Abstract
The molecular basis of the myotonic dystrophy type 1 is the expansion of a CTG repeat at the DMPK locus. The expanded disease-associated repeats are unstable in both somatic and germ lines, with a high tendency towards expansion. The rate of expansion is directly related to the size of the pathogenic allele, increasing the size heterogeneity with age. It has also been suggested that additional factors, including as yet unidentified environmental factors, might affect the instability of the expanded CTG repeats to account for the observed CTG size dynamics over time. To investigate the effect of environmental factors in the CTG repeat instability, three lymphoblastoid cell lines were established from two myotonic dystrophy patients and one healthy individual, and parallel cultures were concurrently expanded in the presence or absence of the mutagenic chemical mitomycin C for a total of 12 population doublings. The new alleles arising along the passages were analysed by radioactive small pool PCR and sequencing gels. An expansion bias of the stepwise mutation was observed in a (CTG)124 allele of a cell line harbouring two modal alleles of 28 and 124 CTG repeats. Interestingly, this expansion bias was clearly enhanced in the presence of mitomycin C. The effect of mitomycin C was also evident in the normal size alleles in two cell lines with alleles of 13/13 and 12/69 repeats, where treated cultures showed new longer alleles. In conclusion, our results indicate that mitomycin C modulates the dynamics of myotonic dystrophy-associated CTG repeats in LBCLs, enhancing the expansion bias of long-pathogenic repeats and promoting the expansion of normal length repeats.
INTRODUCTION
Myotonic dystrophy type 1 (DM1) is an autosomal dominant neuromuscular disorder with a multisystemic phenotype. The molecular basis of DM1 is the expansion of an unstable CTG repeat in the 3′-untranslated region of the myotonic dystrophy protein kinase gene, on chromosome 19q13.3 (1). The size of the expanded allele directly correlates with the severity of the disease (2,3), and the number of CTG repeats progressively increases in successive generations of DM1 families, being the molecular explanation for anticipation (4).
Early Southern blot analysis of expanded CTG repeats revealed their somatic instability (5,6). Such instability was later demonstrated in both somatic and germ cells by using the small pool PCR (SP-PCR) technique (7). Somatic tissues of DM1 patients show mosaicism for CTG repeat size and, it has been found, a high tendency towards expansion throughout their lifetime. Thus the number of CTG repeats in peripheral blood leukocytes of young patients is rather stable whereas high levels of heterogeneity are observed in older patients (6,8). Germ cells from DM1 patients also show a high degree of variation in repeat size, including large contractions (9,10).
Analysis of tissue samples from DM1 subjects revealed that the major factor affecting the rate of expansion is the initial size of the expanded allele, with increased size heterogeneity over time (6,8). Although the model for the somatic instability proposed by Monckton et al. (7), based on the stepwise gain of a small number of repeats with age proceeding through expansion, agrees with these observations, a detailed analysis of the lifelong dynamics of somatic repeat size suggests that the expansion process is affected by other unknown transacting genetic factors (8). Additional in vitro studies using lymphoblastoid cell lines (LBCL) from DM1 patients have shown that two types of mutations arise in the pathogenic CTG repeats: (i) a frequent gain or loss of a small number of repeats, which is in concordance with the stepwise model described by Monckton et al. (7); and (ii) a less frequent large change of the repeat size (11), associated with a growth advantage of the cells bearing longer CTG repeats (12). This later phenomenon has been nominated mitotic drive (12).
The influence of environmental factors on the instability of CTG repeats associated with DM1 has not yet been properly addressed, although it has been hypothesized that such factors could modulate CTG repeat instability (8). Among these environmental factors, mutagenic compounds are good candidates since they can interfere with mechanisms known to be involved in trinucleotide repeat instability such as replication, recombination and DNA repair. In this study, we evaluate the effect of mutagenic stress on the dynamics of CTG repeat instability in mitotically dividing cells. To do so, we have established LBCLs from two DM1 patients and one normal individual to analyse the effect of mitomycin C (MMC) on the CTG repeat size dynamics of both normal length and long pathogenic alleles, along successive cell generations. The variant allele sizes in the heterogeneous population of transformed cells were resolved using the SP-PCR technique together with sequencing gel analysis. Using this procedure we have been able to study the dynamics of the CTG size and detect new alleles arising along the passages, in untreated and MMC-treated LBCL cultures.
