Abstract
Escherichia coli has long been regarded as a model organism in the study of codon usage bias (CUB). However, most studies in this organism regarding this topic have been computational or, when experimental, restricted to small datasets; particularly poor attention has been given to genes with low CUB. In this work, correspondence analysis on codon usage is used to classify E.coli genes into three groups, and the relationship between them and expression levels from microarray experiments is studied. These groups are: group 1, highly biased genes; group 2, moderately biased genes; and group 3, AT-rich genes with low CUB. It is shown that, surprisingly, there is a negative correlation between codon bias and expression levels for group 3 genes, i.e. genes with extremely low codon adaptation index (CAI) values are highly expressed, while group 2 show the lowest average expression levels and group 1 show the usual expected positive correlation between CAI and expression. This trend is maintained over all functional gene groups, seeming to contradict the E.coli–yeast paradigm on CUB. It is argued that these findings are still compatible with the mutation–selection balance hypothesis of codon usage and that E.coli genes form a dynamic system shaped by these factors.
INTRODUCTION
Early observations in Escherichia coli suggested that codon usage among its ribosomal protein genes is not random (1). This observation led Ikemura (2,3) to show that usage of preferred codons in these and other genes was positively correlated with their respective major isoacceptor tRNA levels, and this was explained as an adaptation of highly expressed genes to translational efficiency. These observations were quickly extended to other organisms (4,5), especially yeast (6), where extensive studies on codon usage bias (CUB) have been performed. These studies led to the establishment of an E.coli–yeast paradigm, where highly expressed genes used a preferred set of optimal codons in accordance with their respective major isoacceptor tRNA levels. The case for higher eukaryotes is not as clear cut, and models proposing a balance between translational selection and mutational bias have been proposed to account for the CUB observed in these organisms (7–9). For example, organisms like Drosophila or Caenorhabditis seem to resemble the E.coli–yeast paradigm (10–13), while in other eukaryotes like humans CUB seems to be determined by local genomic GC content.
Despite E.coli being a model for CUB, most studies regarding this topic in this organism have been computational or, when experimental, have been performed on relatively small datasets. Only in yeast, and more recently worm, have the advances of the post-genomic era and microarray technology been applied to the study of CUB (14,15). It is striking that the wealth of information on mRNA expression levels for E.coli (16–19) has not been used to analyse CUB on a whole genome basis for this organism. For example, the traditional view establishes that genes with low CUB are expressed at low levels (20), but to our knowledge, this assumption has not been tested experimentally. Some authors have proposed that the presence of rare codons in some genes is a regulatory strategy to reduce protein levels within the cell (21), while other authors have not found evidence for this and maintain that rare codons are the product of mutational bias (22). However, systematic studies of the expression levels in these types of gene have not been reported, thus a comprehensive study embracing expression levels and CUB in E.coli is required. The objective of this study, then, was to analyse the relationship between mRNA levels obtained from different microarray experiments and CUB in E.coli from a genomic perspective, framing our findings in the context of translational selection and mutational bias.
MATERIALS AND METHODS
The genomic sequence for E.coli K-12 MG1655 was obtained from GenBank accession no. U00096 (23). All open reading frames listed as coding for proteins (confirmed and hypothetical) were considered in this study. Basic data manipulation was performed under Microsoft Excel and Star Office Calc. Statistical analysis was done using the freely available R package (http://www.r-project.org/). Codon adaptation index (CAI) (24), effective number of codons (Nc) (25), whole GC and silent GC content (GC3s), the GRAVY index of hydrophobicity (26), the aromaticity index and correspondence analysis on codon usage (using absolute codon frequencies) were performed using the program CodonW (J.F.Peden, unpublished, available at ftp://molbiol.ox.ac.uk/cu). Correspondence analysis (COA) has been extensively used to analyse codon usage (27) and it will not be described here; for a comprehensive book on this subject we advise the reader to consult M.J. Greenacre (28). mRNA expression levels were obtained from public databases and publications. Three datasets from different sources were chosen for analysis in this study. In the first (17), ExpressDB at http://twod.med.harvard.edu/ExpressDB/, a comparison was made between expression levels in E.coli MG1655 grown to either mid log phase (LP) or stationary phase (SP). In the second study (19), E.coli NCM3416 was cultured in LB or M9 + 0.2% glucose medium, grown to OD600 = 0.8 and transcript abundance and decay were studied. The last dataset was obtained from the ASAP database (29) (http://www.genome.wisc.edu), where three different sets of experimental data for strains MG1655, DH5α and DH10b grown on LB were retrieved. We will refer to this data as the Selinger (17), Berstain (19) and ASAP (29) datasets, respectively. tRNA data was obtained from the Genomic tRNA Database (30) (http://lowelab.ucsc.edu/GtRNAdb/).
In order to test for translational selection, we devised an index for tRNA usage, inspired by the CAI of Sharp and Li (24). We start from the observation that for several organisms tRNA gene copy number (tGCN) correlates strongly and positively with tRNA levels within the cell (2,3,6,31,32). In order to calculate this index, the absolute adaptiveness values for each codon, Wi, are obtained in the following way
Wi = ∑nj = 1(1 – sij)·tGCNij
where tGCNij are the gene copy numbers of the respective isoacceptor tRNAs for the ith codon and sij are selective constraints on the efficiency of the codon–anticodon coupling for the jth tRNA. In this study we set s = 0 for the natural isoacceptor and s = 0.5 for the isoacceptors that mismatch at the wobble position. After constructing a table of all Wi values, the relative adaptiveness value of a codon, wi, can be obtained as
wi = Wi/Wmax
where Wmax is the highest Wi value in the table. Then, the tRNA adaptation index (tAI) for a given coding sequence is calculated as the geometric mean of the wi values for each codon present in that sequence
tAI = (∏Lk = 1wk)1/L
where L is the length in codons of the coding sequence (excluding any stop codons). A more complete description of this index and its applications will be the subject of a separate publication.
