Abstract
As the sole nutrition provided to infants, bioactive molecules dissolved in milk influence the development of our gut microbiota. Accordingly, human milk oligosaccharides (HMOs) are minimally digested by the infant and persist to negatively and positively regulate gut microbiota. Accordingly, infant-type bifidobacteria utilize these soluble carbohydrate oligomers by convergent mechanisms. Bifidobacterium longum subsp. infantis efficiently consumes several small mass HMOs and possesses a large gene cluster and other loci dedicated to HMO metabolism. In contrast, adult-associated bifidobacteria such as the closely related B. longum subsp. longum, are deficient for HMO utilization, although they retain the capacity to ferment plant oligosaccharides and constituent pentose sugars. Thus, the ability to subsist on HMO may demark infant-associated ecotypes as these bifidobacteria may have adapted to colonize the nursing infant.
Milk and the neonatal superorganism
Lactation emerged as a nutritional strategy following the divergence of mammals more than 160 million years ago [1]. Most scientific inquiry into milk’s biological role has focused on the linear transfer of material and energy from mother, furnished for the sole privilege of her progeny. However, as the sole source of exogenous material, the provision of breast milk to the neonate has considerable implications in the development of the infant intestinal microbiota.
Several molecules incorporated into mature milk and colostrum (see Glossary) supplement innate immunity, impacting the composition of the infant microbiota. Antimicrobial factors, several of which are activated by partial digestion of milk, include milk-borne fatty acids and peptides [2, 3]. In addition, milk components such as secretory IgA, lactoferrin, lysozyme, lipoprotein lipase as well as soluble signals modulate local and systemic neonate immunity (reviewed in Ref. [4]). Whereas milk’s inhibition of pathogens is well established, milk may also positively select for beneficial microbes of the gastrointestinal tract (GIT). Indeed there is nascent evidence that mother’s milk supplies bacterial cells and products to potentially inoculate or tune tolerogenic responses in the infant [5, 6].
Paradoxically evading digestion, human milk oligosaccharides (HMOs) do not directly nourish the infant, but are growth factors that enrich for commensals proficient at utilizing these atypical carbohydrates [7, 8]. Accordingly, microbiomes of nursing infants are often enriched for bifidobacteria and thus HMOs are believed to act as bifidogenic oligosaccharides. Numerous nutritional and clinical interventions have sought to identify bifidogenic molecules to promote desirable health outcomes linked to bifidobacteria [9, 10]. This review introduces the concepts and evidence for a mammalian nutritive strategy in which milk influences, in part, the composition of the infant intestinal microbiota.
Early bifidobacterial colonization of the infant
Bifidobacteria belong to the phylum Actinobacteria which encompass Gram-positive bacteria characterized by chromosomes enriched for guanine and cytosine content [11]. In addition, bifidobacteria are non-motile, non-sporulating, and do not produce gas through fermentative metabolism. Whereas most species are strict anaerobes, a few have been characterized as microaerophillic [12]. Bifidobacteria are isolated from several mammalian-associated ecological niches (i.e. the oral cavity, genitourinary tract and GIT) in addition to certain insect and bird GITs as well as sewage [13]. Bifidobacteria derive their name from a characteristic bifid morphology, although they are pleomorphic in vitro depending on culture conditions.
Bifidobacteria are often overrepresented in the breastfed infant microbiome relative to their appearance in adults. This phenomenon has been observed in culture-based studies, and more recently, verified with molecular methodologies including those applied in a large prospective study [14–18]. There are, however, infant microbiomes characterized by a limited bifidobacterial population. In these instances, it is possible that functionally redundant microbes have actively supplanted or opportunistically seized this vacated niche [19, 20]. A recent review details the establishment of gut microbiota in Western infants and potential consequences to infant health [21].
Breastfed infants typically harbor Bifidobacterium longum, Bifidobacterium breve, and/or Bifidobacterium bifidum within their distal GIT microbiome [22]. Though the extent, or mechanism, by which a particular bifidobacterial phylotype dominates a geographical region or correlates to the host genotype is not currently well understood. This issue is also confounded by the inability to distinguish closely related bifidobacterial taxa exclusively by 16S rDNA-based approaches. While B. longum subsp. infantis is primarily isolated from infants, the habitats which B. longum subsp. longum colonize may be more diverse [23]. Furthermore, the definition of a strict infant-type bacterium, one which is consistent from an evolutionarily perspective, is hindered by contemporary practices including cesarean delivery, the supplementation of nursing with formula and exposure of infants to antibiotics and other microbially-active compounds in the first months of life. Nevertheless, in general, infant-type bifidobacteria subsist on milk oligosaccharides or derivatives, congruent with their frequent predominance in the breastfed infant colon [24].
HMO structure and properties
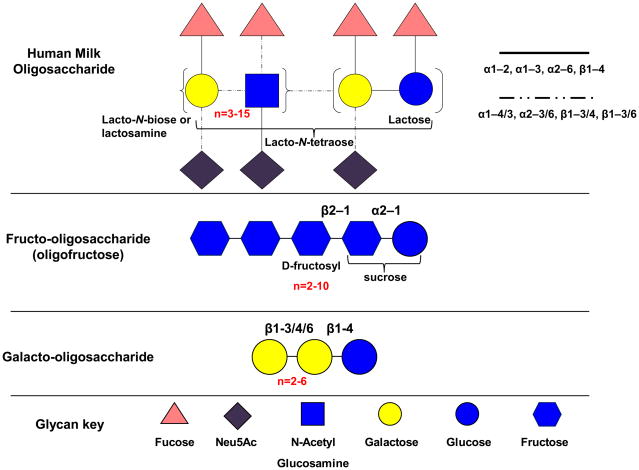
HMOs are a heterogeneous mix of soluble glycans that can often exceed the protein content of human breast milk [25]. The precise concentration of milk oligosaccharides vary by individual and declines over the course of lactation, but they are generally present at ≥ 4 g/L in milk, with higher levels observed in colostrum [26]. Milk oligosaccharides contain N-acetylglucosamine with a degree of polymerization (DP) ≥ 4 and incorporate D-glucose, D-galactoseL-fucose and N-acetylneuraminic acid (Neu5Ac) residues in an assortment of isomeric configurations. Lactose (Galβ1-4Glc) is found at the reducing end with lacto-N-biose I units (LNB; Galβ1-3GlcNAc) or lactosamine (Galβ1-4GlcNAc) in the more rare type II structures. Both LNB and lactosamine are elongated from a β1–3 linkage to the lactosyl terminus, with an additional β1–6 linkage in branched forms. Major variations in DP arise from serial integration of multiple LNB units. Further complexity is provided by terminal fucosylation via α1-2/3/4 linkages and/or α2-3/6 sialylation. Absent fucosidase and sialidase activities, these residues obstruct HMO core structures from microbial fermentation. Milk oligosaccharides have also been characterized in domesticated animals including cow and goat, although they are generally lower in abundance and vary in prevalence of specific oligosaccharide compositions [27, 28]. In addition, animal milk oligosaccharides include N-glycolylneuraminic acid residues in contrast with HMO, as modern humans lost the ability to synthesize this sialic acid[29]. A structural comparison of HMO to other bifidogenic substrates intended to mimic these milk sugars is depicted in Figure 1.
