Abstract
The Rps0 proteins of Saccharomyces cerevisiae are components of the 40S ribosomal subunit required for maturation of the 3′ end of 18S rRNA. Drosophila and human homologs of the Rps0 proteins physically interact with Rps21 proteins, and decreased expression of both proteins in Drosophila impairs control of cellular proliferation in hematopoietic organs during larval development. Here, we characterize the yeast RPS21A/B genes and show that strains where both genes are disrupted are not viable. Relative to the wild type, cells with disrupted RPS21A or RPS21B genes exhibit a reduction in growth rate, a decrease in free 40S subunits, an increase in the amount of free 60S subunits, and a decrease in polysome size. Ribosomal RNA processing studies reveal RPS21 and RPS0 mutants have virtually identical processing defects. The pattern of processing defects observed in RPS0 and RPS21 mutants is not a general characteristic of strains with suboptimal levels of small subunit ribosomal proteins, since disruption of the RPS18A or RPS18B genes results in related but distinct processing defects. Together, these data link the Rps0 and Rps21 proteins together functionally in promoting maturation of the 3′ end of 18S rRNA and formation of active 40S ribosomal subunits.
INTRODUCTION
The RPS0 genes of Saccharomyces cerevisiae are duplicated genes that encode small subunit ribosomal proteins. Deletion of either RPS0 gene reduces growth rate, while deletion of both is lethal (1). The Rps0 proteins are required for the maturation of the 3′ end of 18S rRNA. Specifically, Rps0 proteins are needed for efficient processing of the 20S rRNA precursor to 18S rRNA, a late step in the maturation of 40S ribosomal subunits (2,3).
The yeast Rps0 proteins have over 60% sequence identity with mammalian p40 proteins (1). The mammalian p40 proteins are associated with 40S ribosomal subunits, and both p40 and Rps0 proteins belong to the S2 family of ribosomal proteins (4,5). Members of the S2 family have been identified in all major lines of descent (6). Given the high level of sequence conservation among S2 family members, it is likely that these proteins have conserved functions.
Numerous studies have linked mammalian p40 proteins to tumorigenesis (7–14). The role of the p40 protein in tumor development is generally considered in the context of its function as a putative precursor to the high affinity laminin receptor (15). However, whether p40 functions as a laminin receptor precursor is controversial, and recent developments suggest that the role of p40 in tumorigenesis may be related to its function as a component of the translational machinery (16). Studies in Drosophila have shown that recessive lethal mutations that severely reduce expression of the p40 protein give rise to a tumorous phenotype in hematopoietic organs during larval development (17). A similar phenotype is observed in cells with mutant alleles of the gene encoding ribosomal protein Rps21. Moreover, depletion of both p40 and Rps21 proteins produces a more pronounced tumorous phenotype than depletion of either protein alone (17).
Mutations in Drosophila genes encoding components of the translational machinery generally result in a Minute phenotype characterized by small body size, delayed development, and short slender bristles (18). While dominant mutations in p40 and Rps21 genes that result in haploinsufficiency display Minute phenotypes (17,19), recessive phenotypes brought on by further reductions in expression of these proteins cause excessive cell proliferation and tissue overgrowth. The recessive phenotypes observed in p40 and Rps21 mutants are distinct from phenotypes reported for other ribosomal protein genes, thereby genetically linking the p40 and Rps21 proteins together and setting them apart from other ribosomal components. The observations that both Drosophila and human p40 and Rps21 proteins physically interact provide further evidence linking these two ribosomal proteins together (17,20).
The S.cerevisiae genome contains duplicated genes that encode members of the Rps21 family of ribosomal proteins. Here, we report that the disruption of the RPS21A or RPS21B gene results in a reduction in growth rate and a decrease in the steady-state level of 40S ribosomal subunits. Cells lacking both RPS21A and RPS21B genes are not viable, indicating that the Rps21 proteins are essential. The Rps21 proteins, like the Rps0 proteins, are required for efficient processing of the 20S rRNA precursor at the D cleavage site which gives rise to the mature 3′ end of 18S rRNA. Both Rps21 and Rps0 proteins are also needed for efficient processing at the A2 cleavage site in the internal transcribed spacer 1 (ITS1) region of the rRNA precursor transcript. Thus, in addition to studies that genetically and physically link the Rps21 and Rps0/p40 proteins, these data indicate that the two families of proteins are functionally related.