In order to study possible predictors of mRNA levels, a generalised additive model was fitted to the expression data (33). This is a non-parametric regression model that has the general form
Y = a + ∑pj = 1sj(x¯j) + ε
where Y is the response variable (in this case mRNA levels), x̄j are the predictor variables, sj are a set of smooth spline functions and ε is the random error, assumed to be described by the exponential family. The advantage of this kind of model is that the form of the s functions are very general, freed from restrictive parametric assumptions and the possible predictors can be added or subtracted sequentially to the model in order to test their suitability.
The following codes are used throughout this work to show statistical significance: *, significant P < 0.05; **, very significant P < 0.01; ***, extremely significant P < 0.001.
RESULTS
Quality of the microarray data
The reproducibility of microarray data was evaluated through the computation of correlation coefficients within and among the datasets studied (Table 1). It can be clearly seen that these coefficients vary broadly (they range from 0 to 0.87), indicating the very noisy nature of microarray experiments and their lack of accuracy. The highest correlation coefficients are within datasets from the same source. The data by Selinger shows the least agreement with other datasets, while the data from Bernstein and ASAP seem to agree reasonably well. Also, the number of genes studied varies widely, from less than half the genome (Table 1) to all the known and hypothetical ORFs. The sample of genes analysed by Bernstein seems to be biased towards GC richness. To prove that, a t-test was performed in order to compare mean GC content between the genes analysed in their LB experiment and the whole E.coli genome and it was found that they differ significantly (t = –5.4813***, df = 4569.47); a similar trend was observed for the M9 experiment and also, albeit far less marked, the LP experiment of Selinger (data not shown). This has important implications for the study of CUB and expression since AT-rich genes usually present the lowest CAI values.
Table 1. Pairwise correlation coefficients among gene expression levels from different microarray experiments.
| Bernstein | Selinger | ASAP | |||||
|---|---|---|---|---|---|---|---|
| LBa | M9b | LPc | SPd | MG1655e | DH5αe | DH10be | |
| M9 |
0.66 |
1 |
|
|
|
|
|
| LP |
0.12 |
0.046 |
1 |
|
|
|
|
| SP |
–0.0018 |
0.059 |
0.52 |
1 |
|
|
|
| MG1655 |
0.54 |
0.43 |
0.017 |
–0.039 |
1 |
|
|
| DH5α |
0.62 |
0.46 |
0.088 |
0.025 |
0.65 |
1 |
|
| DH10b | 0.61 | 0.50 | 0.050 | –0.0083 | 0.83 | 0.77 | 1 |
an = 1802.
bn = 2844.
cn = 3726.
dn = 4140.
en = 4289.
Relationship between CAI and mRNA levels
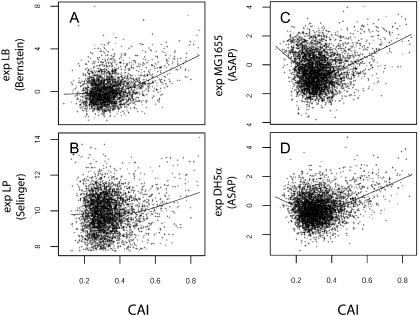
Figure 1 shows the relationship between the CAI values and mRNA expression levels for different E.coli strains from different research groups. We chose this index on the basis that it has been shown to correlate highly to expression levels in yeast (14) and it seems to perform better than other indices, like CBI, Nc or Fop (34). It can clearly be seen that for genes with high CAI values (>0.5) there is a strong correlation between CAI and expression levels. Interestingly, the Lowess fitting shows a negative correlation between genes with low CAI values (<0.3) and their expression levels for MG1655 and DH5α (Fig. 1C and D), and also for DH10b (not shown). This tendency is not seen at all in Figure 1A, although these data are not representative of AT-rich genes. In Figure 1C (Selinger data), although this tendency is similarly not observed, there is a conspicuous set of genes with low CAI values and high expression levels (upper left corner of plot). The drawback of this dataset is that a lower cut-off was selected by these researchers in order to tag the transcripts as ‘detected’ or ‘not-detected’. This cut-off is evident at the bottom of the plot and has the problem of excluding transcripts that are present at very low concentrations within the cell, which in turn influences the lowess regression. The general shape of these plots is independent of the codon bias index being considered (i.e. CBI, Fop, tAI or Nc; data not shown). The datasets of Selinger and Bernstein were excluded from further analysis due to the lack of reproducibility of the former and the unrepresentative gene sample of the latter. From the ASAP data, strain MG1655 was selected for further analysis due to the fact that it is the strain from which the genomic sequence of E.coli K-12 was determined. However, all of the following results can also be obtained by analysing the other two strains.
Figure 1.
Expression levels versus CAI for different microarray datasets. (A) Scatter plot of expression levels for strain NCM3416 grown on LB medium versus CAI. Dataset from Bernstein. The line shown is the Lowess non-parametric local regression adjusted to the dataset. (B) Selinger dataset. Expression levels are in loge(2max) units (for details see 17). (C) ASAP database. Strain MG1655 grown on LB. Expression levels are in log2(transcript level within cell). (D) As (C) but for strain DH5α.