MATERIALS AND METHODS
Lymphoblastoid cell lines and treatment
Peripheral blood samples were obtained from two symptomatic DM1 patients and one healthy individual by venipuncture, and lymphocytes were EBV-transformed into LBCLs. The cells were cultured as we previously reported (13,14). Transformed cells (2 × 106) were cultured in 75 cm2 flasks at a concentration of 1 × 105 cells/ml and allowed to grow until the culture reached 2 × 107 cells (1 × 106 cells/ml). These expanded cultures were passed on as follows: 2 × 106 cells were transferred to new 75 cm2 flasks and maintained until the culture reached 2 × 107 cells and then a new passage was performed. The remaining cells were split in two aliquots, one was kept frozen and the other was used for DNA extraction following a standard phenol-chloroform protocol. This procedure was repeated in each passage and the number of population doublings (PDs) in each passage was calculated.
MMC was diluted to a concentration of 1 mg/ml and stored at 4°C and treatments of the LBCL were performed by adding MMC to the medium to get a final concentration of 5 ng/ml. The mutagen treatment was continuous. The effect of MMC in cell proliferation and viability was evaluated by the Cell proliferation Kit II (XTT) (Boehringer Mannheim) and the fluorescein diacetate-mediated viability assay (15), respectively. We found that 5 ng/ml of MMC was not cytotoxic for any of the three LBCL but reduced the cell growth rate by 15% (data not shown). The effect of MMC on the culture growth dynamics is shown in the Supplementary Material available at NAR Online.
SP-PCR analysis
The concentration of the DNA extracted from the cell lines was measured spectrophotometrically and 5 µg of genomic DNA were digested with EcoRI. In order to estimate the true number of amplifiable genome equivalence (a.g.e.) in a volume of DNA sample, digested DNA was diluted in 10 mM Tris–HCl (pH 7.5), 1 mM EDTA and 5 ng/µl carrier herring sperm DNA by serial 1:10 dilution steps to a final concentration of 5 pg/µl. Then the number of amplifiable molecules was determined by SP-PCR using the final dilution containing 5 pg/µl of digested genomic DNA. At this level of input DNA, some reactions will fail to contain an amplifiable copy of one or the other allele, and observing the number of reactions that amplified one allele or fail to amplify, the true number of a.g.e. in the input DNA was estimated (data not shown). Subsequently the CTG repeat size variations were analysed by performing 40 SP-PCRs using 200 a.g.e. of input DNA in each 25 µl reaction. The SP-PCR conditions previously described by Cobo et al. (16) were used, with some minor modifications. 36% of betaine (Sigma) was added to the reaction tube and the reactions were performed using the DM-93 and DM-103 primers described by Brook et al. (1). The amplified products were labelled by adding 3 µCi [α-33P]dCTP to the SP-PCRs. Amplification conditions were 94°C 3 min for 1 cycle; 96°C 45 s, 68°C 45 s, 70°C 3 min for 28 cycles, with a final extension step at 68°C 1 min, 70°C 10 min, in a PT-100 Thermocycler (MJ Research, USA). Then, 7 µl of the SP-PCR products were resolved on 6% polyacrylamide sequencing gels and visualised by autoradiography. A 50 bp ladder (Amersham Biosciences) end-labelled with [γ-33P]ATP was used as a molecular size marker. The length of the CTG repeats in the long alleles was estimated by densitometry using the Molecular Analyst 1.5 software (Bio-Rad).
Statistical analysis
Statistical comparisons between mutation frequencies were performed with the Fisher’s exact test.
RESULTS
To investigate the effect of MMC on the CTG repeat instability of the DM1 locus, LBCLs were established from two DM1 patients and one normal individual. The sizes of the CTG repeats on these individuals were determined in peripheral blood samples by using standard radioactive PCR (Table 1). After Epstein–Barr virus (EBV) transformation, the lymphoblastoid cells were expanded until 1 × 106 cells/ml and the size of the CTG repeat was again analysed to establish the repeat length at the start of the experimental process. We have designated this stage as passage zero (P0). Due to the heterogeneous size of the expanded CTG repeats in the peripheral blood cells of the two DM1 individuals, the LBCLs derived from their lymphocytes showed a modal pathogenic allele that differs from the modal size determined in the blood sample of the DM1 individuals. Normal size alleles, however, maintained their original size in the cell line (see Table 1). At P0, the cells were diluted to 1 × 105 cells/ml in medium with and without MMC and cultures were allowed to expand up to 1 × 106 cells/ml and then subpassed again. The same process was repeated four times. Following this approach, the molecular analysis of CTG instability was performed after 0, 6 and 12 PDs, both in the presence or absence of mutagenic treatment, by radioactive SP-PCR. To obtain the variant allele distribution in the cell population, at each point of the time-course study, we performed 40 identical SP-PCRs with an input of DNA of 200 a.g.e., which represents a total of 8000 analysed alleles in each PD. Under our SP-PCR conditions, and in the range of the allele size studied (12 to 124 CTGs), the amplification of the normal and the pathogenic alleles showed the same efficiency and, therefore, in the heterozygous LBCLs we considered a 1:1 ratio for normal and disease-causing alleles (4000 of each allele analysed at each point of study).
