Abstract
Background
Detection of viral genome in rejecting cardiac transplant patients has been reported, with coxsackievirus and adenovirus causing premature graft failure. Recently, parvovirus B19 (PVB19) genome in myocardial samples has been increasingly reported but its role in cardiac pathology and effect on transplant graft survival are unknown. The objectives were to determine if changes in the viruses identified in the myocardium represent an epidemiologic shift in viral myocardial disease and whether PVB19 adversely affects transplant graft survival.
Methods
From 9/2002 to 12/2005, 99 children (3 weeks-18 years) with heart transplants had endomyocardial biopsies evaluated for the presence of viral genome utilizing nested PCR. Cellular rejection was assessed by histology of biopsies, while transplant coronary artery disease (TCAD) was diagnosed by coronary angiography or histopathology.
Results
Seven hundred biopsies were evaluated from 99 patients; 121 biopsies had viral genome with 100 (82.6%) positive for PVB19, 24 for Epstein-Barr virus (EBV; 7 positive for PVB19 and EBV), 3 for CMV and 1 for adenovirus. Presence of PVB19 genome did not correlate with rejection score, nor did higher viral copy number. Children with persistent PVB19 infection (>6 months; n=20), had early development of advanced TCAD (p<0.001).
Conclusions
PVB19 is currently the predominant virus detected in heart transplant surveillance biopsies, possibly representing an epidemiologic shift. While cellular rejection does not correlate with the presence or quantity of PVB19 genome in the myocardium, children with chronic PVB19 infection have increased risk for earlier TCAD, supporting the hypothesis that PVB19 negatively affects graft survival.
INTRODUCTION
Cardiac transplantation is an important treatment modality for children with end-stage heart disease due to congenital heart disease or cardiomyopathies, with cardiomyopathies predominating at present. However, transplantation is only palliative as long-term graft survival in children is limited to a median survival of 11.3 years.(1) Graft loss occurs due to complications from acute cellular rejection, transplant coronary artery disease (TCAD), lymphoproliferative disease, and infection.(1)
Viruses have been implicated in the development of graft dysfunction of heart,(1) lung,(2) and renal transplants.(3) The association between viral infections and myocardial disease was first described by Grist and Bell in 1974(4) based on serologic evidence of enteroviral infection with myocarditis. These studies led to the recognition of a potential cause and effect relationship of viruses and cardiac dysfunction. The group B coxsackieviruses were initially identified as the primary viruses in patients with viral myocarditis. This was true throughout the 1970’s and the 1980’s.(4–10) During the 1990’s adenovirus was increasingly reported as an important etiologic agent in myocarditis,(11–13) nonimmune hydrops fetalis and intrauterine growth restriction,(14, 15) as well as in graft loss after heart(16) and lung(1) transplantation. In fact, the frequency of adenovirus genome identification in the myocardium surpassed that of enterovirus during the 1990’s in most studies.(12) More recently (i.e., since 2000), other viruses have become increasingly reported, particularly parvovirus B19 (PVB19). The specific viral pathogens implicated in transplant graft dysfunction have mirrored those found in myocarditis patients.(8, 12, 16)
Parvovirus B19 is a ubiquitous pathogen, with the prevalence of IgG antibodies ranging from 2 to 15% in children 1 to 5 years old, and 15 to 60% in children 6 to 19 years old.,(17–20) but typically causing only mild illness in children. Since its initial description by Cossart et al.,(21) PVB19 has been implicated in such disease states as erythema infectiosum (“fifth disease”), anemia and aplastic crises, hydrops fetalis, arthritis, and vasculitis.(22–24) PVB19 was previously a rarely identified virus in endomyocardial biopsy samples (EMB), but has emerged as an important pathogen in cardiac patients over the past several years.(11, 25–29) Although increasingly detected, the role of PVB19 in heart disease is not well understood. Therefore, we sought to evaluate the epidemiology of viral myocardial disease and determine the implications of PVB19 genome in the endomyocardium of pediatric heart transplant patients.
METHODS
Patients
This ambispective study evaluates the endomyocardial biopsies from heart transplant recipients at Texas Children’s Hospital (TCH) between September 2002 and December 2005. The medical records of all children with a history of heart transplantation followed at TCH from September 2002 to July 2004 were retrospectively reviewed. Children undergoing heart transplantation from July 2004 to December 2005 were prospectively enrolled; children with heart transplants who underwent endomyocardial biopsy during this period were also prospectively enrolled for Parvovirus quantification and concurrent serologic evaluation. The Baylor College of Medicine Institutional Review Board approved the study and informed consent was obtained from each participant, or his or her guardian. Retrospective enrollment was exempted from consent. Serial endomyocardial biopsy specimens from children with heart transplants undergoing routine rejection surveillance or acute rejection evaluation were analyzed. Laboratory and clinical data generated between September 2002 and July 2004 was reviewed retrospectively. The presence of viral genome in EMBs amplified from at least 2 biopsy samples obtained at least six months apart, with identical genotypes, was considered evidence of chronic infection.
Previously published data describing the prevalence of specific viruses identified in the myocardium of biopsy samples at our institution, but in an earlier era, (12, 16) were reviewed to compare to viruses identified in the current era.
Cardiac Catheterization and Endomyocardial Biopsy
Endomyocardial biopsies were obtained from the right ventricular septum by standard percutaneous transvenous femoral or jugular approach using a Cordis bioptome (Cordis, Haan, Germany) modified by Olsen et al.(30) Biopsy samples for PCR analysis were immediately immersed in RNAlater (Qiagen, Valencia, CA) and stored at −80°C. Biopsy samples for histologic analysis were formalin-fixed and paraffin-embedded.