Figure 1.
Basic structures of human milk oligosaccharides and glycan mimetics. HMO possesses much more structural variability in monosaccharide constituents and linkages compared to fructo-oligosaccharides and galacto-oligosaccharides.
Interestingly, only ~200 distinct compositions have been identified in pooled breast milk despite HMO’s immense combinatorial potential [30]. Indeed, the constraints on HMO synthesis is suggestive of an adaptation for structure-specific functions, such as the prevalence of type I (β1–3) linkages in core LNB of human oligosaccharides. Accordingly, HMO structures do vary among maternal genotypes, with the particular oligosaccharide complement present in milk dependent on Lewis blood group and secretor status [31].
In addition to bifidogenic enrichment, HMOs participate in innate immunity to pathogen colonization by saturating the lower GIT with soluble ligands [32]. HMO motifs reflect the same glycan structures found in mucins and epithelial glycoconjugates. Thus the numerically dominant HMOs outcompete preferred host epitopes for ligation by pathogen adhesins. A well-characterized example of this ligand mimicry is provided by α1,2-fucosylated HMOs which mimic the H-2 epitope to minimize Campylobacter jejuni binding and infection in vivo [33]. Similar barriers to autochthonous colonization have been described in the intestinal mucosa [34], the laryngopharynx [35] and the urinary tract since some HMOs are also absorbed and excreted by the infant [36]. In addition, HMOs affect epithelial glycosyltransferase expression which, in turn, modulates the glycan topology presented to microbes [37, 38]. Finally, there is mounting evidence that HMOs initiate signal cascades in immune and other cell types [39, 40]. Further information on HMO structure and function is provided by recent reviews of the topic [41].
The genomics of bifidobacterial oligosaccharide metabolism
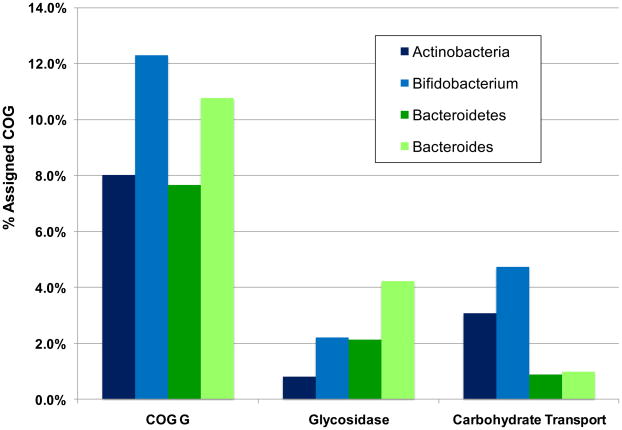
The 20 draft and finished Bifidobacterium genomes (March 2009) have considerably advanced our understanding of bifidobacterial metabolism [42]. Accordingly, both glycosidases and membrane-spanning transporters are essential to avail oligosaccharides or derivatives to the bifidobacterial fructose-6-phosphate phosphoketolase central metabolic pathway. These oligosaccharide-processing genes are typically clustered within conserved modules consisting of upstream regulatory elements, ABC (ATP-binding cassette) transporters with high-affinity carbohydrate binding proteins and one or more glycosyl hydrolase [43–46]. Several of these oligosaccharide clusters contain recently duplicated genes and/or mobile elements suggesting rapid innovation of polysaccharide metabolism to process available carbohydrate substrates. Accordingly, ~12% of bifidobacterial genes assigned a COG have been designated as carbohydrate transport or metabolism-related, as one would predict in a saccharolytic microorganism (Figure 2). Likewise, a sizable fraction of bifidobacterial genomes (~5%) are predicted to encode carbohydrate transport proteins (Figure 2) [47]. Evidently, bifidobacteria seize environmental carbohydrates through their extensive complement of ATP-dependent transporters, cation symporters, phosphotransferase systems as well as other translocation mechanisms [43, 45, 48, 49]. This contrasts starkly with Bacteroides and other members of the GIT consortium that secrete an arsenal of glycosidases to compete for complex carbohydrates [50, 51]. Bifidobacterium animalis subsp. lactis is a notable exception as its genome is relatively deficient for glycosidases, and ABC transporter genes appear independent of an operon structure [46].
Figure 2.
Percent of COG genes assigned carbohydrate processing function in Bifidobacterium and Bacteroides. The phyla Actinobacteria and Bacteroidetes have approximately equivalent percentages of carbohydrate transport and metabolism COG category G proteins, while these are enriched in member genera Bifidobacterium and Bacteroides. Bifidobacterium are enriched for transport-related COGs while Bacteroides possesses an abundance of glycosyl hydrolases.