Correspondence analysis and partition of the E.coli genome into codon usage groups
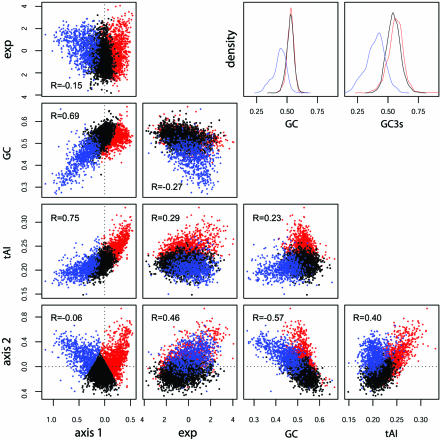
Aiming to study the relationship of codon bias to expression levels, a COA on absolute frequencies of codon usage for all ORFs was performed (Fig. 2). Four principal axes were calculated which account for 33% of the total variation in codon usage. The first axis obtained from this analysis correlates highly with CAI values (R = 0.8475, Kendall’s τ = 0.75***) and with GC content. The second axis also correlates with GC content and silent GC content, and these two axes account for 23% of the observed variation. The third and fourth axes correlate with the GRAVY (R = –0.86, τ = –0.57***) and aromaticity (R = –0.50, τ = 0.35***) indices, respectively. These axes reflect amino acid variation and not codon bias (35); thereafter they were excluded from further analysis. The bottom left corner of Figure 2 shows the distribution of ORFs along the first and second principal axes.
Figure 2.
Scatter plots of COA axis scores versus tAI, GC and expression (strain MG1655, ASAP database). Red, group 1; black, group 2; blue, group 3. (Upper right) The Gaussian kernel density estimates for GC and GC3s content (49). Colours as before.
As an exploratory approach, the data were partitioned into three groups according to their principal axis scores using the CLARA algorithm (36), in the R package. This algorithm finds k representative objects from the sample (called medoids) and then it assigns the rest of the dataset into k clusters according to the similarity of each object to the medoids. For comparison purposes a k value of 3 was selected in order to obtain a partition of the dataset similar to one reported previously (37). As can be seen in Figure 2, ORFs behave in a gradient like manner, so the clustering of the objects in the boundary zones is somewhat arbitrary. Points in the right-most cluster (group 1, n = 1398, 33%) represent overexpressed genes that present high CUB. The central cluster contains most E.coli ORFs (group 2, n = 2164, 50%) and it can be considered to represent typical E.coli genes. The left-most cluster (group 3, n = 727, 17%) contains AT-rich genes, whose codon usage differs significantly from the rest of the genome; many of them are thought to have been horizontally transferred into the E.coli chromosome (37–41). Codon usage tables for the three gene groups were computed but the results are very similar to those that have been published previously (37), thus they are not shown here. Table S1 in Supplementary Material lists all the genes analysed in this study and their corresponding cluster group.
In order to validate these results and taking into account the advice of Perrière and Thioulouse (27), a COA was repeated on relative codon frequency values (RSCU) as defined previously (24). The results obtained were then compared to the ones reported here. We have found that, at least for E.coli, both analyses yield basically the same results when only axes 1 and 2 are analysed (data not shown). When the third and fourth axes are analysed, important differences between them are obtained since the RSCU values effectively eliminate the effect of amino acid usage on the axis distribution. This does not, however, affect the work presented here since only axes 1 and 2 are being analysed. The original analysis is henceforth used, in order to avoid the artifactual deformation of the data that the RSCU values can cause (27).
Further analysis of Figure 2 reveals many interesting characteristics of the three gene groups. First, there is a strong correlation between axis 1 scores and tAI values (R = 0.75, τ = 0.59***); genes on the right-hand side of this axis tend to use codons that recognise the most abundant tRNAs (from a gene copy number perspective), while genes on the left-hand side tend to use less common tRNAs. This axis is also highly correlated to GC content (R = 0.69, τ = 0.41***), although this correlation is higher for group 3 (R = 0.64) than for groups 1 and 2 (R = 0.37 and 0.50, respectively). This axis has largely been considered to represent the main trend in synonymous CUB and is thought to reflect translational selection. As expected there is a positive correlation between group 1 gene expression levels and their first axis scores (R = 0.12, τ = 0.037*), while, surprisingly, groups 2 and 3 show a negative correlation between these two variables (R = –0.14, τ = –0.11*** and R = –0.22, τ = –0.13***, respectively). This is in opposition to previous suggestions (i.e. group 3 genes are not under translational selection, present low synonymous CUB and hence are expressed at low levels). However, the correlation between expression and axis 2 scores is much higher (R = 0.46, τ = 0.29***), more so for group 1 (R = 0.55) than for groups 2 and 3 (R = 0.22 and 0.28, respectively). Expression levels show a negative correlation to GC content (R = –0.41, τ = –0.29***). Axis 2 also shows a negative correlation to GC (R = –0.57, τ = –0.39***), although two different trends can be seen in the scatter plot, which might reflect different mutational trends among gene groups. Finally, it can be seen that groups 1 and 2 show the same distribution of whole GC content among their genes (x̄ = 0.53 for both groups; Fig. 2, upper right corner), group 3 being substantially AT rich (x̄ = 0.44). However, group 1 has a slightly higher GC3s content than group 2 (x̄ = 0.562 and 0.547, respectively), while group 3 displays an avoidance of GC at the silent positions (x̄ = 0.40).