Screening coronary angiography was performed at three months post-transplantation for baseline assessments, then annually. Assessment of coronary arteriopathy was performed by the cardiologist performing the catheterization and reviewed by the director of cardiac transplantation (WJD). The cardiologists evaluating coronary angiography were blinded to the results of PCR data. Coronary vasodilators are not administered as part of routine coronary angiography.
Detection of Viral Genomes by Nested PCR
Total RNA and DNA were isolated simultaneously from frozen tissue samples using Trizol (Invitrogen; Gaithersburg, MD).(13, 31) The presence of amplifiable nucleic acid extracted from each sample was verified by amplifying cellular nucleic acid (G6PDH for RNA and β-actin for DNA) in all cases. Each biopsy sample was screened for a panel of viral genomes: adenovirus, cytomegalovirus (CMV), Epstein-Barr virus (EBV), enterovirus and PVB19. Viral genomes were detected by nested reverse transcription-polymerase chain reaction (RT-PCR: RNA viruses) or PCR (DNA viruses): all analyses were performed in the John Welsh Cardiovascular Diagnostic Laboratory at Baylor College of Medicine, as previously described.(13, 32)
For all analyses, negative and positive control reactions were performed in a manner identical to that of the test reactions, using ultrapure water and viral or human nucleic acid (total cardiac RNA and genomic DNA), respectively, as the template. All samples were analyzed without knowledge of the clinical status of the patient or results of histologic evaluation. All viral positive results were confirmed by repeating the analysis in duplicate and by DNA sequencing of the amplification product.
To confirm the identity of viral amplimers and to identify the parvoviral genotype, the PCR products were purified using a QIAquick purification kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions. The PCR product was sequenced using Big Dye Terminator chemistry (version 3.1) and an ABI3700 DNA analyzer (Applied Biosystems (ABI), Foster City, CA), according to the manufacturer’s instructions. Sequence comparisons were performed by BLAST search of GenBank databases. For genotyping, parvoviral DNA sequences were aligned with and compared to the prototypic sequences for parvovirus types 1 (GenBank # AY386330), 2 (GenBank # AY0442680) and 3 (GenBank # AJ24937) using Sequencher (Gene Codes, Ann Arbor, MI).
Parvovirus B19 Quantification
To quantitate virus copy number, real time quantitative PCR (qPCR) was performed on DNA isolated from biopsy samples that were positive for PVB19. For human genomic DNA quantification, DNA standards were prepared at the following concentrations: 10 ng/μl, 1 ng/μl, 0.1 ng/μl, and 0.01 ng/μl. For PVB19 genomic DNA quantification, the following standards were used: 1000 copies/μl, 100 copies/μl, 10 copies/μl, and 1 copy/μl. The DNA was subjected to real-time PCR (TaqMan®) using a developed assay reagent (PDAR) primer/probe combination to detect the RNase P gene (ABI) or reagents developed in-house, designed using Primer Express (ABI), to quantitate parvoviral DNA. Two negative controls were also included for each reaction set. Two and one half microliters of DNA was added to 8.75μl of distilled water, 12.5μl of 2X Mastermix (ABI), and 1.25μl of virus primers and probe or PDAR. The PCR reaction was performed by heating at 50°C for 2 minutes, then at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds then 60°C for 1 minute. This was performed on an Applied Biosystems 7500 Real Time PCR system, using the ABI Sequence Detection Software, version 1.2.3.
Histology
For the evaluation of myocardial inflammation and cellular rejection score, histologic assessment of sections of formalin-fixed, paraffin-embedded endomyocardial biopsies were performed according to the grading criteria of the International Society for Heart and Lung Transplantation(33) by a pediatric cardiovascular pathologist (DLK), blinded to PCR results.
Whole blood studies
DNA was extracted from whole blood samples collected at the time of endomyocardial biopsy retrieval using the MagnaPure Compact Robot (Roche Applied Sciences, Indianapolis, IN). Parvoviral genomes were detected by nested PCR, as described above.
Outcomes
After biopsy studies were performed, the patients’ medical records were evaluated for episodes of acute graft rejection, advanced TCAD, and graft loss (retransplantation or death). Acute graft rejection was based on the patient’s clinical presentation as assessed by the transplant physician, including signs, symptoms and echocardiographic data, and confirmed by histopathology grading of the endomyocardial biopsy. Acute rejection was also determined during routine surveillance biopsy in otherwise asymptomatic patients. Rejection episodes were treated with pulse steroid therapy plus short-term or long-term modifications to the immunosuppressive regimen. The diagnosis of TCAD was based on coronary angiography or on histopathology assessment of the transplant graft at explant or autopsy. Severity of TCAD was graded per the criteria established by the Cardiac Transplant Research Database(34) (CTRD). Patients were diagnosed with advanced TCAD if they met criteria for moderate or severe TCAD.
Statistical Analysis
A t-test or Mann-Whitney test was used to compare means or medians respectively, and Fishers’ Exact test was used to compare proportions. Analysis for freedom from an outcome event (virus-positive biopsy, acute rejection episode, advanced TCAD, graft loss) was performed using Kaplan-Meier analysis, and the survival curves were compared using a log rank test. Univariate Cox regression was performed to compare groups with respect to time to TCAD while controlling for age at transplant. Multivariate Cox regression was performed adjusting for recipient age, gender, and number of acute rejection episodes in the first year after transplant. Data are presented as mean ± standard deviation. Statistical significance was achieved for p < 0.05. SPSS v.14 and STATA v.8.2 software were used for statistical analysis.