Bifidobacterial metabolism of HMO core structures
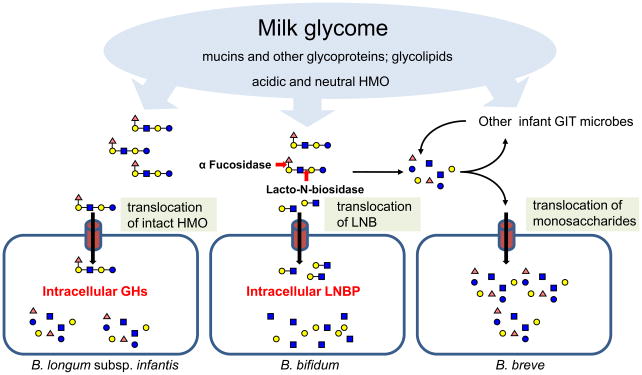
Select bifidobacteria, those typically isolated from infants, are proficient at capturing and utilizing HMO as a sole carbon source [24, 52–54]. The infant commensal B. longum subsp. infantis achieves high cell density on this purified substrate and is regarded as the archetypical HMO consumer [45]. HMO consumption is conserved in the B. longum subsp. infantis lineage, whereas other infant-type bifidobacteria exhibit more strain-specific phenotypic variation [24]. This variation is manifested in B. bifidum ATCC 29521 which degrades HMO, although it does not consume portions of the hydrolyzed oligosaccharide. Yet other bifidobacteria which readily utilize monosaccharides liberated from HMO are incapable of cleaving intact HMO as was observed for B. breve ATCC 27539 [24, 53]. This phenotypic diversity hints at niche partitioning within the infant GIT consortium and potential protocooperation among bifidobacterial phylotypes. While unable to directly access HMO carbon, certain bacteria may release oligosaccharide-bound monosaccharides to the benefit of scavenging heterologous consortium members (e.g. B. breve) (Figure 3). Accordingly, a mixed-species transcriptome of the breastfed infant microbiome is generally enriched for bifidobacterial carbohydrate utilization, suggesting that milk sugars are actively metabolized by phylotypes incapable of utilizing intact HMO under in vitro isolation [55].
Figure 3.
Putative modes of accessing milk glycans in the infant gut by bifidobacteria. B. longum subsp. infantis captures intact HMO, while B. bifidum secretes extracellular enzymes prior to translocating lacto-N-biose degradation products. B. breve utilizes HMO monosaccharides cleaved by extracellular enzymes secreted by heterologous members of the consortium.
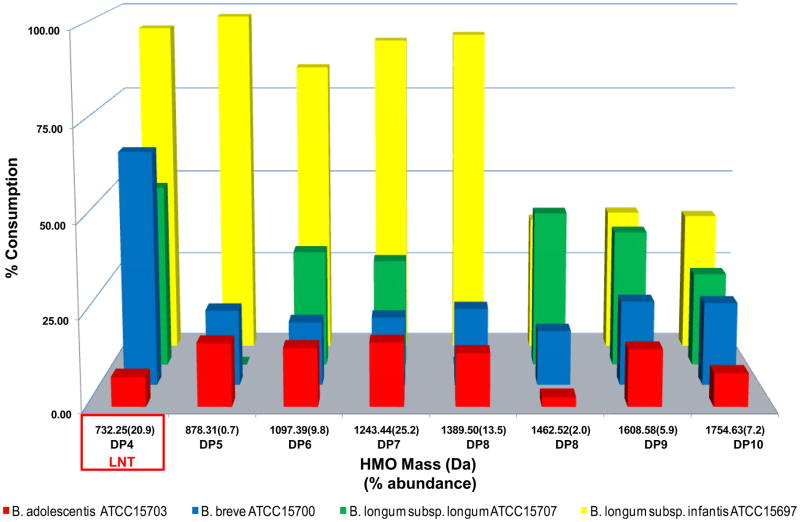
To further resolve the HMO utilization phenotype, mass spectrometry has been employed to glycoprofile the specific HMO masses consumed in axenic fermentations [30]. Most HMO utilizing bifidobacteria metabolize only a single composition corresponding to lacto-N-tetraose (LNT; Galβ1-3GlcNAc β1-3 Galβ1-4Glc) [24, 54]. The LNT tetrasaccharide is a core structure found invariably in higher molecular weight HMO. While B. longum subsp. infantis consumes LNT to extinction, B. longum subsp. longum, B. breve (Figure 4) and B. bifidum [24]exhibits a more modest degradation of this molecule. Thus unmodified LNT (i.e. non-fucosylated or sialylated) is susceptible to enzymatic degradation by several representative bifidobacteria. Interestingly, adult-type bifidobacteria, B. adolescentis and B. animalis do not degrade LNT or other HMO species. This may be due in part to the absence of an identifiable lacto-N-biose phosphorylase gene (EC 2.4.1.211) in these species.
Figure 4.
Phenotypic variation of HMO utilization in bifidobacteria. The consumption of HMO as a sole carbon source data is derived from Ref. [24], and the relative abundances were originally presented in Ref. [54]. Abbreviations: LNT, Lacto-N-tetraose-like.
Moreover, type I glycans such as HMO incorporate repeating LNB, a disaccharide that can be synthesized in vitro by enzymes derived from B. bifidum and B. longum subsp. longum [56]. Human milk differs from most other animal milks by the predominance of LNB as the repeating unit instead of N-acetlyllactosamine (Galβ1-4GlcNAc), which has led some to postulate that LNB is the essential bifidogenic factor delivered in human breast milk [57]. LNB promotes bifidobacterial growth in species that are able to utilize it as a sole carbon source [58]. In these instances, LNB is a proxy for intact HMO molecules as it is not abundant in milk as a soluble dissacharide, and lacks the full structural diversity, and thus function, of HMO. Nevertheless, synthetic LNB offers a significant advance in HMO research as large-scale oligosaccharide purification from fluid milk remains a challenge. Indeed infant-type bifidobacteria have been demonstrated to consume purified LNB whereas adult-type phylotypes generally do not [59].