Expression levels across functional groups
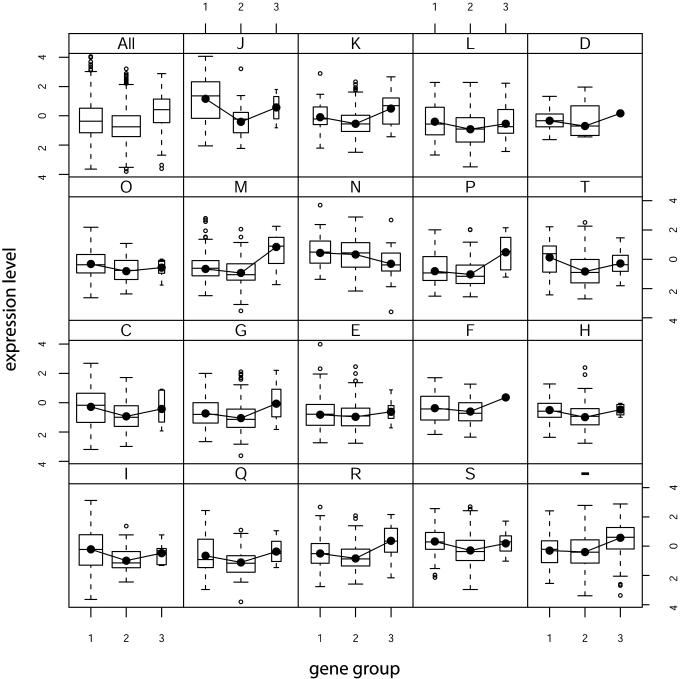
Figure 3 shows a series of box plots of expression levels partitioned among gene classes and cluster of orthologous groups (COGs) classes. It can clearly be seen that group 3 genes present the highest average expression levels (x̄ = 0.33, SD = 1.10), followed by group 1 (x̄ = –0.24, SD = 1.24) and finally group 2 (x̄ = –0.65, SD = 1.05), although group 1 presents the uppermost extreme outliers. Interestingly this V-shaped trend is maintained across all COGs classes with the exception of class N (cell motility and secretion) and it is statistically significant (F2, 4232 = 244, ***P < 2.2 × 10–16). It is interesting to notice that for some COGs classes, group 1 genes present higher average expression levels than group 3; special mention should be given to class J (translation, ribosomal structure and biogenesis). It is also striking that most group 3 genes (69%) belong to COGs classes with poorly known or unknown function (R and S) or do not belong to any COGs class at all (Table S2 in Supplementary Material).
Figure 3.
Box plots of expression level (strain MG1655, ASAP database) versus gene group partitioned among COGs. Boxes, 25% quartile, median and 75% quartile; whiskers, observations no greater than 1.5 times the interquartile range; empty dots, outliers; box widths are proportional to the square root of the number of observations in a given class. Lines join the mean values (black dots) for each group. (All) all genes; (J) translation, ribosome structure and biogenesis; (K) transcription; (L) DNA replication, recombination and repair; (D) cell division and chromosome partitioning; (O) post-translational modification, protein turnover, chaperones; (M) cell envelope biogenesis, outer membrane; (N) cell motility and secretion; (P) inorganic ion transport and metabolism; (T) signal transduction mechanisms; (C) energy production and conversion; (G) carbohydrate transport and metabolism; (E) amino acid transport and metabolism; (F) nucleotide transport and metabolism; (H) coenzyme metabolism; (I) lipid metabolism; (Q) secondary metabolites biosynthesis, transport and catabolism; (R) general function prediction only; (S) function unknown; –, not in COGs.
It should be emphasised here that the words ‘overexpressed’ to qualify group 1 signify that these genes tend to have expression values that are above the median for the whole genome (m = –0.46). Truly highly expressed genes are a subset of this group (since they share the same codon usage properties with the rest of the group) and would represent the right-uppermost genes depicted in the bottom left corner of Figure 2. A neat codon driven definition of highly expressed genes that comprises 8% of the genome may be found in the literature (42), in which this group is labelled as putatively highly expressed. Comparing those genes against the expression values analysed in this study reveals that these genes are indeed highly expressed (x̄ = 0.38) and their average expression is just higher than that of group 3.
Non-parametric regression analysis on mRNA expression levels
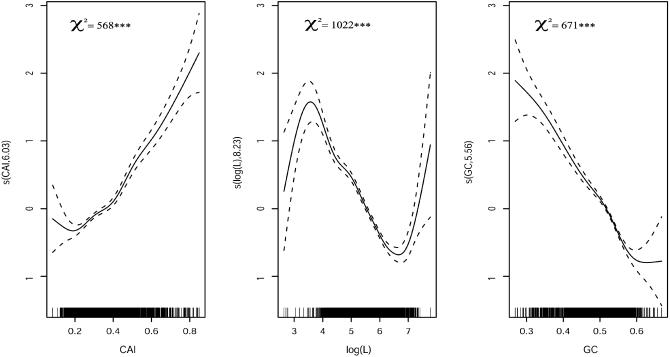
In order to identify what factors might be affecting gene expression and to carry out the analysis in greater depth, a non-parametric regression analysis on mRNA transcript levels was performed. As many variables as possible were included in the model as possible predictors in order to isolate the effects of specific variables whilst taking the rest of the variables into account. These variables were tAI, CAI, CBI, Fop, GC, GC3s, protein length (L) and the GRAVY and aromaticity indices. The best codon bias predictors were CAI and CBI, Fop and tAI being only slightly worse (overall adjusted R2 drops by less than 1%); CAI was arbitrarily kept for the rest of the analysis. GC is a better predictor than GC3s (adjusted R2 drops by 2% when GC is replaced by GC3s). The axis scores computed from COA were also tested, but all of them performed worse than the biological variables (data not shown). The contributions of GRAVY and aromaticity to the model were very small (∼1%), although highly significant (***P < 0.001). Table 2 shows the drop in adjusted R2 with elimination of any term in the model. The idea of this procedure is that in deleting each variable from the model and re-testing it statistically, the contribution of each variable to gene expression can be gauged and good predictors isolated. The elimination of log(L) causes the greatest reduction in R2, showing that this is the best predictor of mRNA concentration within the cell, followed by GC content and CAI. These findings are shown in Figure 4.