RESULTS
Patients and Viral PCR Results
Between September 2002 and December 2005, 99 consecutive patients were enrolled in the study. Seventy-four of the patients were prospectively enrolled. Sixty-four were male (65%) and 35 were female (35%). The mean age at heart transplantation was 6.0 ± 5.6 years (median age 3.6 yrs; 22 days to 18.3 years). The indications for heart transplant were congenital heart disease (n = 40), dilated cardiomyopathy (n = 34), restrictive cardiomyopathy (n = 15), hypertrophic cardiomyopathy (n = 1), and cardiac re-transplantation (n = 9). The standard immunosuppressive regimen utilized in our institution includes triple drug therapy with cyclosporine or tacrolimus, prednisone, and azathioprine or mycophenolate mofetil. Since 1999, tacrolimus became the calcineurin inhibitor of choice, with cyclosporine used in patients with adverse effects to tacrolimus. The antiproliferative agent (azathioprine or mycophenolate) was discontinued in 41% of the patients during the course of the study due to persistent leukopenia. No induction therapy is provided.
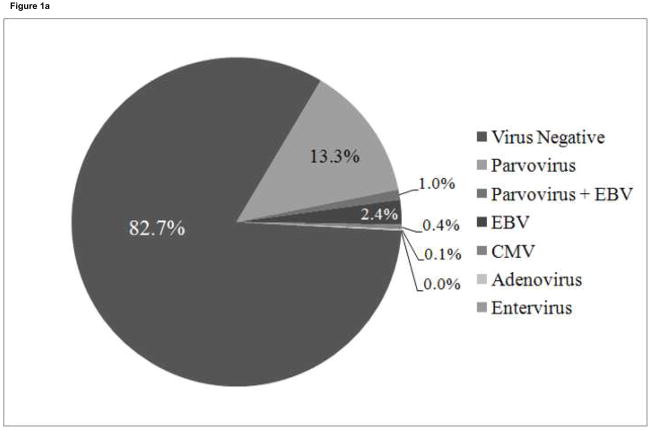
A total of 700 endomyocardial biopsies from the 99 patients were analyzed for the presence of viral genome by PCR. Viral genome was amplified in 121 biopsies (17.3%) from 45 patients (45%) (Figure 1a). PVB19 was the most common viral genome amplified and was detected in 100 biopsies (100/121, 82.6%) from 35 patients (35%)(Figure 1b). Seven of the 100 parvovirus-positive biopsies from 6 patients were also positive for EBV. Twenty-four biopsies were positive for EBV (24/121, 19.8%) from 13 patients (including the 7 samples positive for parvovirus), 3 were positive for CMV (3/121, 2.5%) from 3 patients, and 1 was positive for adenovirus (1/121, 0.8%). No samples were positive for enterovirus. PVB19 genome persisted for longer than 6 months in 20 of the 35 parvovirus-positive patients, with 2 to 8 PVB19 positive biopsies during the study period. Fifteen parvovirus-positive patients were positive on only one biopsy. All PVB19 amplicons were genotyped as type 1 parvovirus, with minor nucleotide variations detected between patients,(35) which has previously been the most common genotype detected in children.(24)
Figure 1. Distribution of viral genomes.
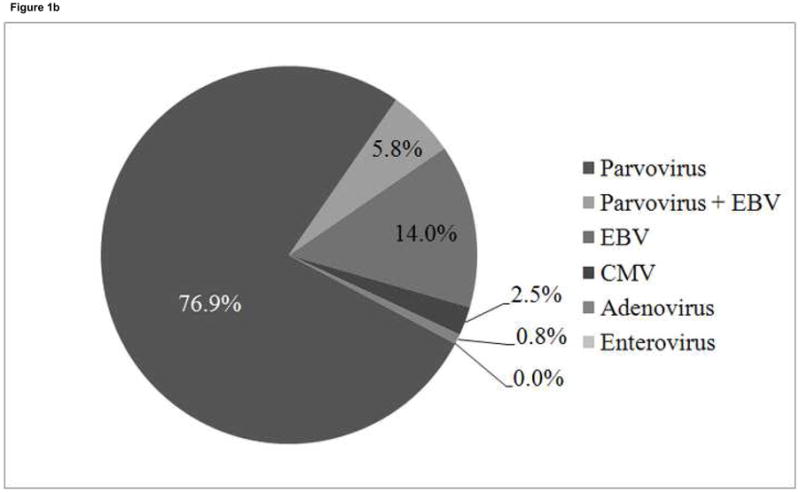
1a. Distribution of virus types in 700 biopsies from 99 patients.
1b. Frequency of virus types in 121 virus-positive biopsies from 45 patients. Total frequency of parvovirus B19 was 83%; total frequency of EBV was 20%
The patient age at transplant differed between PVB19 positive patients and negative patients. The mean age at transplant for PVB19 positive patients was 8.0 ± 5.5 years and that of PVB19 negative patients was 5.0 ± 5.5 years (p = 0.01). There was no difference in biopsy ISHLT score, patient gender, or pre-transplant diagnosis between patient groups.