MATERIALS AND METHODS
Yeast and bacterial strains
The yeast strains used in this study were YMW1 (MATa/MATα ade2-1/ade2-1 ade3Δ 22/ade3Δ 22 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3/leu2-3 trp1-1/trp1-1 ura3-1/ura3-1), W3031B (MATα ade2-1 can1-100 his3-11,15 leu2-3 trp1-1 ura3-1), W3031A (MATa ade2-1 can1-100 his3-11,15 leu2-3 trp1-1 ura3-1), 1–111x12 (MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3/leu2-3 trp1-1/trp1-1 ura3-1/ura3-1 RPS21A/rps21A:: kanMX6), W8-3dx (MATα ade2-1 can1-100 his3-11,15, ura3-1 leu2-3 trp1-1 rps0A::URA3) and W8-3dy (MATa ade2-1 can1-100 his3-11,15 ura3-1 leu2-3 trp1-1 rps0B:: HIS3). A diploid homozygous for an RPS21B disruption (BY4743 clone 37002 MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/met15Δ0 ura3Δ0/ura3Δ0 rps21B::kanMX4/rps21B::kanMX4) and haploids containing either an RPS18A (clone 4284 MATa his3Δ1 leu2Δ0 met15Δ0ura3Δ0 rps18A:: kanMX4) or RPS18B (clone 547 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rps18B::kanMX4) disruption generated by the Saccharomyces genome deletion project consortium were obtained from Research Genetics. Media used in cultivating yeast were YPD [1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose] and synthetic [0.67% (w/v) yeast nitrogen base without amino acids and 2% (w/v) glucose]. Where appropriate, nutrients were added to synthetic media in amounts specified by Sherman (21). Diploids were sporulated on solid sporulation media [1% (w/v) potassium acetate, 0.1% (w/v) yeast extract, 0.05% (w/v) glucose and 2% (w/v) agar). Additional nutrients were added to sporulation media in 25% of the amounts used in synthetic media. The Escherichia coli strain used in this study was XL1-Blue (Stratagene, La Jolla, CA).
DNA manipulations
The complete RPS21A open-reading frame was replaced with the heterologous dominant G418 resistance gene, kanMX6, using the PCR-based disruption strategy of Wach et al. (22,23). Briefly, the pFAK6a-kanMX6 plasmid was used as a template in a PCR where the primers contained regions complementary to the pFA6-kanMX6 plasmid were flanked by regions homologous to sequences in the 5′ and 3′ flanking regions of the RPS21A gene. The RPS21A/kanMX6 primers were 5′-GGCGTAAATTAACCACATACAAACCATAGAAACATCATACCAAAATGCGTACGCTGCAGGTCGAC-3′ and 5′-CTGAACGATATGAAGCTTAAATCGAATCCTCTTTTTCTTTTCTCTTAATCGATGAATTCGAGCTCG-3′. Underlined sequences in the oligonucleotides indicate sequences complementary to pFA6-MCS in the plasmid pFA6-kanMX6 (23). The PCR product was transformed into the diploid strain YMW1 and transformants were selected for resistance to G418. Transformants were sporulated and tetrads were dissected. Genomic DNA was isolated from haploid spores and PCR was used to identify cells where rps21A:: kanMX6 had replaced the chromosomal RSP21A gene (data not shown). Strains where the RPS21B gene was disrupted were derived from a homozygous diploid obtained as part of the Saccharomyces genome deletion project. Haploid spores with disrupted alleles of RPS21A and RPS21B were outcrossed to a W303 nuclear background. RPS21A/B alleles were genotyped using a PCR-based strategy using oligonucleotides to regions of the RPS21A/B genes that flanked the kanMX cassettes. The oligonucleotides used in genotyping RPS21A and RPS21B alleles were: RPS21A forward, 5′-CATAAGAGGATTTGATTTCTTTCCA-3′; RPS21A reverse, 5′-GCAAAACTTTAAACAGAATAGCCAA-3′; RPS21B forward, 5′-ATTACC TTTTGACGACAATGTGTTT-3′; RPS21B reverse, 5′-GTGCTATTTCAAGGATAACGAAAAA-3′.