Table 2. Adjusted R2 versus term dropped in the generalised additive model analysis for mRNA levels.
| Term dropped | Adjusted R2a | GCV scorea |
|---|---|---|
| None |
0.41 |
0.827 |
| Log(L) |
0.29 |
0.990 |
| GC |
0.32 |
0.950 |
| CAI |
0.34 |
0.922 |
| GRAVY |
0.41 |
0.833 |
| Arom | 0.40 | 0.827 |
aModel [mRNA] = α + s1(CAI) + s2[log(L)] + s3(GC) + s4(GRAVY) + s5(arom) + ε ∼ i.i.d. N(0,δ2).
Figure 4.
Non-parametric regression of mRNA levels versus CAI, GC and protein length (L). Each predictor variable is plotted against their respective spline function s(variate, estimated degrees of freedom); dotted lines are plotted at 2 SE above and below the estimated spline.
DISCUSSION
To our knowledge, this is the first time a comprehensive study embracing whole genome expression data and CUB for the whole E.coli genome has been performed, and it has yielded some very interesting results. The preliminary analysis on the quality of microarray data shows that these kinds of experiments are inherently noisy and of low reproducibility. Our results agree very well with the findings of Coghlan and Wolfe (14) in their study of three expression level datasets in yeast, where they found correlations of between 0.50 and 0.68 among different experiments. The quality of microarray data seems to be a very important factor in this kind of analysis; large variances may reduce the significance of statistical tests and might hide interesting trends in complex data. This might be the case for the datasets by Bernstein and Selinger, where unrepresentative samples were used. This precludes any meaningful analysis of the behaviour of group 1, 2 and 3 genes against expression level when these data are used.
A thorough analysis of low codon bias genes and their expression levels
The relationship between CAI and mRNA levels seen in Figure 1 also agrees with the findings of Coghlan and Wolfe (14) in yeast for genes with substantially high CAI values. However, these authors intentionally excluded genes with very low CAI values from their analysis, considering that they might not be under the influence of translational selection, preventing a comparison between the relationship of CAI and expression for these genes. It would be interesting to see if the V-shaped trend observed between CAI and expression for strains MG1655, DH5α and DH10b in E.coli is also present in yeast. A quick analysis made by Akashi (13) suggests the contrary (fig. 1 in that review), showing a smooth increase in average mRNA level with major codon usage for all genes with detected transcripts. To our knowledge, an extensive analysis between CUB and expression at the protein level has not been done on a proteomic scale. In a paper by Eyre-Walker (43) a plot of CAI versus protein level for 46 genes in E.coli is presented (fig. 3 in the mentioned publication) and the same trend observed for mRNA levels is observed for genes with moderately high CAI values (>0.4), but no analysis was done for proteins with CAI values significantly smaller than 0.3 where the stronger negative correlation between expression and codon bias is seen in our plots. We are currently performing experiments on selected group 3 genes to verify if they present high protein levels within the cell.
Correspondence analysis reveals puzzling trends in the organisation of the E.coli genome
COA and cluster analysis have been common techniques in the study of CUB (37,44–46), serving as powerful tools to detect hidden trends in codon usage data. An interesting fact is that the first axis obtained from COA and other principal component techniques (33) has long been thought to reflect the effects of translational selection and thereafter to correlate with expression levels, but to our knowledge few authors have taken the care to test this assumption. Coghlan and Wolfe (14), in their work on yeast, found that axis 1 is not as good a predictor of mRNA levels as the codon bias indices studied; however, in an interesting work on Xenopus laevis (47) a strong correlation between EST frequencies and axis 2 (instead of axis 1) scores was found. The case for E.coli is quite puzzling; although the correlation between axis 1 and tAI or CAI is high, its relationship with expression is far from linear, and is worsened by the fact that genes with very low CAI values have expression levels much higher than expected. Another striking feature of gene groups is that their expression levels behave in a similar way over all COGs with the exception of class N, indicating that this trend is independent of protein function. The behaviour of group 3 genes assigned to COG class N might be explained if for example there are fewer horizontally transferred genes in this particular instance. Why this should be the case is puzzling at present, as the functions of many of these genes remain to be definitively assigned. Whether these are functions that are likely to have been horizontally transferred or whether any of them are essential to E.coli would make for an interesting future line of research.
Although tAI is not as good a predictor of tRNA levels as CAI or other CUB indices, it has certain desirable advantages. First, the set of adaptive values selected are objective and do not depend on the selection of an arbitrary set of highly expressed genes; this is especially useful when no experimental data on expression is available for an organism. Second, this index might be used to test for co-adaptation between the tRNA genomic gene pool and codon usage, which can be considered strong evidence in favour of translational selection. This ‘co-adaptation’ phenomenon has already been observed in yeast and Caenorhabditis elegans (48,49) and alongside our findings in E.coli serves to corroborate this general picture. A more extensive study on how to apply this index in order to detect translational selection and co-adaptation between codon usage and tRNA gene number is now underway in this group.