Table 1. Size of the CTG repeat associated with DM1 in peripheral blood cells and their corresponding lymphoblastoid cell lines (LBCLs) of two DM1 patients and one normal individual.
| Individual | Age (years) | Genotype in blooda (CTG)n | LBCL genotype at P0a (CTG)n |
|---|---|---|---|
| DM1-1 | 31 | 114/28 | 124/28 |
| DM1-2 | 30 | 70/13 | 69/12 |
| CM-1 | 27 | 13/13 | 13/13 |
aFor the expanded allele, only the number of CTGs of the modal allele is indicated.
Mutations at the DM1-pathogenic CTG alleles
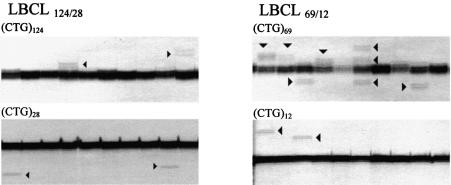
The SP-PCR approach, together with sequencing gel analysis, was applied to study the size-distribution of variant long alleles in two DM1 LBCLs, LBCL69/12 and LBCL124/28. The cells were concurrently grown in the presence or absence of MMC. The SP-PCR is a technique that allows for the resolution of individual alleles by limiting the amount of input DNA in the PCR (7,17). However, the use of too little amplifiable DNA would limit the detection of infrequent mutant alleles, unless many PCRs are performed. On the other hand, due to the PCR stutter effect when large trinucleotide repeats are amplified (18), small variations in allele size would not be detected while larger amounts of input DNA are used. Therefore, in our experimental conditions where 200 a.g.e. of input DNA were used, small changes in the CTG repeat size of the two analysed large alleles (69 and 124 CTG repeats) were not resolved, whereas rare gross mutations were efficiently detected by performing 40 SP-PCRs (Fig. 1).
Figure 1.
Representative examples of the SP-PCR analysis in the DM1 LBCLs using 200 a.g.e. of input DNA per reaction. Each line represents an SP-PCR. Variations of the normal and pathogenic alleles are indicated (arrowheads).
The SP-PCR analysis showed that the two LBCLs with long pathogenic alleles (69 and 124 CTG repeats), growing in standard conditions (without MMC), presented two types of spontaneous CTG repeat mutations: small changes in the number of CTG repeats and large size changes in the repeat number. The small size changes were manifested as a wide band containing a modal allele. The modal allele sizes and the upper and lower limits of these bands were determined as an indication of the allele size heterogeneity, being 69 ± 5 repeats for the (CTG)69 allele and 124 ± 6 repeats for the (CTG)124 allele at P0. This type of mutation was constant along the passages and was detected in all the SP-PCRs performed. It is unlikely that PCR stutter could account for this type of small change in repeat size, since this experimental artefact would have an effect on repeat size no larger than a three repeat shift, as recently reported (18). Nevertheless, whereas in the LBCL69/12 cell line the modal (CTG)69 allele remains the same along the passages, in the LBCL124/28 cell line the modal (CTG)124 allele at P0 showed a gradual shift to slightly longer CTG tracks along the passages, being the new modal alleles 126 and 129 CTG tracks long after 6 and 12 PD, respectively (see Fig. 2). This shift was detected in all 40 SP-PCRs performed at each point of study, and it is clear evidence of a stepwise mutation mechanism with an expansion bias. In addition, new alleles with large size changes were also detected as discrete bands outside the wide major band (see Fig. 1). Based on the number of SP-PCRs, the relative frequency of such mutations with respect to the modal allele was determined, and their frequency distributions are represented in Figure 2. Large contractions and expansions were observed in both alleles and at all PD. Considering that 4000 expanded alleles per cell line were analysed at each point of study by using 200 a.g.e. of input DNA in each of the 40 SP-PCR performed, we have estimated the spontaneous frequency of large size changes at 0, 6 and 12 PD. As no differences were observed between PD, data were pooled and the overall spontaneous frequency of large mutations was 4.6 × 10–3 and 2.2 × 10–3 for the (CTG)69 allele and for the (CTG)124 allele, respectively.