Blood analysis for parvovirus B19 genome
Twenty-three blood samples from 14 patients with a corresponding EMB that was positive for PVB19 were available for PCR study. The blood was drawn during the catheterization that procured the PVB19 positive biopsy. Two blood samples (8.7%) from 2 different patients had PCR evidence of PVB19 in their blood. The remaining blood samples were negative suggesting that most patients (86%) with PVB19 in the EMB were not systemically infected at the time of EMB. Seventeen patients with negative biopsies for PVB19 had blood samples available. All 17 control patients were negative for PVB19 indicating no systemic infection at the time of EMB.
OUTCOME
Acute Graft Rejection
Of the 700 sets of biopsy samples collected for histologic assessment, 690 were considered adequate for histology scoring by the pediatric cardiac pathologist. Of the 100 PVB19 positive biopsies, 14 had a histologic score of IIIA or greater (14%), our threshold for modifying the immunosuppression regimen. Of the 590 virus negative cases, 73 (12.4%) had grade IIIA rejection or higher. The presence of PVB19 genome in the biopsy sample was not associated with histologic acute rejection (Fisher’s exact test, p = 0.65).
Thirty-two PVB19 positive biopsies were available for qPCR analysis. Five biopsies (15.6%) were from patients with histologic evidence of acute rejection (ISHLT score ≥ IIIA). The mean parvovirus copy number per biopsy sample with acute rejection was 42.1 ± 68.5 copies/ng DNA; median 3.74 (0.44–185.75) copies/ng DNA (mean 1397.7 ± 793.5 copies/biopsy; median 2000). The mean copy number in biopsies without rejection was 49.6 ± 111.5 copies/ng DNA; median 9.33 (0.32–606.88) copies/ng DNA (mean 2348.9 ± 4515.2 copies/biopsy; median 866). There was no quantitative difference in parvovirus copy number between biopsies with histologic evidence of acute rejection versus those without rejection (p=0.58).
Long-term Graft Survival
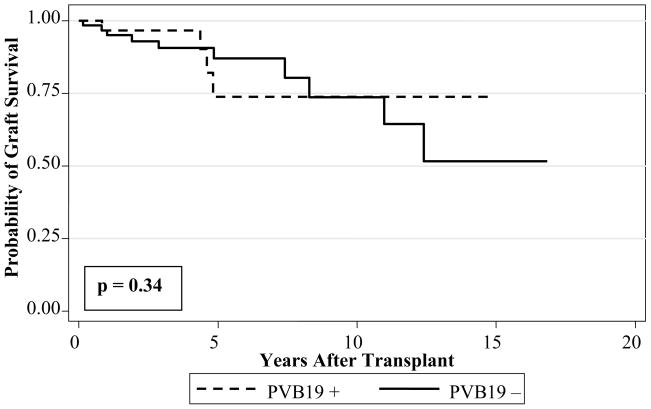
Fourteen patients experienced graft loss (retransplantation or death). Ten were patients with no history of parvovirus in the EMB and 4 had a history of PVB19 genome in the EMB. The most common cause of graft loss was TCAD (n = 7), of whom 3 were PVB19 positive. Other causes of graft loss or death included acute cellular rejection (n = 3), systemic infection (n = 2), arrhythmia (n = 1), and pulmonary vein stenosis (n = 1). The presence of PVB19 genome in the EMB was not associated with cardiac graft loss (log rank, p = 0.94) (Figure 2). When patients with chronic PVB19 in the EMB (n=20) were compared to PVB19 negative patients, there was no statistically significant association with graft loss (log rank, p = 0.34) (Figure 2).
Figure 2.
Kaplan-Meier analysis of graft survival in pediatric cardiac transplant patients with parvovirus B19 amplified from biopsy samples (PVB19 +) compared to patients with no history of parvovirus B19 genome in biopsy samples (PVB19 −)
Transplant Coronary Artery Disease
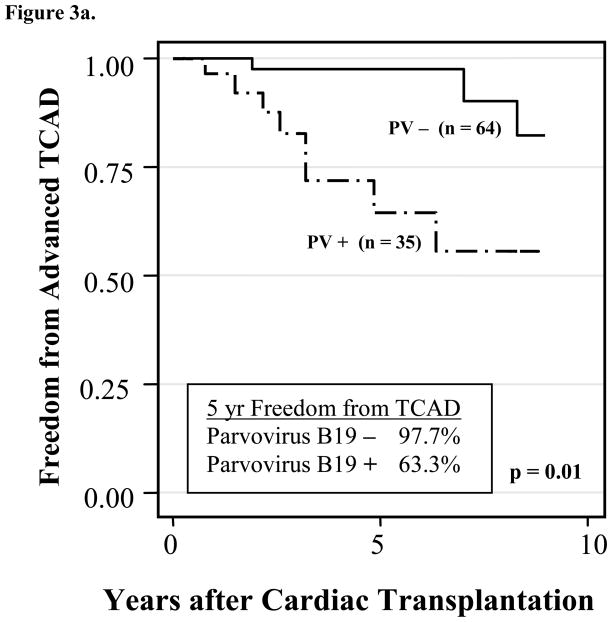
Advanced TCAD was present in 13 patients. Eight of these patients (61.5%) had PVB19 positive biopsies. Chronic PVB19 positive patients accounted for 5 of the 8 PVB19 positive patients. The presence of PVB19 genome in the EMB was positively associated with early development of advanced TCAD (log rank, p = 0.01) (Figure 3a). PVB19 positive patients developed TCAD earlier (3.3 ± 1.8 years from date of transplant) than PVB19 negative patients (8.0 ± 4.0 years) (p = 0.01). Using Cox regression to control for patient age at transplantation, the relative risk of developing TCAD in PVB19 positive patients was 5.3 times that of virus negative patients (p = 0.01, 95%CI: 1.54–18.14). The cumulative probability of developing advanced TCAD within the first year after the first PVB19-positive biopsy was 21% (95%CI 8.5–47%); within two years was 45% (95%CI 21–78%). For PVB19-positive patients, the time from the first PVB19 positive biopsy to the recognition of TCAD was 0.7 ± 0.7 years. As the total number of patients in this analysis is small, these results should be interpreted with caution.