Polysome analysis
Polysomes were prepared and fractionated on 7–47% sucrose gradients as described by Baim et al. (24). Centrifugation was generally carried out at 21 000 r.p.m. for 15 h in an SW28.1 rotor (Beckman Instruments, Fullerton, CA). Sucrose gradients were fractionated and absorbance at 254 nm was monitored using an ISCO model 185 gradient fractionator and a UA-5 absorbance detector. Data were digitized using UN-SCAN-IT software (Silk Scientific, Orem, UT).
Northern analysis
Total RNA was prepared from yeast cells by the hot phenol method (25). Cytoplasmic RNAs were prepared as described by Udem and Warner (26) with the exception that cells were not treated with cycloheximide to prevent polysome run-off. RNA was fractionated on 1.5% formaldehyde–agarose gels and transferred to a Zeta-probe (Bio-Rad, Hercules, CA) membrane. Membranes were hybridized with the following oligonucleotides: 1, 5′-CCAGATAACTATCTTAAAAG-3′; 2, 5′-TGATCCTTCCGCAGGTTCACCTACGGAAAC-3′; 3, 5′-GAAATCTCTCACCGTTTGGAATAGC-3′; 4, 5′-ATGAAAACTCCACAGTG-3′; 5, 5′-CCAGTTACGAAAATTCTTG-3′; 6, 5′-GGCCAGCAATTTCAAGTTA-3′. The regions of the rRNA repeat that hybridize with these oligonucleotides are shown in Figure 3. Membranes were also hybridized with an oligonucleotide complementary to the U3 snoRNA, 5′-GGATTGCGGACCAAGCTAA-3′. Oligonucleotide probes were labeled at their 5′ ends with [γ-32P]ATP (New England Nuclear, Boston, IL) using T4 polynucleotide kinase (New England Biolabs, Beverly, MA). Membranes were subjected to phosphoimage analysis (PhosphorImager SF; Molecular Dynamics, Sunnydale, CA).
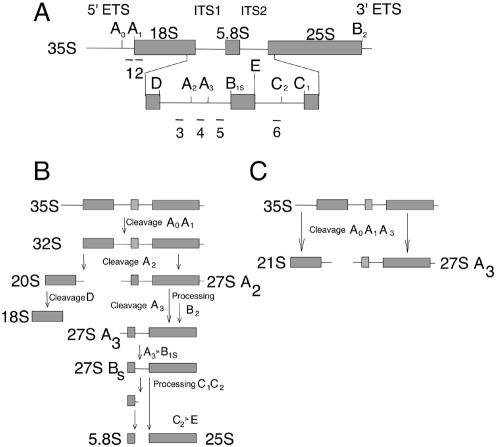
Figure 3.
The pre-rRNA processing pathway in S.cerevisiae (43). (A) Structure of the 35S rRNA precursor. Mature rRNA species are represented by boxes. External and internal transcribed sequences are shown as thin lines. Numbered letters indicate processing sites and lines numbered under the 35S precursor represent oligonucleotide probes described in Materials and Methods. (B) The major rRNA processing pathway in yeast. Horizontal arrows represent exonucleolytic cleavages; other cleavage steps are endonucleolytic. (C) Pathway used in strains defective in A2 processing.
RESULTS
The S.cerevisiae genome contains duplicated genes that encode members of the eukaryotic family of Rps21 proteins. The yeast Rps21A/B proteins are composed of 87 amino acids and have over 98% sequence identity. The yeast proteins have over 55% sequence identity with mammalian and Drosophila melanogaster Rps21 proteins (17). This protein family appears restricted to the eukaryotic lineage and is distinct from the ribosomal protein S21 family in eubacteria.