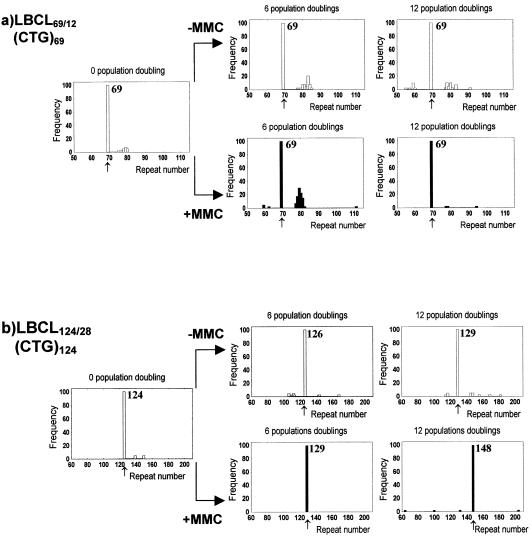
Figure 2.
Distribution of the pathogenic DM1 alleles in the LBCL69/12 and LBCL124/28 cultures grown in the presence or absence of MMC up to 12 PD. Modal alleles and mutant alleles resulting from large contractions and expansions along the passages are shown. The frequency of mutant alleles is calculated considering the number of SP-PCRs that showed new bands with respect to modal allele bands. Modal alleles are indicated with an arrow. Number shows the modal CTG repeat size.
MMC enhances the expansion bias of the (CTG)124 allele
When the LBCL69/12 and the LBCL124/28 cultures were concurrently expanded in the presence of MMC, the same type of small changes observed in the long CTG repeats in the absence of MMC were found, i.e. along the passages a width SP-PCR band indicates the heterogeneity of the allele size around the modal allele. The heterogeneity for the (CTG)69 and (CTG)124 alleles was in the same range of the control experiment. As shown in Figures 2 and 3, the major effect of the MMC was an increase in the shift towards expansion of the (CTG)124 modal allele along the passages, compared to the control culture. Thus, the modal allele at P0 shifts from 124 to 129 and 148 CTG repeats after 6 and 12 PD, respectively. These results clearly suggest that MMC accelerated the expansion bias of the pathogenic allele in successive cell generations. In contrast, a statistically significant decrease in the number of large changes in the allele size was observed in the presence of MMC, with respect to control cultures. Thus, of 4000 alleles analysed after 12 PD, 22 and three alleles were large size variants of the (CTG)69 allele in control and MMC-treated cultures, respectively (P < 0.001). In the case of the (CTG)124 allele, we found 10 and four of this type of variant allele in the absence or presence of MMC, respectively (P < 0.01). A feasible explanation for this fact is that a selection against those highly MMC-damaged cells is taking place along the passages, with a consequent elimination of the infrequent types of cells in the population.
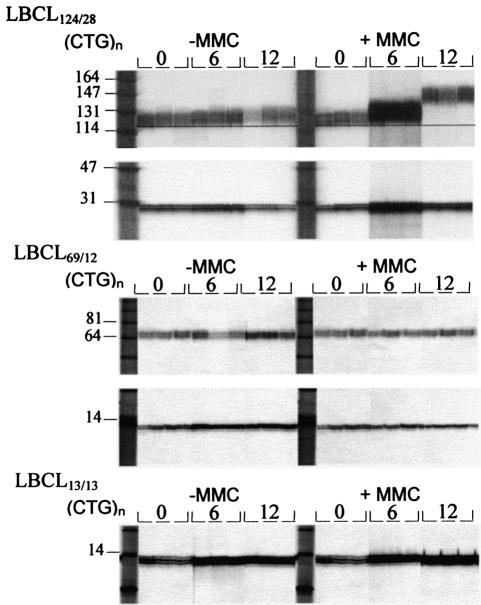
Figure 3.
Evolution of the (CTG)124 and (CTG)28 allele sizes in the three lymphoblastoid cell lines along the culture passages in the presence or absence of MMC. The number of cell population doublings, 0, 6 and 12, is indicated. Each line represents an SP-PCR.