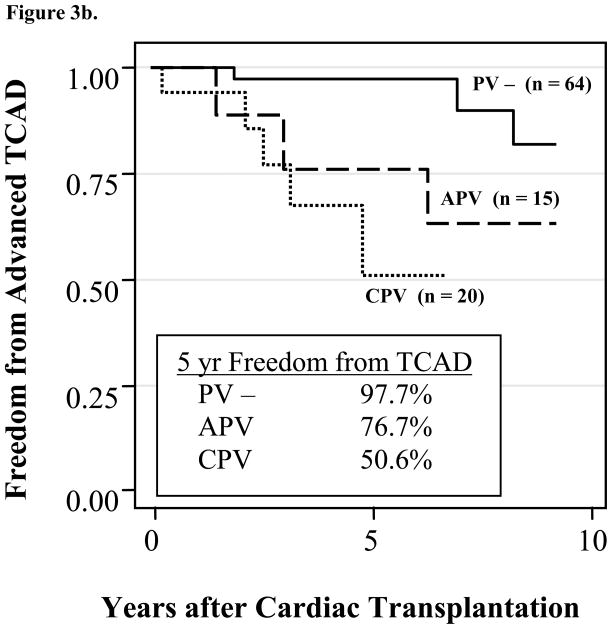
Figure 3.
Figure 3a. Kaplan-Meier analysis of freedom from the development of advanced transplant coronary artery disease in pediatric cardiac transplant patients with PVB19-positive versus PVB19-negative EMBs. (p=0.01)
Figure 3b. Kaplan-Meier analysis of freedom from advanced transplant coronary artery disease in pediatric cardiac transplant patients with chronic parvovirus B19 (CPV) versus no history of parvovirus B19 (PV −) (p < 0.001) and acute parvovirus B19 (APV) versus PV − (p = 0.27)
When patients with chronic PVB19 were compared to PVB19 negative patients, a significant association between chronic PVB19 and early advanced TCAD was identified (log rank, p < 0.001) (Figure 3b). Multivariate analysis demonstrates that chronic PVB19 is an independent risk factor for earlier TCAD (p=0.007, hazard ratio 8.7, 95%CI 1.79–43.17) after adjusting for recipient age, recipient gender, and number of acute rejection episodes in the first year after transplantation. The patient cohort was insufficient to determine a relationship between acute PVB19 and TCAD, nor a difference between acute and chronic PVB19 patients.
DISCUSSION
Viral myocardial disease has been shown to be an important factor in the development of idiopathic dilated cardiomyopathy,(10) myocarditis, and cardiac transplant rejection. Furthermore, some viruses, specifically adenovirus and enteroviruses, have been shown to have a significant prognostic influence on the survival of heart transplant recipients.(16)
Epidemiology of viral genome identification
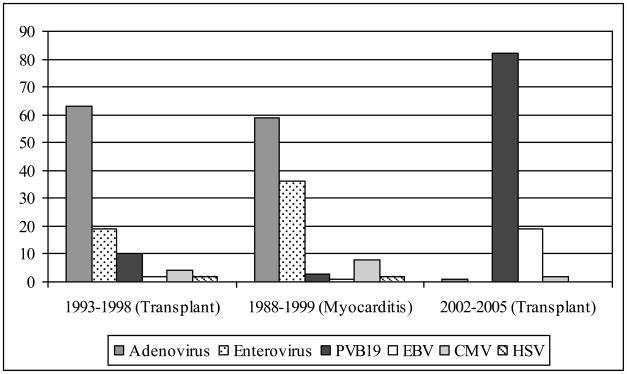
Our institution routinely evaluates all EMBs for viral genome, commencing in 1993.(14) Between 1993 and 1998, Shirali et al.(16) reported that from 553 consecutive biopsy samples from 149 heart transplant patients, 48 biopsies (8.7%) from 34 patients (23%) were positive for virus (Figure 4). From these 48 virus-positive biopsies, adenovirus was identified in 30 (63%), enterovirus in 9 (19%), parvovirus in 5 (10%), CMV in 2 (4%), herpes simplex virus type 1 in 1 (2%), and EBV in 1 (2%). The median age at heart transplantation was 2.3 months (range, 0.1 months to 18 years). In a parallel study evaluating the viral types identified in patients with myocarditis and dilated cardiomyopathy from 1988 to 1999, Bowles et al.(12) reported similar proportions of amplified viral genome.
Figure 4. Epidemiology of Virus Types Identified in Endomyocardial Biopsy Samples.