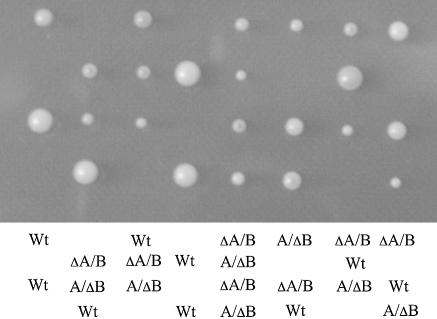
The RPS21 genes were disrupted either alone or together to determine if these genes are required for yeast cell growth and division. A diploid strain harboring heterozygous alleles RPS21A/rps21A::kanMX6 and RPS21B/rsp21B::kanMX4 was sporulated and tetrads dissected. Figure 1 shows that disruption of either RPS21 gene reduces the growth rate relative to the wild type, and that double mutants fail to germinate.
Figure 1.
Strains with disrupted alleles of RPS21A and RPS21B are not viable. A diploid strain heterozygous for RPS21A/rps21A::kanMX6 and RPS21B/rps21B::kanMX4 was sporulated, tetrads dissected, and the resulting haploid spores grown on YPD plates at 30°C. The relevant genotypes of viable strains are designated: Wt, RSP21A RPS21B; ΔA, rps21A::kanMX6 RPS21B; ΔB, RPS21A rps21B::kanMX4. Genotypes were determined by PCR analysis using oligonucleotides described in Materials and Methods.
The effects of the RPS21A/B disruptions are similar to phenotypes reported for strains harboring disrupted alleles of either RPS0A or RPS0B and other essential ribosomal proteins (1). In Drosophila, reduced expression of Rps21 and p40 (the Drosophila homolog of Rps0A/B) enhances the tumorous phenotype beyond that observed with a reduction in either protein alone (17). Therefore, we sought to determine whether the yeast RPS21 and RPS0 genes displayed genetic interactions. Diploid strains heterozygous for RPS21A/rps21A:: kanMX6 and RPS0A/rps0A::URA3 alleles or RPS21A/rps21A::kanMX6 and RPS0B/rps0B::HIS3 alleles were sporulated and tetrads dissected. Growth rates of haploid progeny revealed that the effects of the RPS0A and RPS0B disruptions on growth rate were generally more severe than that of the RPS21A disruption (Table 1). Table 1 also shows that combining the RPS21A disruption with either RPS0A or RPS0B disruptions did not reveal a stronger phenotype than the RPS0 disruptions alone, indicating that unlike the case in Drosophila, mutant alleles of RPS21 and RPS0 family members in yeast do not enhance one another phenotypically.
Table 1. Growth rate analysis of wild-type and single RPS0/RPS21A mutants relative to double mutants.
| Relevant genotypea | nb | Relative growth ratec | Standard deviationd | P-value versus double mutantse |
|---|---|---|---|---|
| RPS0A RPS0B RPS21A | 7 | 0.18 | 0.02 | 0.009; 0.001 |
| RPS0A RPS0B Δrps21A | 7 | 0.14 | 0.02 | 0.29; 0.008 |
| Δrps0A RPS0B RPS21A | 3 | 0.13 | 0.01 | 0.6; NDf |
| Δrps0A RPS0B Δrps21A | 3 | 0.13 | 0.03 | ND; ND |
| RPS0A Δrps0B RPS21A | 4 | 0.11 | 0.01 | ND; 0.36 |
| RPS0A Δrps0B Δrps21A | 4 | 0.11 | 0.02 | ND; ND |
aOnly relevant alleles of RPS0A, RPS0B and RPS21A are listed.
bNumber of independent strains examined.
cRelative growth rates are derived from the slopes of linear growth functions plotting lnA600 versus time. Values listed represent means of the data sets for a particular genotype.
dStandard deviations from the mean for each relative growth rate listed.
eP-values for comparisons between the genotype listed and either of the two double mutants. The first number listed is a comparison with Δrps0A RPS0B Δrps21A, the second number with RPS0A Δrps0B Δrps21A.
fNot determined.