Effect of MMC on the stability of normal length DM1 alleles
At the same time that the pathogenic alleles were analysed, the distribution of normal alleles in the DM1 LBCL69/12 and the LBCL124/28 cell lines was explored, together with the LBCL13/13 obtained from a healthy individual of the same age. Performing 40 SP-PCRs with an input of DNA of 200 a.g.e., we were able to detect rare alleles emerging along the passages, including those small variations in repeat number. This was due to the fact that our sequencing gel conditions led to a higher resolution on the short CTG tracks range. The distribution of the normal alleles in the three cell lines expanded in the presence or absence of MMC is shown in Figure 4, including the relative frequencies of the rare alleles with respect to the modal normal alleles of 12, 13 and 28 CTG repeats. While both contractions and expansions of the (CTG)12 and (CTG)13 alleles were detected, for the (CTG)28 allele only contractions were found. The same procedure used in the analysis of large changes in the pathogenic alleles was applied to estimate the frequency of rare alleles emerging from the normal DM1 alleles in the three cultures. Thus, at each experimental point, a total of 4000 normal alleles were analysed in the LBCL69/12 and LBCL124/28 cell lines and 8000 normal alleles in the LBCL13/13 cell line. Because no differences between PD were observed, the estimated overall frequencies of spontaneous mutation (untreated cultures) of the normal alleles were 1.1 × 10–3 and 8.3 × 10–4 in the LBCL69/12 and LBCL124/28 cultures, respectively. These frequencies are significantly different and greater than the frequency of 2.9 × 10–4, found in the LBCL13/13 culture (P < 0.01 and P < 0.05, respectively).
Figure 4.
Distribution of the normal-length DM1 alleles in the LBCL13/13, LBCL69/12 and LBCL124/28 cultures, grown in the presence or absence of MMC up to 12 PD. The frequency of mutant alleles is calculated considering the number of SP-PCRs that showed new bands with respect to modal allele band.
Regarding the effect of MMC in the short alleles, we have to point out that the majority of alterations found were expansions into the normal range size. As shown in Figure 4a and b, a clear difference was observed in the length distributions of new alleles found in the MMC-treated LBCL13/13 and LBCL69/12 cell lines; i.e. in these cultures, after 6 and 12 PDs, new long alleles to the normal size range were apparent. Most of these new alleles have a number of repeats that exceeds twice the number of repeats of the most represented allele (range 27–29 repeats). In addition, the frequency of new long alleles found in concurrent LBCL13/13 cultures in the absence or presence of MMC, after 6 and 12 PD, is significantly higher (6.3 × 10–5 versus 7.5 × 10–4, respectively; P < 0.01). In the LBCL69/12 culture, a significant effect of MMC was also observed when we considered the frequency of expansion mutations in the normal size range (1.3 × 10–4 in control versus 1.0 × 10–3 in MMC-treated cells; P < 0.05). In contrast, only contractions were observed in the (CTG)28 allele of the LBCL124/28 cell line in all PD irrespective of MMC-treatment (see Fig. 4c).
DISCUSSION
Somatic CTG repeat instability associated with DM1 has a strong bias towards expansion that depends on the size of the repeat and the age of the affected individuals (6). In addition, previous studies on the repeat size dynamics hypothesized that environmental factors might affect the expansion process (8). In this study we have evaluated the effect of MMC on the instability of the CTG repeat of DM1 alleles of both normal and pathogenic length. The analysis of the CTG size dynamics was carried out in LBCLs from two DM1 patients and one normal individual. This analysis was performed along successive cell generations in a time-course study, using radioactive SP-PCR and sequencing gel analysis to resolve small variations in the CTG repeat size.
The instability of the expanded CTG alleles found in the untreated DM1 LBCLs cultures agrees with previously reported data on DM1 LBCLs (11,12). Thus, frequent small changes in the CTG repeat size (contractions or expansions of five to six repeats) were observed along the passages, in accordance with the stepwise mutation model proposed by Monckton et al. (7) and Khajavi et al. (12) to explain the in vivo and in vitro size heterogeneity of this repeat, respectively. Moreover, and as shown in Figure 3, the sensitive method of analysis used allowed us to detect a gradual increase of the (CTG)124 allele size in successive cell generations, revealing the expansion bias of the stepwise mutation of this allele in short-term cultures. Such an effect was not observed in the (CTG)69 allele. A possible explanation for the difference found between the two pathogenic alleles could be the extension time of the cultures, so that longer cultures would be necessary to detect a presumed expansion bias of the (CTG)69 allele. Alternatively, a threshold in the number of CTG repeats might exist to display a bias towards expansion of the stepwise mutations occurring over time. Based on our results, this threshold would lie between 69–124 repeats, which includes the protomutation range (50–80 repeats), considered relatively stable in vivo somatic cells (19,20). In a similar study carried out in LBCLs (12) the authors reported expansion bias of alleles longer than 180 CTG repeats, which are not comparable to the 69 repeats allele of the present investigation. Martorell et al. (8) did not detect changes in the size of DM1 alleles of <200 repeats over time in vivo, although the resolution of their analysis procedure (Southern blot) would have limited the detection of small changes on repeat number. In addition, our data show no effect of MMC in the instability of the (CTG)69 allele (see Fig. 2), supporting our hypothesis of the presence of a size threshold in the expansion bias of the stepwise mutations.