The cluster analysis yielded very similar results to the ones reported by Médigue et al. (37). Their classes II, I and III are, respectively, equivalent to groups 1, 2 and 3 in the analysis presented in this manuscript. These classes II, I and III are almost identical to the groups 1, 2 and 3 in this manuscript in terms of characterisation of their functional composition as well as in their codon usage. However, we disagree with these authors on two points: (i) we think that E.coli genes cannot be unambiguously split into three classes as stated (50) because they behave in a gradient like manner and classification of genes in the boundary areas is somewhat arbitrary, indeed, it is very hard to establish which genes are foreign solely from the point of view of codon usage; (ii) the same pattern of codon preferences is not observed in the three groups: while group 1 utilise codons that have GC-rich ends (Fig. 2, density plots), group 3 genes apparently avoid these, so for certain amino acids the pattern of codon preference is inverted. Cluster analysis is a useful exploratory technique from a statistical point of view and it provides a general means to characterise biological trends into discrete entities, but this type of analysis cannot be regarded as conclusive.
The main factors that shape the E.coli genome are translational selection and mutational bias
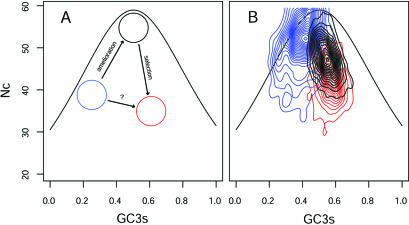
An important note of caution is needed here: although the correspondence axes are uncorrelated they are not independent, and this is evident in the V-shaped distribution of genes in this bivariate representation (Fig. 2). This plot informs us of two hidden factors, one polarising group 3 versus group 2 and the other polarising group 2 versus group 1. What is important is that the correlations among expression and axis scores is in reality a correlation between expression and some linear combination of these hidden factors, and this should be taken into account in order to understand the behaviour of expression levels versus gene groups. So, what are the forces shaping the E.coli genome? The main factors that are thought to determine codon usage are translational selection and mutational bias (7,8), and these factors can be called upon again to account for the trends in COA and expression levels observed in this study. To understand how these factors might be shaping the E.coli genome, the following points must be taken into account: (i) group 3 genes have long been considered to have been acquired through horizontal transfer (37), explaining why they are so AT rich and have very low CAI values; (ii) it has been suggested that foreign genes ameliorate (38), i.e. present biased mutational trends that steadily change their GC content towards that of the host; (iii) the effect of translational selection is to restrict the diversity of codons used in highly expressed genes, thereafter reducing the ‘effective number of codons’ in this set of genes (25). Considering these points, it can be seen that the ‘perpendicular’ forces of amelioration and selection must be responsible for the V-shaped distribution of genes in the codon landscape (Fig. 2, axis 1 versus axis 2); the right horn represents selection pulling overexpressed genes (group 1) away from the main gene core (group 2) and the left horn represents the force of amelioration, driving group 3 genes inexorably towards the main core. An analogous and more biological way of interpreting this phenomenon is to analyse classic Nc plots (25); Figure 5 shows this plot for a hypothetical organism (Fig. 5A) and for actual E.coli genes (Fig. 5B). In the hypothetical case, a group of recently acquired, AT-rich genes would move along the left-hand side of the ‘hill’ driven by biased mutational rates towards the ‘normal’ set of genes, while a set of overexpressed genes would be split from the main core under the action of selection and would move downwards, reducing their overall Nc values as an adaptation to transcriptional optimisation. If, by some measure, the main core is also under the (albeit weaker) effect of translational selection, this group would move downwards, stabilising its position somewhere in the middle between the top of the hill and the overexpressed group. Analysing the real plot for E.coli sheds much light on this issue. First, it can be seen that group 2, although presenting the lower average expression levels, is already transcriptionally optimised because their Nc values are much smaller than expected according to their GC3s content. Group 1, as expected, presents the lowest Nc values. Group 3 is positioned near the top, with Nc values close to their expected values and with a conspicuous spur of genes towards the left which might represent the more recently acquired genes.
Figure 5.
Classic Nc plots for E.coli genes. (A) Hypothetical plot depicting three gene groups: one group of AT-rich, recently acquired genes (blue circle), one group of ‘normal’ genes, under no translational selection with moderate GC content (black circle), and one group of genes under strong translational selection and moderate GC content (red circle). The arrows indicate the hypothetical pathways any given gene might follow in this bivariate landscape under the forces of mutation (as amelioration) or selection. (B) Contour plots for the three gene groups considered in this study. Red cloud, group 1; black cloud, group 2; blue cloud, group 3. These clouds represent the gene population density in the Nc–GC3s landscape, obtained from a Gaussian bivariate density kernel estimate (49).