Data from our institution published by Shirali et al.(16) (1993–1998 (Transplant)) and Bowles et al.(12) (1988–1999 (Myocarditis)) compared to the current era (2002–2005 (Transplant))
These earlier studies describing the epidemiology of viral myocardial disease reported a predominance of either enteroviruses or adenovirus as the major pathogen, with PVB19 found in less than 1% of endomyocardial biopsies.(16, 29, 32) However, since 2002 multiple studies report a higher prevalence of parvovirus genomes in myocardial samples from patients with myocarditis or dilated cardiomyopathy, with the frequency of detection between 7 and 44%.(11, 26, 27, 36) We report an 82.6% incidence of PVB19 in patients that have identified viral genome from a transplant biopsy (14.3% of all biopsies). Our data support the findings of other reports, and comparison to earlier data generated at our institution indicates a shift in etiology from enterovirus and adenovirus to PVB19, thereby signifying a change in the epidemiology of viral myocardial disease (Figure 4).
Given the common finding of PVB19 antibodies in the general population,(17–19) concerns have been raised about whether finding PVB19 in endomyocardial biopsy tissue is indicative of a disease process. Kuethe et al.(37) evaluated 100 patients who underwent open-heart surgery and found PVB19 DNA in 85% of patients. Key differences are apparent as Kuethe et al. studied a population of patients greater than 45 years of age, and sought findings related to myocarditis and the development of cardiomyopathy. The predominant PVB19 type was serotype 2. Our population is less than 19 years of age and are all heart transplant patients. The primary PVB19 serotype in our patients was serotype 1, attributed to more severe disease.(38) Moreover, inflammation and evidence of acute rejection were more prevalent with earlier studies documenting adenovirus as the virus identified(16), whereas earlier evidence of transplant vasculopathy was notable with the PVB19 positive patients in this study. Additionally, as detection of PVB19 has improved with more sophisticated PCR techniques, our lab has utilized the same methodology since 1996, and has only seen the marked increase in PVB19 identification since 2002.
Outcomes of Transplanted Children
Adenovirus was shown to adversely affect heart transplant graft survival in the prior era,(16) and the mechanism of graft loss was via increased cellular rejection. Our study did not find a correlation between acute rejection and PVB19 infection. This is also reported in renal transplant recipients with PVB19 infection.(39) Additionally, we were unable to find a relationship between viral load (PVB19 genome copy number) and histologic rejection. Moreover, only the typical genotype 1 parvovirus was detected suggesting that the PVB19 recognized does not represent a different or unique strain affecting the myocardium. Therefore, it is unlikely that there is a relationship between genotype and outcome or persistence.
The presence of PVB19 genome in the transplant graft was associated with earlier, advanced TCAD. While TCAD is associated with graft loss, it is important to recognize that the increased identification of PVB19 is a recent phenomenon and may not have had sufficient time to demonstrate an effect on graft survival. As studies of patients with PVB19 myocarditis have demonstrated that the virus is not detected in cardiomyocytes but rather in endothelial cells of the vasculature,(40) we speculate that PVB19 may be involved in the endothelial cell dysfunction associated with TCAD. This implicates a different pathophysiologic mechanism of transplant dysfunction and underscores the need to further characterize this mechanism as a means to improve therapeutic options. As TCAD is the leading long-term cause of death in pediatric heart transplant recipients,(41) this finding suggests that PVB19 may negatively impact heart transplant graft survival in the future.
Conclusions
Viral myocardial disease continues to be an important factor in pediatric heart transplant dysfunction. We have shown that viral myocardial disease is shifting with respect to the organisms involved. We demonstrated that the epidemiology of viral myocardial disease has changed from the predominance of first enteroviruses and then adenovirus, to PVB19 in the current era. We further hypothesized that PVB19 would influence transplant dysfunction in a similar way to previously reported viral pathogens. Instead, we found that the presence of PVB19 does not relate to acute cellular rejection, but appears to influence earlier development of TCAD.
Limitations
As diagnostic methodologies have changed considerably over the past 2–3 decades, it is possible that the lower sensitivity of prior testing could have yielded false negative results. While we can confidently assert that enteroviruses and adenoviruses were important pathogens in the past, and that currently they do not significantly contribute to viral myocardial disease, it is more difficult to establish that parvovirus was not detected in the past.
Two biopsy samples obtained six months apart were used to define the presence of chronic, or persistent, viral infection. It is possible that the absence of viral genome following a positive biopsy could represent sampling error. This is a known variance of the EMB procedure. It is probable that persistent infection would be identified with further, follow-up procedures.