A less frequent type of change of the (CTG)124 and the (CTG)69 alleles, which corresponds to size changes exceeding five to six repeats, the modal allele size was also found along the passages, displaying discrete bands on the sequencing gels (see Fig. 1). Unlike previous studies that reported more contractions than expansions (11,12), our data show that this type of variant allele included a similar number of contractions and expansions.
The clear effect of MMC on the (CTG)124 allele instability is intriguing. MMC increased the spontaneous expansion bias of this allele along the passages resulting in a shift of 24 CTG repeats in 12 PD (from 124 to 148 repeats). One may speculate that either the (CTG)148 allele already existed at the starting point or that this mutation arose by chance in the culture. However, independently of MMC, and regardless of the possibility that mitotic drive could increase a possible likelihood of the cells with the (CTG)148 allele to take over the rest of the cell population through the passages, the probability of this happening by chance would be very low, considering the short term cultures established. In addition, it should be considered that these rare cells would suffer a cell drift in successive cell generations and eventually disappear from the cell population, although a counter cell drift by mitotic drive is also expected. Therefore, this indicates that MMC plays a direct role in the observed repeat expansion. MMC produces DNA interstrand cross-links that are repaired by an error prone pathway at replication, consisting in the combination of nucleic excision repair and a lesion bypass process (21). The bypass process is responsible for the MMC S-phase dependent mutagenesis. Thus, new variant alleles could be induced by a replication-dependent stepwise mechanism in the presence of MMC. The MMC dose (5 ng/ml) used in our experiments had no effect on cell viability although it induced a 15% delay in the cell cycle and a consequent increase in the cell doubling time. As hypothesized before (11) this could be an important factor in DM1 repeat mutation. Accordingly, young patients with congenital DM1 do not manifest repeat heterogeneity regardless of the rapid cell divisions that take place in embryogenesis (22). Taking into account the above observations, we propose that MMC induces new variant alleles in culture cells and that the genetically heterogeneous cell population would be the substrate for the selection of those cells bearing larger alleles, ultimately displacing the original cell population in few passages. This model is supported by recent published data suggesting an association between high proliferating rate and large CTG repeats (12).
The spontaneous rates of instability of the non-pathogenic DM1 alleles in the LBCL69/12, LBCL124/28 and LBCL13/13 cell lines were in the same order of magnitude that the overall rate of instability of microsatellite sequences reported in humans (23). However, the spontaneous mutation frequency of these normal alleles in the cell lines derived from DM1 patients was statistically higher that the mutation frequency of the allele in the cell line derived from a healthy individual. Although we do not have any feasible explanation for this finding, growth rate differences should be considered in interpreting our results. The population doubling time of the DM1 cell lines LBCL69/12 and LBCL124/28 was 3 and 3.5 days, respectively, compared to 2 days for the LBCL13/13 cell line. Therefore the relative lower growth rate of the DM1 cell lines would have allowed additional time for spontaneous replication-independent mutations to occur. In addition, new alleles were observed in the presence of MMC, mainly with expansion of the CTG repeat, some of them reaching 29 repeats. The trinucleotide repeat instability induced by MMC is mainly independent of mismatch repair, as we have recently reported (24). Therefore these variant alleles could be generated by either recombination-dependent or -independent repair pathways involved in DNA interstrand cross-link repair (21,25). The MMC induced alleles lie within the long modal distribution of the normal size range and, therefore, could have significant importance in generating new premutational alleles to maintain the high incidence of DM1 in the population (15,26).
