016/s0165-2478(98)00047-9. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrosse B, Treboute C, Brelot A, Alizon M. Cooperation of the V1/V2 and V3 domains of human immunodeficiency virus type 1 gp120 for interaction with the CXCR4 receptor. J Virol. 2001;75(12):5457–5464. doi: 10.1128/JVI.75.12.5457-5464.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemasson I, Housset V, Calas B, Devaux C. Antigenic analysis of HIV type 1 external envelope (Env) glycoprotein C2 region: implication for the structure of Env. AIDS Res Hum Retroviruses. 1995;11(10):1177–1186. doi: 10.1089/aid.1995.11.1177. [DOI] [PubMed] [Google Scholar]
- Miller MA, Cloyd MW, Liebmann J, Rinaldo CR, Jr, Islam KR, Wang SZ, Mietzner TA, Montelaro RC. Alterations in cell membrane permeability by the lentivirus lytic peptide (LLP-1) of HIV-1 transmembrane protein. Virology. 1993;196(1):89–100. doi: 10.1006/viro.1993.1457. [DOI] [PubMed] [Google Scholar]
- Miyauchi K, Komano J, Yokomaku Y, Sugiura W, Yamamoto N, Matsuda Z. Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion. J Virol. 2005;79(8):4720–4729. doi: 10.1128/JVI.79.8.4720-4729.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, McKeating JA, Weiss RA, Sattentau QJ. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250(4984):1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- Morrison HG, Kirchhoff F, Desrosiers RC. Effects of mutations in constant regions 3 and 4 of envelope of simian immunodeficiency virus. Virology. 1995;210(2):448–455. doi: 10.1006/viro.1995.1361. [DOI] [PubMed] [Google Scholar]
- Orloff SL, Bandea CI, Kennedy MS, Allaway GP, Maddon PJ, McDougal JS. Increase in sensitivity to soluble CD4 by primary HIV type 1 isolates after passage through C8166 cells: association with sequence differences in the first constant (C1) region of glycoprotein 120. AIDS Res Hum Retroviruses. 1995;11(3):335–342. doi: 10.1089/aid.1995.11.335. [DOI] [PubMed] [Google Scholar]
- Owens RJ, Dubay JW, Hunter E, Compans RW. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci U S A. 1991;88(9):3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowell JF, Stanhope PE, Siliciano RF. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol. 1995;155(1):473–488. [PubMed] [Google Scholar]
- Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296(3):911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14(4):465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296(3):921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- Shang L, Yue L, Hunter E. Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell-cell fusion and virus infection. J Virol. 2008;82(11):5417–5428. doi: 10.1128/JVI.02666-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Gojobori T. How can human and simian immunodeficiency viruses utilize chemokine receptors as their coreceptors? Gene. 2000;259(1–2):199–205. doi: 10.1016/s0378-1119(00)00432-7. [DOI] [PubMed] [Google Scholar]
- Smyth RJ, Yi Y, Singh A, Collman RG. Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate. J Virol. 1998;72(5):4478–4484. doi: 10.1128/jvi.72.5.4478-4484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2001;17(18):1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- Tencza SB, Miller MA, Islam K, Mietzner TA, Montelaro RC. Effect of amino acid substitutions on calmodulin binding and cytolytic properties of the LLP-1 peptide segment of human immunodeficiency virus type 1 transmembrane protein. J Virol. 1995;69(8):5199–5202. doi: 10.1128/jvi.69.8.5199-5202.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterreitmeier S, Fuchs A, Schaffler T, Heym RG, Frishman D, Langosch D. Phenylalanine promotes interaction of transmembrane domains via GxxxG motifs. J Mol Biol. 2007;374(3):705–718. doi: 10.1016/j.jmb.2007.09.056. [DOI] [PubMed] [Google Scholar]
- White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk T, Pfeiffer T, Bukovsky A, Moldenhauer G, Bosch V. Glycoprotein incorporation and HIV-1 infectivity despite exchange of the gp160 membrane-spanning domain. Virology. 1996;218(1):269–274. doi: 10.1006/viro.1996.0190. [DOI] [PubMed] [Google Scholar]
- Willey RL, Martin MA. Association of human immunodeficiency virus type 1 envelope glycoprotein with particles depends on interactions between the third variable and conserved regions of gp120. J Virol. 1993;67(6):3639–3643. doi: 10.1128/jvi.67.6.3639-3643.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69(9):5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Yue L, Shang L, Hunter E. Truncation of the Membrane-spanning Domain of Human Immunodeficiency Virus Type I Envelope Glycoprotein Defines Elements Required for Fusion, Incorporation and Infectivity. J Virol. 2009 doi: 10.1128/JVI.00914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, O'Leary J, Burda S, Gorny MK, Kim M, Mascola J, McCutchan F. Serotyping of primary human immunodeficiency virus type 1 isolates from diverse geographic locations by flow cytometry. J Virol. 1995;69(6):3807–3815. doi: 10.1128/jvi.69.6.3807-3815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]