In apparent contrast with B. longum subsp. infantis, B. bifidum acquires HMO-bound LNB with extracellular LNB liberating enzymes as depicted in Figure 3. Accordingly, B. bifidum JCM1254 secretes an extracellular 1,2-α-L-fucosidase (AfcA) and 1-3/4-α-L-fucosidase (AfcB) which cleave terminal fucosyl linkages, permitting further degradation of the LNB core structure to proceed [60–62]. Subsequent to defucosylation, a lacto-N-biosidase (EC 3.2.1.140) liberates LNB from lacto-N-tetraose and other HMO compositions lacking fucosylated or sialylated LNB moieties [63]. Once released, LNB is translocated across the cell membrane by an ABC transporter associated with an LNB-specific SBP [64, 65]. Interestingly, B. longum subsp. longum possesses an endo-α-N-acetylgalactosaminidase (EC 3.2.1.97) which liberates galacto-N-biose (GNB) from O-linked mucin glycoproteins [66]. Indeed, the presence of both a endo-α-N-acetylgalactosaminidase and fucosidase has been linked to the B. bifidum mucin degradation phenotype, with expression of both genes induced in the presence of porcine mucin [67]. An intracellular phosphorylase cleaves the LNB-GNB disaccharide derived from HMO or mucin [57, 68]. Finally, a modified Leloir pathway feeds galactose into the central fructose-6-phosphate phosphoketolase pathway to generate cellular ATP [69].
B. longum subsp. infantis consumes a range of short chain HMO
B. longum subsp. infantis has evolved within a host whose sole source of exogenous nutrition is mother’s milk [45]. As a potential consequence, B. longum subsp. infantis exhibits vigorous growth on several neutral small mass oligosaccharides that are secreted coincident with the start of lactation, whereas other bifidobacteria only partially consume LNT, if at all (Figure 4) [52, 54]. Although hypothesized, the extent to which bifidobacteria utilize the less abundant sialylated milk oligosaccharides is not currently well understood.
Subsistence on a broad assortment of milk oligosaccharides is now known to be a chromosomally encoded trait [45]. B. longum subsp. infantis possesses several HMO-active gene clusters encoding sialidases (EC 3.2.1.18) and fucosidases (EC 3.2.1.51), which are expressed to process sialylated and fucosylated glycans, respectively [54]. This hydrolytic capability broadens its oligosaccharide utilization range to include terminally substituted HMO by enabling access to LNT or LNB core structures. These HMO-active gene clusters are scattered throughout the B. longum subsp. infantis genome and are conspicuously absent from the subspecies longum genome. Of particular significance is the identification of a contiguous (43 kbp) genomic tract consisting of HMO-related genes in B. longum subsp. infantis [42]. As with other bacterial genomes, genetic linkage suggests a greater likelihood of lateral gene transfer preceding fixation within the subspecies infantis clade. Likewise, there is an increased probability for transfer of this locus to other members of the GIT consortium. Moreover, the orientation of genes in a concerted transcriptional direction may be indicative of co-regulation. Finally, and most notable with regards to milk utilization, gene co-localization is a hallmark of participation in a common metabolic operation. This HMO cluster, and other milk-active glycolytic clusters, is not unique to the fully sequenced type strain of B. longum subsp. infantis ATCC 15697, as confirmed by sequencing of additional subspecies infantis genomes [45].
The 43 kbp HMO cluster encodes glycosyl hydrolases active on the four milk oligosaccharide glycosidic linkages (i.e. α-fucosidase, α-sialidase, β-galactosidase and β-N-hexosaminidase). These four glycosidases are interspersed amid an array of ABC transporters and their associated solute binding proteins (SBPs) predicted to bind oligosaccharides. Interestingly, these SBPs exhibit a pronounced sequence divergence relative to other family 1 SBPs observed in ATCC 15697 as well as B. longum subsp. longum NCC2705 and B. adolescentis ATCC15703. The importance of oligosaccharide transport is further accentuated by the predicted intracellular localization of subspecies infantis glycosidases, which generally lack identifiable transmembrane domains, secretion signals or Gram-positive cell wall anchor motifs. This is consistent with the glycoprofile which clearly demonstrates that ATCC 15697 preferably utilizes HMO compositions in vitro that are DP ≤8 with a molecular mass < 1400 Da (Figure 4). This restriction may be due to transporter specificity or steric factors that inhibit translocation of large substrates across the cell envelope. The precise identities of oligosaccharide structural isomers consumed are an area of active research.
Many glycosidases, transporters and catabolic enzymes predicted to be active on HMO and derivatives were detected by proteomics while growing on HMO. This partially verified their participation in HMO catabolism. Furthermore, in vivo metabolic flux analysis has demonstrated that metabolites derived from HMO and sialic acid Neu5Ac are metabolized via the fructose-6-phosphate phosphoketolase pathway in B. longum subsp. infantis ATCC 15697 [45].
Divergence of carbohydrate utilization in B. longum
Significant metabolic disparities are expected between bacterial species, particularly those isolated from dissimilar habitats. Somewhat surprisingly, a conspecific genome comparison between B. longum subsp. longum and subsp. infantis has revealed a vastly divergent intraspecies strategy for carbohydrate acquisition [45]. Subspecies infantis oligosaccharide catabolic clusters are generally dedicated to mammalian glycan metabolism, many of which are fucosylated and/or sialylated. Evidently, xylose and arabinose metabolism has been supplanted by sialic acid and fucose catabolism, as pentose metabolic gene clusters present in subspecies longum are absent or degraded in subspecies infantis..
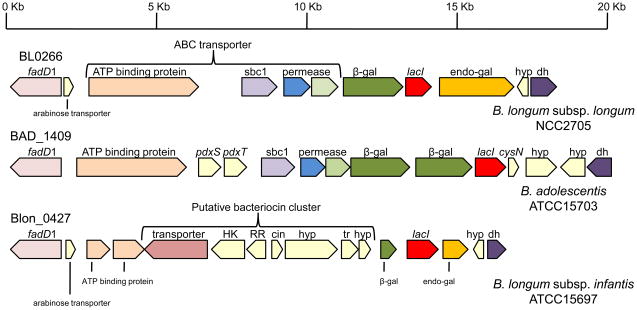
The inability of subspecies infantis to ferment arabinogalactan or constituent arabinose residues provides a functional manifestation of this reorientation to mirror its host’s primary dietary polysaccharide [23]. In B. longum subsp. longum, however, an extracellular endogalactanase liberates galactotrisaccharides from arabinogalactan, an arabinose-substituted galactan produced by various plants such as potato and soybean. Interestingly, in the analogous chromosomal locus, B. longum subsp. infantis has a severely truncated endogalactanase gene (Blon_0440) located adjacent to a similarly degraded β-galactosidase in the absence of the sugar ABC transporter (Figure 5) [43]. Evidently, these genes became expendable subsequent to the divergence of subspecies infantis from longum, as the endogalactanase appears to have been acquired following the divergence of the progenitors of B. longum and B. adolescentis.