In conclusion, we have found that MMC modulates the dynamics of DM1-associated CTG repeats in mitotically dividing human cells, enhancing the expansion bias of long-pathogenic alleles and promoting the instability of normal length alleles. To our knowledge, this is the first description of an environmental factor affecting DM1-associated trinucleotide repeat instability. Although our in vitro results in somatic cells cannot be directly extrapolated to the in vivo situation, our findings suggest a novel mechanism of trinucleotide repeat instability with important clinical implications. Thus, mutagenic stress induced by environmental agents could accelerate the anticipation process and also increase the severity of the disease in symptomatic individuals due to an increased instability of the trinucleotide repeats in their somatic cells. It will be interesting to prove in mouse models whether mutagenic stress is involved in the anticipation process, by promoting the in vivo germ line expansion of trinucleotide repeats, and in the in vivo heterogeneity and mosaicism in the number of repeats.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr L. Martorell (Sant Pau Hospital, Barcelona, Spain) for helpful comments and technical advice. This work was partially funded by La Fundació La Marató de TV3 (project 1999-85), the Generalitat de Catalunya (SGR-00197-2002), the Spanish Ministry of Health and Consumption (projects FIS PI020145, FIS PI020648 and FIS-Red G03/073), the Spanish Ministry of Science and Technology (projects PM 1999-0067, SAF2001-5138 and SAF2003-020328) and The European Union (project FIGH-CT-2002-00217). E.P. and L.F.-L. were supported by predoctoral fellowships awarded by the MCYT. J.S. is supported by a ‘Ramón y Cajal’ project entitled ‘Genome stability and DNA repair’ awarded by the MCyT and co-financed by the UAB.
REFERENCES
- 1.Brook J.D., McCurrach,M.E., Harley,H.G., Buckler,A.J., Church,D., Aburatani,H., Hunter,K., Stanton,V.P., Thirion,J.P., Hudson,T. et al. (1992) Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell, 68, 799–808. [DOI] [PubMed] [Google Scholar]
- 2.Harley H.G., Brook,J.D., Rundle,S.A., Crow,S., Reardon,W., Buckler,A.J., Harper,P.S., Housman,D.E. and Shaw,D.J. (1992) Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature, 355, 545–546. [DOI] [PubMed] [Google Scholar]
- 3.Martorell L., Martinez,J.M., Carey,N., Johnson,K. and Baiget,M. (1995) Comparison of CTG repeat length expansion and clinical progression of myotonic dystrophy over a five year period. J. Med. Genet., 32, 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harley H.G., Rundle,S.A., MacMillan,J.C., Myring,J., Brook,J.D., Crow,S., Reardon,W., Fenton,I., Shaw,D.J. and Harper,P.S. (1993) Size of the unstable CTG repeat sequence in relation to phenotype and parental transmission in myotonic dystrophy. Am. J. Hum. Genet., 52, 1164–1174. [PMC free article] [PubMed] [Google Scholar]
- 5.Buxton J., Shelbourne,P., Davies,J., Jones,C., Van Tongeren,T., Aslanidis,C., de Jong,P., Anvret,M., Riley,B. et al. (1992) Detection of an unstable fragment of DNA specific to individuals with myotonic dystrophy. Nature, 355, 547–548. [DOI] [PubMed] [Google Scholar]
- 6.Wong L.J., Ashizawa,T., Monckton,D.G., Caskey,C.T. and Richards,C.S. (1995) Somatic heterogeneity of the CTG repeat in myotonic dystrophy is age and size dependent. Am. J. Hum. Genet., 56, 114–122. [PMC free article] [PubMed] [Google Scholar]
- 7.Monckton D.G., Wong,L.J., Ashizawa,T. and Caskey,C.T. (1995) Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum. Mol. Genet., 4, 1–8. [DOI] [PubMed] [Google Scholar]
- 8.Martorell L., Monckton,D.G., Gamez,J., Johnson,K.J., Gich,I., de Munain,A.L. and Baiget,M. (1998) Progression of somatic CTG repeat length heterogeneity in the blood cells of myotonic dystrophy patients. Hum. Mol. Genet., 7, 307–312. [DOI] [PubMed] [Google Scholar]
- 9.Martorell L., Monckton,D.G., Gamez,J. and Baiget,M.(2000) Complex patterns of male germline instability and somatic mosaicism in myotonic dystrophy type 1. Eur. J. Hum. Genet., 8, 423–430. [DOI] [PubMed] [Google Scholar]