Acknowledgments
Grant Support
National Institutes of Child Health and Human Development, Pediatric Scientist Development Program - K12-HD00850 (JPB, JAT), and funds provided by the Abby Glaser Children’s Heart Fund (JAT) Texas Children’s Foundation Chair in Pediatric Cardiac Research (JAT) John Welsh Cardiovascular Diagnostic Laboratory (JAT, KRB, NEB)
Footnotes
The authors had full access to the data and take responsibility for its integrity.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boucek MM, Waltz DA, Edwards LB, et al. Registry of the International Society for Heart and Lung Transplantation: ninth official pediatric heart transplantation report--2006. J Heart Lung Transplant. 2006;25:893–903. doi: 10.1016/j.healun.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 2.Bridges ND, Spray TL, Collins MH, Bowles NE, Towbin JA. Adenovirus infection in the lung results in graft failure after lung transplantation. J Thorac Cardiovasc Surg. 1998;116:617–23. doi: 10.1016/S0022-5223(98)70168-0. [DOI] [PubMed] [Google Scholar]
- 3.Ramos E, Vincenti F, Lu WX, et al. Retransplantation in patients with graft loss caused by polyoma virus nephropathy. Transplantation. 2004;77:131–3. doi: 10.1097/01.TP.0000095898.40458.68. [DOI] [PubMed] [Google Scholar]
- 4.Grist NR, Bell EJ. A six-year study of coxsackievirus B infections in heart disease. J Hyg (Lond) 1974;73:165–72. doi: 10.1017/s0022172400023998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kandolf R. Molecular biology of viral heart disease. Herz. 1993;18:238–44. [PubMed] [Google Scholar]
- 6.Riecansky I, Schreinerova Z, Egnerova A, Petrovicova A, Bzduchova O. Incidence of Coxsackie virus infection in patients with dilated cardiomyopathy. Cor Vasa. 1989;31:225–30. [PubMed] [Google Scholar]
- 7.Novikov Iu I, Stulova MA, Lavrova IK. The role of Coxsackie B viruses in the etiology of dilated cardiomyopathy. Ter Arkh. 1989;61:97–102. [PubMed] [Google Scholar]
- 8.Calabrese F, Valente M, Thiene G, et al. Enteroviral genome in native hearts may influence outcome of patients who undergo cardiac transplantation. Diagn Mol Pathol. 1999;8:39–46. doi: 10.1097/00019606-199903000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Tracy S, Wiegand V, McManus B, et al. Molecular approaches to enteroviral diagnosis in idiopathic cardiomyopathy and myocarditis. J Am Coll Cardiol. 1990;15:1688–94. doi: 10.1016/0735-1097(90)92846-t. [DOI] [PubMed] [Google Scholar]
- 10.Friman G, Wesslen L, Fohlman J, Karjalainen J, Rolf C. The epidemiology of infectious myocarditis, lymphocytic myocarditis and dilated cardiomyopathy. Eur Heart J. 1995;16(Suppl O):36–41. doi: 10.1093/eurheartj/16.suppl_o.36. [DOI] [PubMed] [Google Scholar]
- 11.Pankuweit S, Moll R, Baandrup U, Portig I, Hufnagel G, Maisch B. Prevalence of the parvovirus B19 genome in endomyocardial biopsy specimens. Hum Pathol. 2003;34:497–503. doi: 10.1016/s0046-8177(03)00078-9. [DOI] [PubMed] [Google Scholar]
- 12.Bowles NE, Ni J, Kearney DL, et al. Detection of viruses in myocardial tissues by polymerase chain reaction. evidence of adenovirus as a common cause of myocarditis in children and adults. J Am Coll Cardiol. 2003;42:466–72. doi: 10.1016/s0735-1097(03)00648-x. [DOI] [PubMed] [Google Scholar]
- 13.Pauschinger M, Bowles NE, Fuentes-Garcia FJ, et al. Detection of adenoviral genome in the myocardium of adult patients with idiopathic left ventricular dysfunction. Circulation. 1999;99:1348–54. doi: 10.1161/01.cir.99.10.1348. [DOI] [PubMed] [Google Scholar]
- 14.Towbin JA, Griffin LD, Martin AB, et al. Intrauterine adenoviral myocarditis presenting as nonimmune hydrops fetalis: diagnosis by polymerase chain reaction. Pediatr Infect Dis J. 1994;13:144–50. doi: 10.1097/00006454-199402000-00013. [DOI] [PubMed] [Google Scholar]
- 15.Van den Veyver IB, Ni J, Bowles N, et al. Detection of intrauterine viral infection using the polymerase chain reaction. Mol Genet Metab. 1998;63:85–95. doi: 10.1006/mgme.1997.2651. [DOI] [PubMed] [Google Scholar]
- 16.Shirali GS, Ni J, Chinnock RE, et al. Association of viral genome with graft loss in children after cardiac transplantation. N Engl J Med. 2001;344:1498–503. doi: 10.1056/NEJM200105173442002. [DOI] [PubMed] [Google Scholar]
- 17.Araujo FM, Koch MC, Araujo AR. Prevalence of parvovirus B19 infection in Portugal. Blood Coagul Fibrinolysis. 1995;6:687. doi: 10.1097/00001721-199510000-00013. [DOI] [PubMed] [Google Scholar]
- 18.Bhattarakosol P, Pancharoen C, Kowitdamrong E, Thammaborvorn R, Mungmee V. Prevalence of parvovirus B19 infection in Thai young adults. Southeast Asian J Trop Med Public Health. 2003;34:585–8. [PubMed] [Google Scholar]
- 19.Cohen BJ, Buckley MM. The prevalence of antibody to human parvovirus B19 in England and Wales. J Med Microbiol. 1988;25:151–3. doi: 10.1099/00222615-25-2-151. [DOI] [PubMed] [Google Scholar]