Figure 5.
Endo-galactanase locus in B. longum subsp. longum NCC2705 and variations in other bifidobacteria. The gene cluster is conserved in B. adolescentis ATCC15703 with the exception of the endo-galactanase. The closely related B. longum subsp. infantis ATCC15697 is missing or possesses degraded homologs of this cluster. Gene fragments are denoted in smaller text below the genes. The locus number for the fadD1 gene is listed above. Abbreviations: sbp1, solute binding protein family 1; β-gal, β-galactosidase; endo -gal, endo-galactosidase; hyp, hypothetical protein; dh, dehydrogenase; HK, histidine kinase; RR, response regulator; cin, bacteriocin; tr, transport related; fadD1, long-chain acyl-CoA synthetase; pdxS, pyridoxine biosynthesis gene; pdxT, glutamine amidotransferase; lacI, lacI family transcriptional regulator; cysN, sulfate adenylyltransferase subunit 1.
Furthermore, B. longum subsp. longum possesses an arabinose isomerase (araA) and L-ribulose-5-phosphate 4-epimerase (araD), both of which are absent in B. longum subsp. infantis. In B. longum subsp. longum these genes enable conversion of L-arabinose to L-xylulose-5-phosphate with the aid of an L-ribulokinase. A recent non-orthologous displacement appears responsible for arabinose gene decay in B. longum subsp. infantis. Remarkably, a fucosidase and a permease (Blon_0425 and Blon_0426) are situated in place of the three arabinose catabolic genes, with a vestigial kinase fragment to demarcate the ancestral arabinose cluster’s location [42]. This strongly suggests selection for fucosylated mammalian glycan metabolism in lieu of arabinose in a bifidobacterial clade that ceased to encounter plant-derived sugars during its evolution. The apparent selection that has culled arabinose catabolism from subspecies infantis has likely encouraged a similar purge in other infant-type bifidobacteria. Both B. bifidum NCIMB 41171 and B. breve DSM 20213 are missing this arabinose cluster while preserving a small fragment of araD in a similar genomic context to indicate its ancestral presence, although fucose-related genes have not been inserted in its place [70].
Furthermore, among bifidobacteria that ferment pentoses, xylose is isomerized (XylA; EC 5.3.1.5) and phosphorylated (XylB; EC 2.7.1.17) prior to entry into central metabolism as xylulose-5-phosphate. Consistent with its xylose-negative phenotype, the B. longum subsp. infantis ATCC 15697 genome is devoid of a recognizable xylose isomerase (xylA) in clear contrast to other bifidobacteria, including subsp. longum, which localizes xylA upstream of a gene cluster dedicated to xylose metabolism including xylB [43, 45]. As was evident in the loss of arabinose metabolism, xylA is absent from the infant-types B. bifidum NCIMB 41171 and B. breve DSM 20213 [70]. In B. longum subsp. infantis ATCC 15697, the severely truncated xylB gene is likely non-functional (Blon_0572) as only a third of the 5′ terminus is retained while absent from the B. bifidum NCIMB 41171 genome altogether.
Milk oligosaccharide mimetics
Numerous applications have been developed to mimic the bifidogenic properties of milk oligosaccharides, though they lack the inherent complexity of HMOs in terms of monosaccharide composition, linkage diversity and structural organization (Figure 1). Nevertheless, there has been much attention given to commercial bifidogenic substrates, as alternatives to HMO, since in vitro and clinical evidence support their bifidogenic efficacy [71–73].
Milk oligosaccharide mimetics include fructo-oligosaccharides (FOS) commonly extracted as inulin from chicory or other plant sources. FOS is broadly bifidogenic and is utilized by most bifidobacteria, in contrast to HMO’s observed specificity [53]. Galacto-oligosaccharides (GOS) are enzymatically synthesized from dairy galactose (reviewed in Ref. [74]). Similar to FOS, their structural potential is markedly less than that of milk oligosaccharides albeit more diverse than linear FOS fructans. The basic structure of GOS incorporates lactose at the reducing end which is typically elongated with up to six galactose residues ([Gal(β1-3/4/6]1–6)Gal(β1-4)Glc). There is some confusion concerning the relative contribution of naturally occurring GOS to the human milk glycome [75, 76]. While limited short-chain galactosyllactose (Gal(β1-3/4/6)Gal(β1-4)Glc) has been detected at millimolar concentrations in breast milk, this is 3–4 orders of magnitude lower than the total concentration of the HMO pool [77]. There is little evidence for high DP (≥4) GOS in human milk, despite an entrenched, and often repeated, misperception.
It remains unclear, however, if these oligosaccharide analogs retain similar immunological and pathogen deflection functions and whether they selectively increase bifidobacterial populations to the exclusion of undesirable genera. Importantly, nutraceutical oligosaccharides do not reflect the genomic and physiological links between infant-type bifidobacteria and HMO; instead they target the bifidobacterial population non-specifically.
Milk oligosaccharide utilization by other bacteria
Although the B. longum subsp. infantis HMO consumption phenotype is predicated on intracellular glycosidases and solute transporters, the presence of similar genes in other species may not be indicative of a coevolutionary relationship with milk. To wit, several genera such as soil Streptomyces, do not encounter HMO in their ecological niche, but secrete a wide range of glycosidases which may enable milk glycan utilization upon in vitro assay. A less definitive case is provided by mucolytic gut commensals such as Bacteroides, since Bacteroides fragilis and Bacteroides vulgatus have been observed to ferment HMO in vitro (D. Mills, unpublished data). Bacteroides typically dominate the adult distal GIT, and colonize infants more variably and at lower concentrations than bifidobacteria [15]. Whether early colonizing Bacteroides utilize HMO in vivo or subsist on other infant or milk glycoconjugates is not yet known. One may speculate that bifidobacteria successfully compete with Bacterioides in the infant colon by virtue of a superior ability to sequester soluble HMO or degradation products, although a thorough vetting of this hypothesis remains to be performed.