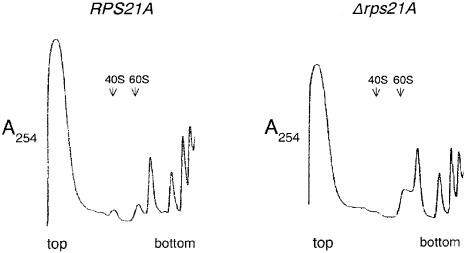
To determine if the decrease in growth rate in RPS21 mutants is due to a defect in some aspect of ribosome synthesis or function, we examined polysome profiles in these strains. Figure 2 shows that relative to the wild type, the strain containing the rps21A::kanMX6 allele has a reduced level of free 40S subunits, a reduction in the amount of large polysomes, and an increase in the amount of free 60S subunits. Similar results are observed for strains with a disrupted RPS21B allele (included in Figure 7). These data indicate that the decreased growth rates of strains containing disrupted alleles of RPS21A and RPS21B are linked to reductions in the amount of 40S ribosomal subunits.
Figure 2.
Haploid strains containing the rps21A::kanMX6 allele have reduced amounts of 40S subunits relative to wild type. Cell extracts were prepared and fractionated by sucrose gradient centrifugation as described by Baim et al. (24). Gradients were fractionated and absorbance at 254 nm was monitored using an ISCO model 185 density gradient fractionator and UA-5 absorbance detector. Data were digitized using UN-SCAN-IT software.
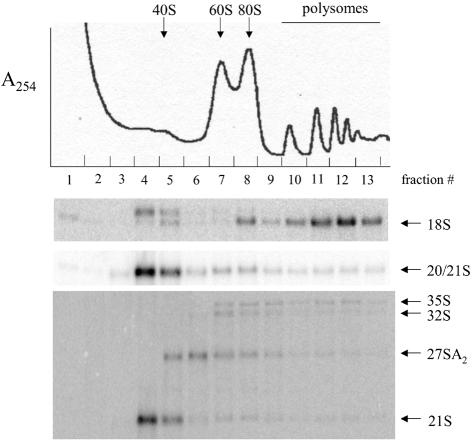
Figure 7.
Northern blot of sucrose gradient fractions from rps21B::kanMX4 cell extracts. Gradients were fractionated as in Figure 2. RNAs were isolated from sucrose gradient fractions, separated on 1.5% argarose– formaldehyde gels, and transferred to a Zeta-probe as described by Ford et al. (2). Membranes were hybridized with oligonucleotide 2 (top), oligonucleotide 3 (middle) and oligonucleotide 4 (bottom).
Previous studies have shown that the Rps0 proteins are required for maturation of 18S rRNA in the yeast rRNA processing pathway (2). To determine whether the Rps21 proteins are also required for maturation of 18S rRNA, northern analysis was performed on strains derived from a tetrad exhibiting a tetratype segregation pattern of heterozygous RPS21A/rps21A::kanMX6 and RPS0B/rps0B::HIS3 alleles. Total RNA from each strain was fractionated on formaldehyde–agarose gels, transferred to a solid support, and hybridized with oligonucleotides, described in Figure 3. Oligonucleotide 2, which hybridizes to mature 18S rRNA, shows a reduction in signal by 20–40% in RPS21A and RPS0B mutants relative to the wild type (Fig. 4A), consistent with the lower levels of 40S subunits observed in strains with disrupted alleles of RPS21A and RPS0B. A similar decrease in 18S rRNA is seen in the double mutant. Oligonucleotide 4, which hybridizes between the A2 and A3 cleavage sites within ITS1, shows a decreased signal for 27A2 rRNA in mutants relative to the wild type, and a corresponding increase in a species that is most likely 21S rRNA (Fig. 4B). 21S rRNA extends from the 5′ end of mature 18S rRNA to the A3 cleavage site (Fig. 3). This assignment was confirmed by showing that oligonucleotides upstream of the A1 cleavage site and downstream of A3 did not hybridize efficiently with this species (Fig. 4C and D). These data indicate that cleavage at the A2 site in ITS1 is affected by the RSP0B and RPS21A disruptions.