Regression analysis is a powerful technique for understanding the behaviour of expression levels
Considering the above discussion it is easy to understand why CAI and other CUB indices fail to predict expression levels in group 3 genes. After all, what determines mRNA levels are promoter strength and mRNA stability (under suitable regulatory conditions due to growth phase, media, etc.) and not codon usage, the latter being a subsequent adaptation of highly expressed genes to translational efficiency. Nassal et al. (51) describe an interesting example where two versions of the opsin gene (one made up of major E.coli codons and one made up of GC-rich ones) have indistinguishable protein levels in vivo when appropriate leader sequences are present in the constructs. Bacterial genomes are compact, conspicuously lacking long intergenic regions, introns, repetitive sequences and pseudogenes; there are strong selective pressures acting on genome size. If group 3 genes are present in the E.coli genome it is because they should offer some kind of selective advantage to their host, their expression levels being determined by their regulatory sequences; these genes should go through a slow process of amelioration before they can be transcriptionally optimised. So, why do expression levels increase with lower CAI values? Regression analysis presents a powerful way of providing an answer to this question (see for example Fig. 4). CAI is positively correlated to GC content, but GC content is negatively correlated to expression levels, thus a clear model appears: while expression levels are reduced with lower CAI values, this effect is overcome by an increase in expression due to higher AT content. The relationship between expression and GC content is strikingly puzzling. Konu and Li (52) found a positive correlation between expression and GC3s in rodents, which contrasts with the findings from E.coli presented herein. However, analysis of the rodent data relies strongly upon the elimination of one outlier from the data and it might have been appropriate to use non-parametric correlation tests, which are robust against non-linearity and non-normality. A histogram of expression values is presented (fig. 1 of that publication) where it can be clearly seen that these data are highly skewed and far from normal, invalidating all the calculated P values, which are of marginal significance. Regretably, Coghlan and Wolfe (14) did not study the correlation between expression and GC content in their work on yeast, precluding any useful comparison with E.coli. We are studying the correlation of GC and other factors with expression levels in Saccharomyces cerevisiae, however, our preliminary analyses indicate that this is not the case in this organism.
As a general conclusion, the three groups form a dynamic system shaped by mutation and selection. Group 2 represent the average E.coli genes, group 1 contains genes under strong translational selection that are splitting from the E.coli genetic core, while group 3 genes are fusing steadily with this core driven by mutational forces. It would be interesting to align group 1, 2 and 3 genes between E.coli strains and phylogenetically related microorganisms and see where ancestral sequences lie in the Nc plot. This might give insight into the mutation–selection model, and the evolutionary path of these genes along this adaptive landscape could be further investigated.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Post L.E. and Nomura,M. (1980) DNA sequences from the str operon of Escherichia coli. J. Biol. Chem., 255, 4660–4666. [PubMed] [Google Scholar]
- 2.Ikemura T. (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol., 146, 1–21. [DOI] [PubMed] [Google Scholar]
- 3.Ikemura T. (1981) Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Evol., 151, 389–409. [DOI] [PubMed] [Google Scholar]
- 4.Sharp P.M. and Li,W. (1986) An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol., 24, 28–38. [DOI] [PubMed] [Google Scholar]
- 5.Anderson S.G.E. and Kurland,C.G. (1990) Codon preferences in free-living microorganisms. Microbiol. Rev., 54, 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennetzen J.L. and Hall,B.D. (1982) Codon selection in yeast. J. Biol. Chem., 257, 3026–3031. [PubMed] [Google Scholar]
- 7.Bulmer M. (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics, 149, 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharp P.M., Stenico,M., Peden,J.F. and Lloyd,A.T. (1993) Codon usage: mutational bias, translational selection, or both? Biochem. Soc. Trans, 21, 835–841. [DOI] [PubMed] [Google Scholar]
- 9.Laurent D. (2002) Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev., 12, 640–649. [DOI] [PubMed] [Google Scholar]
- 10.Stenico M., Lloyd,A.T. and Sharp,P.M. (1994) Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational bias. Nucleic Acids Res., 22, 2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duret L. and Mouchiroud,D. (1999) Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila and Arabidopsis. Proc. Natl Acad. Sci. USA, 96, 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akashi H. and Eyre-Walker,A. (1998) Translational selection and molecular evolution. Curr. Opin. Genet. Dev., 8, 688–693. [DOI] [PubMed] [Google Scholar]
- 13.Akashi H. (2001) Gene expression and molecular evolution. Curr. Opin. Genet. Dev., 11, 660–666. [DOI] [PubMed] [Google Scholar]
- 14.Coghlan A. and Wolfe,K.H. (2000) Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast, 16, 1131–1145. [DOI] [PubMed] [Google Scholar]
- 15.Castillo-Davis C.I. and Hartl,D.L. (2002) Genome evolution and developmental constraint in Caenorhabditis elegans. Mol. Biol. Evol., 19, 728–735. [DOI] [PubMed] [Google Scholar]
- 16.Richmond C.S., Glasner,J.D., Mau,R., Jin,H. and Blattner,F.R. (1999) Genome-wide expression profiling in Escherichia coli k-12. Nucleic Acids Res., 27, 3821–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selinger D.W., Cheung,K.J., Mei,R., Johansson,E.M., Richmon,C.S., Blattner,F.R., Lockhart,D.J. and Church,G.M. (2000) RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol., 18, 1262–1268. [DOI] [PubMed] [Google Scholar]