- 20.Heegaard ED, Petersen BL, Heilmann CJ, Hornsleth A. Prevalence of parvovirus B19 and parvovirus V9 DNA and antibodies in paired bone marrow and serum samples from healthy individuals. J Clin Microbiol. 2002;40:933–6. doi: 10.1128/JCM.40.3.933-936.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cossart YE, Field AM, Cant B, Widdows D. Parvovirus-like particles in human sera. Lancet. 1975;1:72–3. doi: 10.1016/s0140-6736(75)91074-0. [DOI] [PubMed] [Google Scholar]
- 22.Brown KE, Young NS. Human parvovirus B19 infections in infants and children. Adv Pediatr Infect Dis. 1997;13:101–26. [PubMed] [Google Scholar]
- 23.Young NS, Brown KE. Parvovirus B19. N Engl J Med. 2004;350:586–97. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann HW, Knoll A, Kuster RM, Modrow S. Frequent infection with a viral pathogen, parvovirus B19, in rheumatic diseases of childhood. Arthritis and rheumatism. 2003;48:1631–8. doi: 10.1002/art.10979. [DOI] [PubMed] [Google Scholar]
- 25.Pankuweit S, Lamparter S, Schoppet M, Maisch B. Parvovirus B19 genome in endomyocardial biopsy specimen. Circulation. 2004;109:e179. doi: 10.1161/01.CIR.0000124881.00415.59. [DOI] [PubMed] [Google Scholar]
- 26.Tschope C, Bock CT, Kasner M, et al. High prevalence of cardiac parvovirus B19 infection in patients with isolated left ventricular diastolic dysfunction. Circulation. 2005;111:879–86. doi: 10.1161/01.CIR.0000155615.68924.B3. [DOI] [PubMed] [Google Scholar]
- 27.Donoso Mantke O, Nitsche A, Meyer R, Klingel K, Niedrig M. Analysing myocardial tissue from explanted hearts of heart transplant recipients and multi-organ donors for the presence of parvovirus B19 DNA. J Clin Virol. 2004;31:32–9. doi: 10.1016/j.jcv.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Murry CE, Jerome KR, Reichenbach DD. Fatal parvovirus myocarditis in a 5-year-old girl. Hum Pathol. 2001;32:342–5. doi: 10.1053/hupa.2001.22743. [DOI] [PubMed] [Google Scholar]
- 29.Schowengerdt KO, Ni J, Denfield SW, et al. Association of parvovirus B19 genome in children with myocarditis and cardiac allograft rejection: diagnosis using the polymerase chain reaction. Circulation. 1997;96:3549–54. doi: 10.1161/01.cir.96.10.3549. [DOI] [PubMed] [Google Scholar]
- 30.Olsen EG. Endomyocardial biopsies. Int J Cardiol. 1983;3:240–3. doi: 10.1016/0167-5273(83)90043-8. [DOI] [PubMed] [Google Scholar]
- 31.Martin AB, Webber S, Fricker FJ, et al. Acute myocarditis. Rapid diagnosis by PCR in children. Circulation. 1994;90:330–9. doi: 10.1161/01.cir.90.1.330. [DOI] [PubMed] [Google Scholar]
- 32.Francalanci P, Chance JL, Vatta M, et al. Cardiotropic viruses in the myocardium of children with end-stage heart disease. J Heart Lung Transplant. 2004;23:1046–52. doi: 10.1016/j.healun.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 33.Billingham ME, Cary NR, Hammond ME, et al. A working formulation for the standardization of nomenclature in the diagnosis of heart and lung rejection: Heart Rejection Study Group. The International Society for Heart Transplantation. J Heart Transplant. 1990;9:587–93. [PubMed] [Google Scholar]
- 34.Costanzo MR, Naftel DC, Pritzker MR, et al. Heart transplant coronary artery disease detected by coronary angiography: a multiinstitutional study of preoperative donor and recipient risk factors. Cardiac Transplant Research Database. J Heart Lung Transplant. 1998;17:744–53. [PubMed] [Google Scholar]
- 35.Erdman DD, Durigon EL, Wang QY, Anderson LJ. Genetic diversity of human parvovirus B19: sequence analysis of the VP1/VP2 gene from multiple isolates. The Journal of general virology. 1996;77:2767–74. doi: 10.1099/0022-1317-77-11-2767. [DOI] [PubMed] [Google Scholar]
- 36.Kuhl U, Pauschinger M, Noutsias M, et al. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation. 2005;111:887–93. doi: 10.1161/01.CIR.0000155616.07901.35. [DOI] [PubMed] [Google Scholar]
- 37.Kuethe F, Lindner J, Matschke K, et al. Prevalence of parvovirus B19 and human bocavirus DNA in the heart of patients with no evidence of dilated cardiomyopathy or myocarditis. Clin Infect Dis. 2009;49:1660–6. doi: 10.1086/648074. [DOI] [PubMed] [Google Scholar]
- 38.Kuhl U, Lassner D, Pauschinger M, et al. Prevalence of erythrovirus genotypes in the myocardium of patients with dilated cardiomyopathy. J Med Virol. 2008;80:1243–51. doi: 10.1002/jmv.21187. [DOI] [PubMed] [Google Scholar]
- 39.Ki CS, Kim IS, Kim JW, et al. Incidence and clinical significance of human parvovirus B19 infection in kidney transplant recipients. Clin Transplant. 2005;19:751–5. doi: 10.1111/j.1399-0012.2005.00415.x. [DOI] [PubMed] [Google Scholar]
- 40.Bultmann BD, Klingel K, Sotlar K, et al. Fatal parvovirus B19-associated myocarditis clinically mimicking ischemic heart disease: an endothelial cell-mediated disease. Hum Pathol. 2003;34:92–5. doi: 10.1053/hupa.2003.48. [DOI] [PubMed] [Google Scholar]
- 41.Ross M, Kouretas P, Gamberg P, et al. Ten- and 20-year survivors of pediatric orthotopic heart transplantation. J Heart Lung Transplant. 2006;25:261–70. doi: 10.1016/j.healun.2005.09.011. [DOI] [PubMed] [Google Scholar]