Conclusions and future directions
Milk defines infant nutrition and, by extension, is integral to the natural history of the earliest symbionts to colonize Homo sapiens. Indigestible milk oligosaccharides negatively regulate the infant microbiota via ligand mimicry, in addition to positively selecting for bifidobacteria. Accordingly, infant-type bifidobacteria possess the requisite gene suite active on mammalian glycans to enable hydrolysis of atypical HMO structures. In particular, B. longum subsp. infantis imports and subsequently hydrolyzes HMO with sialidases and fucosidases to present LNT and other core glycan domains for further processing [45, 53]. The presence of a large gene cluster dedicated to HMO metabolism is consistent with this subspecies’s efficient utilization of an assortment of small mass milk oligosaccharides. This utilization trait contrasts markedly with B. longum subsp. longum, which does not metabolize milk oligosaccharides to the same degree, although it retains the capacity to ferment plant oligosaccharides and their constituent pentose sugars [23, 24]. The selective pressures that have restricted plant oligosaccharide metabolism in infant-type bifidobacteria are as profound as those that actuated milk glycan utilization in the same lineages. Moreover, convergent strategies to utilize HMO by B. breve and B. bifidum have likely evolved independently subsequent to associating with mammals. In addition to evaluating mechanisms that contribute to these physiological disparities, it is of great interest to discriminate traits shared through lateral gene transfer from those that were innovated vertically. This would help describe infant-associated ecotypes within the genus Bifidobacterium, as those that have likely adapted to subsist on milk molecules.
Reconciling a causal linkage of in vivo bifidobacterial colonization to the act of nursing currently transcends available methodologies, although barriers that have thwarted genetic manipulations of bifidobacteria have begun to be addressed [78]. In addition, reliable genetic tools would help further resolve the metabolic definition of the HMO utilization phenotype. Separation or synthesis of gram quantities of pure HMO species is critical in this regard, along with a systems-level understanding of metabolic regulation and function in HMO consuming organisms.
While it is tempting to classify bacterial oligosaccharide metabolism according to binary models such as intracellular versus extracellular utilization, mammalian versus plant glycan preference, Bifidobacterium versus Bacteroides and so forth, such discrete schema do not satisfactorily describe the emergent biology inherent to the infant microbiota. As with many biological systems, the boundaries delineating microbial actors and their biochemical actions positioned in syntrophic networks are amorphous and frustrate rigid classification. Thus it is somewhat surprising that milk’s influence on the infant microbiota is so readily identifiable in a particular symbiont rather than only detectable in the aggregate microbiome. This provocatively suggests a vital role for B. longum subsp. infantis, particularly if it is determined to be one of several functionally redundant core members of the infant microbiome. That this phylotype, and other infant-associated bifidobacteria, specialize to the degree that they are unable to persist past weaning is a question fundamental to evolution of human development.
Clearly milk is one of several principle factors that dictate the population structure of the infant microbiome. As such, microbial population fluxes observed during weaning are likely due to the alleviation of suppressive antimicrobials in milk, removal of growth factors and the addition of foods that encourage microbial diversity. The evolved codependence between humans and our earliest symbionts may ensure the proper development of the superorganism, and is expected to have profound and persistent implications to host homeostasis throughout life. Assuming that milk’s evolutionary vector is generally oriented towards promoting infant fitness, orchestration of the neonate’s microbiota via oligosaccharides and other molecules may have been an adaptive consequence.
Acknowledgments
D.A.S. was supported by a pre-doctoral training grant (NIH-NIGMS T32-GM08799). Contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or NIH. This publication was made possible in part by grant support from the University of California Discovery Grant Program, the California Dairy Research Foundation, USDA NRI-CSREES Award 2008–35200-18776, and by NIH-NICID awards 5R01HD059127 & 1R01HD061923 (D.A.M.).
Glossary
- ATP-binding cassette (ABC) transporter
these hydrolyze ATP to catalyze solute translocation
- Bifidogenic oligosaccharide
a human-indigestible carbohydrate oligomer that is used as a fermentation substrate by bifidobacteria and enriches the representation of these commensals in the distal GIT
- Clusters of orthologous groups of proteins (COG)
protein family classification based on orthology in three or more phylogenetic lineages
- Colostrum
yellowish fluid secreted by mammals during the first few days of lactation prior to the production of true milk. This is particularly abundant in growth factors, immunological molecules and oligosaccharides among other bioactive molecules
- Core microbiome
the set of shared microbial genes in a given niche which is invariable across human microbiomes
- Degree of polymerization (DP)
the number of monosaccharide residues in a given oligosaccharide composition
- Ecotype
a genetically distinct population within a species that has adapted to a specific habitat
- Glycoprofile
mass spectrometry employed to monitor the abundances of a range of oligosaccharide masses. Often used with regards to microbial consumption of oligosaccharides
- Lewis blood group
human blood typing system based on two primary Lewis antigens. Depending on which one is present, this antigen strongly correlates with secretion of blood type antigens in fluids by those termed secretors, or not, in non-secretor phenotypes
- Microbiota
a microbial community that has been established in a given habitat. Typically used to refer to the microbial population associated with a healthy individual or tissue
- Microbiome
the aggregate genomes present in a particular microbiota
- Neonate
a newborn infant within its first 28 days of life.[GT1]
- Niche partitioning
the process by which selection has segregated competing phylotypes into different patterns of resource use
- Phylotype
a phylogenetic cluster of microbes which share a common ancestor and transcended a threshold of molecular divergence from closely-related lineages
- Protocooperation
an interaction between two phylotypes that benefits both and not necessarily required for growth or survival of either
- Syntrophy