- 10.Jansen G., Willems,P., Coerwinkel,M., Nillesen,W., Smeets,H., Vits,L., Howeler,C., Brunner,H. and Wieringa,B. (1994) Gonosomal mosaicism in myotonic dystrophy patients: involvement of mitotic events in (CTG)n repeat variation and selection against extreme expansion in sperm. Am. J. Hum. Genet., 54, 575–585. [PMC free article] [PubMed] [Google Scholar]
- 11.Ashizawa T., Monckton,D.G., Vaishnav,S., Patel,B.J., Voskova,A. and Caskey,C.T. (1996) Instability of the expanded (CTG)n repeats in the myotonin protein kinase gene in cultured lymphoblastoid cell lines from patients with myotonic dystrophy. Genomics, 36, 47–53. [DOI] [PubMed] [Google Scholar]
- 12.Khajavi M., Tari,A.M., Patel,N.B., Tsuji,K., Siwak,D.R., Meistrich,M.L., Terry,N.H. and Ashizawa,T. (2001) Mitotic drive of expanded CTG repeats in myotonic dystrophy type 1 (DM1). Hum. Mol. Genet., 10, 855–863. [DOI] [PubMed] [Google Scholar]
- 13.Callen E., Samper,E., Ramirez,M.J., Creus,A., Marcos,R., Ortega,J.J., Olive,T., Badell,I., Blasco,M.A. and Surralles,J. (2002) Breaks at telomeres and TRF2-independent end fusions in Fanconi anemia. Hum. Mol. Genet., 11, 439–444. [DOI] [PubMed] [Google Scholar]
- 14.Surrallés J., Ramirez,M.J., Marcos,R., Natarajan,A.T. and Mullenders,L.H. (2002) Clusters of transcription-coupled repair in the human genome. Proc. Natl Acad. Sci. USA, 99, 10571–10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Straus G. (1991) Non-random cell killing in cryopreservation: implications for performance of the battery of leukocyte tests (BLT), I. Toxic and immunotoxic effects. Mutat. Res., 252, 1–15. [DOI] [PubMed] [Google Scholar]
- 16.Cobo A., Martinez,J.M., Martorell,L., Baiget,M. and Johnson,K. (1993) Molecular diagnosis of homozygous myotonic dystrophy in two asymptomatic sisters. Hum. Mol. Genet., 2, 711–715. [DOI] [PubMed] [Google Scholar]
- 17.Jeffreys A.J., Tamaki,K., MacLeod,A., Monckton,D.G., Neil,D.L. and Armour,J.A. (1994) Complex gene conversion events in germline mutation at human minisatellites. Nature Genet., 6, 136–145. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y., Monckton,D.G., Siciliano,M.J., Connor,T.H. and Meistrich,M.L. (2002) Age and insertion site dependence of repeat number instability of a human DM1 transgene in individual mouse sperm. Hum. Mol. Genet., 117, 791–798. [DOI] [PubMed] [Google Scholar]
- 19.Barcelo J.M., Mahadevan,M.S., Tsilfidis,C., MacKenzie,A.E. and Korneluk,R.G. (1993) Intergenerational stability of the myotonic dystrophy protomutation. Hum. Mol. Genet., 2, 705–709. [DOI] [PubMed] [Google Scholar]
- 20.Martorell L., Monckton,D.G., Sanchez,A., Lopez De Munain,A. and Baiget,M. (2001) Frequency and stability of the myotonic dystrophy type 1 premutation. Neurology, 56, 328–335. [DOI] [PubMed] [Google Scholar]
- 21.Zheng H., Wang,X., Warren,A.J., Legerski,R.J., Nairn,R.S., Hamilton,J.W. and Li,L. (2003) Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol. Cell. Biol., 23, 754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mankodi A. and Thornton,C.A. (2002) Myotonic syndromes. Curr. Opin. Neurol., 15, 545–552. [DOI] [PubMed] [Google Scholar]
- 23.Weber J.L. and Wong,C. (1993) Mutation of human short tandem repeats. Hum. Mol. Genet., 2, 1123–1128. [DOI] [PubMed] [Google Scholar]
- 24.Fernàndez-López L., Piñeiro,E., Marcos,R., Velásquez,A. and Surrallés,J. (2003) Induction of instability of normal length trinucleotide repeats within human disease genes. J. Med. Genet., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole R.S., Levitan,D. and Sinden,R.R. (1976) Removal of psoralen interstrand cross-links from DNA of Escherichia coli: mechanism and genetic control. J. Mol. Biol., 103, 39–59. [DOI] [PubMed] [Google Scholar]
- 26.Imbert G., Kretz,C., Johnson,K. and Mandel,J.L. (1993) Origin of the expansion mutation in myotonic dystrophy. Nature Genet., 4, 72–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.