- 18.Khil P.P. and Camerini-Otero,R.D. (2002) Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol. Microbiol., 44, 89–105. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein J.A., Khodursky,A.B., Lin,P.H., Lin-Chao,S. and Cohen,S.N. (2002) Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-colour fluorescent DNA microarrays. Proc. Natl Acad. Sci. USA, 99, 9697–9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bulmer M. (1990) The effect of context on synonymous codon usage in genes with low codon usage bias. Nucleic Acids Res., 18, 2869–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kronigsberg W. and Codson,G.N. (1983) Evidence for use of rare codons in the danG gene and other regulatory genes of Escherichia coli. Proc. Natl Acad. Sci. USA, 80, 687–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharp P.M. and Li,W. (1986) Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare’ codons. Nucleic Acids Res., 24, 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Pernal,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Christopher,K.R., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1462. [DOI] [PubMed] [Google Scholar]
- 24.Sharp P.M. and Li,W. (1986) The codon adaptation index—a measure of directional synonymous codon usage bias and its potential applications. Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright F. (1990) The ‘effective number of codons’ used in a gene. Gene, 87, 23–29. [DOI] [PubMed] [Google Scholar]
- 26.Kyte J. and Doolittle,R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed] [Google Scholar]
- 27.Perrière G. and Thioulouse,J. (2002) Use and misuse of correspondence analysis in codon usage studies. Nucleic Acids Res., 30, 4548–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenacre M.J. (1993) Correspondence Analysis in Practice. Academic Press, London, UK. [Google Scholar]
- 29.Glasner J.D., Liss,P., Plunkett,G.,III, Darling,A., Prasad,T., Rusch,M., Byrnes,A., Gilson,M., Biehl,B., Blattner,F.R. et al. (2003) ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res., 31, 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowe T.M. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurland C.G., (1993) Major codon preference: theme and variations. Biochem. Soc. Trans, 22, 841–846. [DOI] [PubMed] [Google Scholar]
- 32.Kanaya S., Yamada,Y., Kudo.Y. and Ikemura,T. (1999) Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene, 238, 143–155. [DOI] [PubMed] [Google Scholar]
- 33.Hastie T.J. and Tibshirani,R.J. (1990) Generalized Additive Models. Chapman and Hall, London, UK. [Google Scholar]
- 34.Comeron J.M. and Aguadé,M. (1998) An evaluation of measures of synonymous codon usage bias. J. Mol. Evol., 47, 268–274. [DOI] [PubMed] [Google Scholar]
- 35.Lobry J.R. and Gautier,C. (1994) Hydrophobicity, expressivity and aromaticity are the major trends of amino-acid usage in 999 Escherichia coli chromosome-encoded genes. Nucleic Acids Res., 22, 3174–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufman L. and Rousseeuw,P.J. (1990) Finding Groups in Data: An Introduction to Cluster Analysis. John Wiley & Sons, New York, NY. [Google Scholar]
- 37.Médigue C., Rouxel,T., Vigier,P., Hénaut,A. and Danchin,A. (1991) Evidence for horizontal gene transfer in Escherichia coli speciation. J. Mol. Evol., 222, 851–856. [DOI] [PubMed] [Google Scholar]
- 38.Ochman H. and Lawrence,J.G. (1996) Phylogenetics and the amelioration of bacterial genomes. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella, Cellular and Molecular Biology. ASM Press, Washington, DC, Vol. II, pp. 2627–2637. [Google Scholar]
- 39.Whittam T.S. (1996) Genetic variation and evolutionary processes in natural populations of Escherichia coli. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella, Cellular and Molecular Biology. ASM Press, Washington, DC, Vol. II, pp. 2708–2720. [Google Scholar]
- 40.Lawrence J.G. and Ochman,H. (1998) Molecular archaeology of the Escherichia coli genome. Proc. Natl Acad. Sci. USA, 95, 9413–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Vallve S., Romeu,A. and Palau,J. (2000) Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res., 10, 1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karlin S., Mrázek,J., Campbell,A. and Kaiser,D. (2001) Characterizations of highly expressed genes of four fast-growing bacteria. J. Bacteriol., 187, 5025–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eyre-Walker A. (1996) Synonymous codon bias is related to gene length in Escherichia coli: selection for translational accuracy? Mol. Biol. Evol., 13, 864–872. [DOI] [PubMed] [Google Scholar]
- 44.Grantham R., Gautier,C. and Gouy,M. (1980) Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res., 8, 1893–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grantham R., Gautier,C., Gouy,M., Mercier,R. and Pavé,A. (1980) Codon catalogue usage and the genome hypothesis. Nucleic Acids Res., 8, r49–r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharp P.M., Tuohy,T.M.F. and Mosurski,K.R. (1986) Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res., 14, 5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Musto H., Cruveiller,S., D’Onofrio,G., Romero,H. and Bernardi,G. (2001) Translational selection on codon usage in Xenopus laevis. Mol. Biol. Evol., 18, 1703–1707. [DOI] [PubMed] [Google Scholar]
- 48.Percudani R., Pavesi,A. and Ottonello,S. (1997) Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol., 268, 322–330. [DOI] [PubMed] [Google Scholar]
- 49.Duret L. (2000) tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet., 16, 287–289. [DOI] [PubMed] [Google Scholar]
- 50.Hénaut A. and Danchin,A. (1996) Analysis and predictions from Escherichia coli sequences, or E. coli in silico. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella, Cellular and Molecular Biology. ASM Press, Washington, DC, Vol. II, pp. 2047–2066. [Google Scholar]
- 51.Nassal M., Mogi,T., Karnik,S.S. and Khorana,H.G. (1987) Structure–function studies on bacteriorhodopsin. J. Biol. Chem., 262, 9264–9270. [PubMed] [Google Scholar]
- 52.Konu Ö. and Li,M.D. (2002) Correlations between mRNA expression levels and GC contents of coding and untranslated regions of genes in rodents. J. Mol. Evol., 54, 35–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.