a relationship by which a microbial phylotype is dependent on another to provide a nutritional requirement
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Oftedal OT. The mammary gland and its origin during synapsid evolution. J Mammary Gland Biol Neoplasia. 2002;7:225–252. doi: 10.1023/a:1022896515287. [DOI] [PubMed] [Google Scholar]
- 2.Phadke SM, et al. Antimicrobial peptides in mucosal secretions: the importance of local secretions in mitigating infection. J Nutr. 2005;135:1289–1293. doi: 10.1093/jn/135.5.1289. [DOI] [PubMed] [Google Scholar]
- 3.Thormar H, Hilmarsson H. The role of microbicidal lipids in host defense against pathogens and their potential as therapeutic agents. Chem Phys Lipids. 2007;150:1–11. doi: 10.1016/j.chemphyslip.2007.06.220. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence RM, Pane CA. Human breast milk: current concepts of immunology and infectious diseases. Curr Probl Pediatr Adolesc Health Care. 2007;37:7–36. doi: 10.1016/j.cppeds.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Martin R, et al. Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res Microbiol. 2007;158:31–37. doi: 10.1016/j.resmic.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Perez PF, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics. 2007;119:e724–732. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- 7.Gnoth MJ, et al. Human milk oligosaccharides are minimally digested in vitro. J Nutr. 2000;130:3014–3020. doi: 10.1093/jn/130.12.3014. [DOI] [PubMed] [Google Scholar]
- 8.Engfer MB, et al. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr. 2000;71:1589–1596. doi: 10.1093/ajcn/71.6.1589. [DOI] [PubMed] [Google Scholar]
- 9.Moro G, et al. A mixture of prebiotic oligosaccharides reduces the incidence of atopic dermatitis during the first six months of age. Arch Dis Child. 2006;91:814–819. doi: 10.1136/adc.2006.098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arslanoglu S, et al. Early dietary intervention with a mixture of prebiotic oligosaccharides reduces the incidence of allergic manifestations and infections during the first two years of life. J Nutr. 2008;138:1091–1095. doi: 10.1093/jn/138.6.1091. [DOI] [PubMed] [Google Scholar]
- 11.Ventura M, et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawasaki S, et al. Response of the microaerophilic Bifidobacterium species, B. boum and B. thermophilum, to oxygen. Appl Environ Microbiol. 2006;72:6854–6858. doi: 10.1128/AEM.01216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turroni F, et al. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek. 2008;94:35–50. doi: 10.1007/s10482-008-9232-4. [DOI] [PubMed] [Google Scholar]
- 14.Harmsen HJM, et al. Analysis of intestinal flora development in breast-fed and formula-fed Infants by using molecular identification and detection methods. Journal of Pediatric Gastroenterology and Nutrition. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 15.Penders J, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 16.Yoshioka H, et al. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983;72:317–321. [PubMed] [Google Scholar]
- 17.Favier CF, et al. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002;68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mariat D, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer C, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98:229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 22.Matsuki T, et al. Genus- and species-specific PCR primers for the detection and identification of bifidobacteria. Current Issues in Intestinal Microbiology. 2003;4:61–69. [PubMed] [Google Scholar]
- 23.Mattarelli P, et al. Proposal to reclassify the three biotypes of Bifidobacterium longum as three subspecies: Bifidobacterium longum subsp. longum subsp. nov., Bifidobacterium longum subsp. infantis comb. nov. and Bifidobacterium longum subsp. suis comb. nov. Int J Syst Evol Microbiol. 2008;58:767–772. doi: 10.1099/ijs.0.65319-0. [DOI] [PubMed] [Google Scholar]
- 24.LoCascio RG, et al. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microbial Biotechnology. 2009;2:333–342. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coppa GV, et al. Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics. 1993;91:637–641. [PubMed] [Google Scholar]
- 26.Asakuma S, et al. Variation of major neutral oligosaccharides levels in human colostrum. Eur J Clin Nutr. 2008;62:488–494. doi: 10.1038/sj.ejcn.1602738. [DOI] [PubMed] [Google Scholar]
- 27.Tao N, et al. Bovine milk glycome. J Dairy Sci. 2008;91:3768–3778. doi: 10.3168/jds.2008-1305. [DOI] [PubMed] [Google Scholar]
- 28.Urashima T, et al. Oligosaccharides of milk and colostrum in non-human mammals. Glycoconj J. 2001;18:357–371. doi: 10.1023/a:1014881913541. [DOI] [PubMed] [Google Scholar]
- 29.Chou HH, et al. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:11751–11756. doi: 10.1073/pnas.95.20.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ninonuevo MR, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 31.Kunz C, et al. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 32.Newburg DS. Neonatal protection by an innate immune system of human milk consisting of oligosaccharides and glycans. J Anim Sci. 2009;87:26–34. doi: 10.2527/jas.2008-1347. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz-Palacios GM, et al. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278:14112–14120. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- 34.Newburg DS, et al. Innate protection conferred by fucosylated oligosaccharides of human milk against diarrhea in breastfed infants. Glycobiology. 2004;14:253–263. doi: 10.1093/glycob/cwh020. [DOI] [PubMed] [Google Scholar]
- 35.Andersson B, et al. Inhibition of attachment of Streptococcus pneumoniae and Haemophilus influenzae by human milk and receptor oligosaccharides. J Infect Dis. 1986;153:232–237. doi: 10.1093/infdis/153.2.232. [DOI] [PubMed] [Google Scholar]
- 36.Obermeier S, et al. Secretion of 13C-labelled oligosaccharides into human milk and infant’s urine after an oral [13C]galactose load. Isotopes Environ Health Stud. 1999;35:119–125. doi: 10.1080/10256019908234084. [DOI] [PubMed] [Google Scholar]
- 37.Angeloni S, et al. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology. 2005;15:31–41. doi: 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- 38.Vanmaele RP, et al. Role of lactosyl glycan sequences in inhibiting enteropathogenic Escherichia coli attachment. Infect Immun. 1999;67:3302–3307. doi: 10.1128/iai.67.7.3302-3307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velupillai P, Harn DA. Oligosaccharide-specific induction of interleukin 10 production by B220+ cells from schistosome-infected mice: a mechanism for regulation of CD4+ T-cell subsets. Proc Natl Acad Sci U S A. 1994;91:18–22. doi: 10.1073/pnas.91.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eiwegger T, et al. Human milk--derived oligosaccharides and plant-derived oligosaccharides stimulate cytokine production of cord blood T-cells in vitro. Pediatr Res. 2004;56:536–540. doi: 10.1203/01.PDR.0000139411.35619.B4. [DOI] [PubMed] [Google Scholar]
- 41.Bode L. Recent advances on structure, metabolism, and function of human milk oligosaccharides. J Nutr. 2006;136:2127–2130. doi: 10.1093/jn/136.8.2127. [DOI] [PubMed] [Google Scholar]
- 42.Liolios K, et al. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schell MA, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JH, et al. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics. 2008;9:247. doi: 10.1186/1471-2164-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sela DA, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barrangou R, et al. Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J Bacteriol. 2009;191:4144–4151. doi: 10.1128/JB.00155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ventura M, et al. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol. 2009;7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- 48.Maze A, et al. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2007;73:545–553. doi: 10.1128/AEM.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parche S, et al. Sugar transport systems of Bifidobacterium longum NCC2705. J Mol Microbiol Biotechnol. 2007;12:9–19. doi: 10.1159/000096455. [DOI] [PubMed] [Google Scholar]
- 50.Xu J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 51.Ramsay AG, et al. Cell-associated alpha-amylases of butyrate-producing Firmicute bacteria from the human colon. Microbiology. 2006;152:3281–3290. doi: 10.1099/mic.0.29233-0. [DOI] [PubMed] [Google Scholar]
- 52.Ward RE, et al. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72:4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward RE, et al. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res. 2007;51:1398–1405. doi: 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- 54.LoCascio RG, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 55.Klaassens ES, et al. Mixed-species genomic microarray analysis of fecal samples reveals differential transcriptional responses of bifidobacteria in breast- and formula-fed infants. Appl Environ Microbiol. 2009;75:2668–2676. doi: 10.1128/AEM.02492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishimoto M, Kitaoka M. Practical preparation of lacto-N-biose I, a candidate for the bifidus factor in human milk. Biosci Biotechnol Biochem. 2007;71:2101–2104. doi: 10.1271/bbb.70320. [DOI] [PubMed] [Google Scholar]
- 57.Kitaoka M, et al. Novel putative galactose operon involving lacto-N-biose phosphorylase in Bifidobacterium longum. Appl Environ Microbiol. 2005;71:3158–3162. doi: 10.1128/AEM.71.6.3158-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiyohara M, et al. Prebiotic effect of Lacto-N-biose I on bifidobacterial growth. Biosci Biotechnol Biochem. 2009;73:1175–1179. doi: 10.1271/bbb.80697. [DOI] [PubMed] [Google Scholar]
- 59.Xiao JZ, et al. Distribution of in vitro fermentation ability of Lacto-N-Biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Appl Environ Microbiol. 2009;76:54–59. doi: 10.1128/AEM.01683-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katayama T, et al. Molecular cloning and characterization of Bifidobacterium bifidum 1,2-alpha-L-fucosidase (AfcA), a novel inverting glycosidase (glycoside hydrolase family 95) J Bacteriol. 2004;186:4885–4893. doi: 10.1128/JB.186.15.4885-4893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagae M, et al. Structural basis of the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase from Bifidobacterium bifidum. J Biol Chem. 2007;282:18497–18509. doi: 10.1074/jbc.M702246200. [DOI] [PubMed] [Google Scholar]
- 62.Ashida H, et al. Two distinct alpha-L-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19:1010–1017. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 63.Wada J, et al. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol. 2008;74:3996–4004. doi: 10.1128/AEM.00149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki R, et al. Structural and thermodynamic analyses of solute-binding protein from Bifidobacterium longum specific for core 1 disaccharide and lacto-N-biose I. J Biol Chem. 2008;283:13165–13173. doi: 10.1074/jbc.M709777200. [DOI] [PubMed] [Google Scholar]
- 65.Wada J, et al. Purification, crystallization and preliminary X-ray analysis of the galacto-N-biose-/lacto-N-biose I-binding protein (GL-BP) of the ABC transporter from Bifidobacterium longum JCM1217. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:751–753. doi: 10.1107/S1744309107036263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujita K, et al. Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum. J Biol Chem. 2005;280:37415–37422. doi: 10.1074/jbc.M506874200. [DOI] [PubMed] [Google Scholar]
- 67.Ruas-Madiedo P, et al. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl Environ Microbiol. 2008;74:1936–1940. doi: 10.1128/AEM.02509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishimoto M, Kitaoka M. Identification of the putative proton donor residue of lacto-N-biose phosphorylase (EC 2.4.1.211) Biosci Biotechnol Biochem. 2007;71:1587–1591. doi: 10.1271/bbb.70064. [DOI] [PubMed] [Google Scholar]
- 69.Nishimoto M, Kitaoka M. Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum. Appl Environ Microbiol. 2007;73:6444–6449. doi: 10.1128/AEM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Markowitz VM, et al. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 2008;36:D528–533. doi: 10.1093/nar/gkm846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kruse HP, et al. Effects of inulin on faecal bifidobacteria in human subjects. Br J Nutr. 1999;82:375–382. doi: 10.1017/s0007114599001622. [DOI] [PubMed] [Google Scholar]
- 72.Roberfroid MB, et al. The bifidogenic nature of chicory inulin and its hydrolysis products. J Nutr. 1998;128:11–19. doi: 10.1093/jn/128.1.11. [DOI] [PubMed] [Google Scholar]
- 73.Gibson GR, et al. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology. 1995;108:975–982. doi: 10.1016/0016-5085(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 74.Boehm G, Stahl B. Oligosaccharides from milk. J Nutr. 2007;137:847S–849S. doi: 10.1093/jn/137.3.847S. [DOI] [PubMed] [Google Scholar]
- 75.Macfarlane GT, et al. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J Appl Microbiol. 2008;104:305–344. doi: 10.1111/j.1365-2672.2007.03520.x. [DOI] [PubMed] [Google Scholar]
- 76.Boehm G, et al. Prebiotic carbohydrates in human milk and formulas. Acta Paediatr Suppl. 2005;94:18–21. doi: 10.1111/j.1651-2227.2005.tb02149.x. [DOI] [PubMed] [Google Scholar]
- 77.Sumiyoshi W, et al. Galactosyllactoses in the milk of Japanese women: changes in concentration during the course of lactation. Journal of Applied Glycoscience. 2004;51:341–344. [Google Scholar]
- 78.O’Connell Motherway M, et al. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2008;74:6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]