Figure 4.
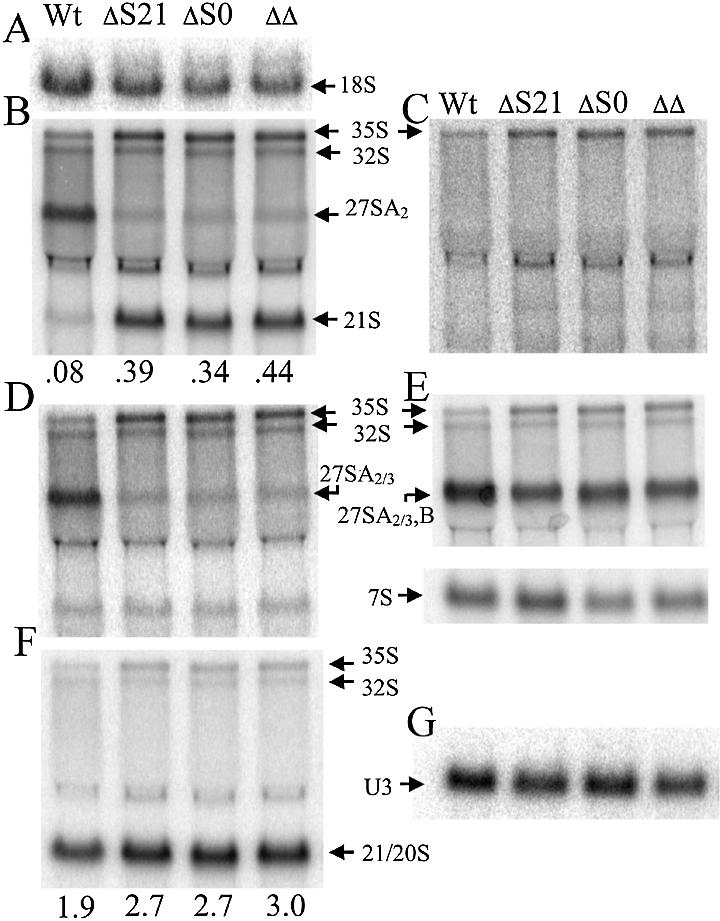
Northern blot analysis of rRNA processing in strains with mutant and wild-type alleles of RPS21A and RPS0B. Total RNA was prepared from strains derived from a tetrad exhibiting a tetratype segregation pattern after sporulation and dissection of a diploid strain heterozygous for RPS21A/rps21A::kanMX6 and RPS0B/rps0B::HIS3. RNA was fractionated by electrophoresis on a 1.5% agarose–formaldehyde gel, transferred to a Zeta-probe (Bio-Rad), and hybridized with oligonucleotides shown in Figure 3A, or to U3 snoRNA. (A) Oligonucleotide 2; (B) oligonucleotide 4; (C) oligonucleotide 1; (D) oligonucleotide 5; (E) oligonucleotide 6; (F) oligonucleotide 3; (G) oligonucleotide to U3 snoRNA. Relevant phenotypes listed above each lane are designated: Wt, RPS21A RPS0B; ΔS21, rps21A:: kanMX6 RPS0B; ΔS0, RPS21A rps0B::HIS3; ΔΔ, rps21A::kanMX6 rps0B:: HIS3. (B), (F) and (G) were subject to phosphorimage analysis and data collected on 35/32S, 21S, 21/20S and U3 snoRNA signals. The values for 21S and 21/20S signals in (B) and (F), respectively, were normalized through the 35/32S signals in their respective lanes. The values reported below each lane represent the 21S (B) and 21/20S (F) normalized for loading against the values obtained in (G) for U3 snoRNA.
In the absence of A2 cleavage, processing within ITS1 occurs at the A3 site generating 27SA3 rRNA, which is rapidly processed by the exosome to 27SBS (27). The rapid processing of 27SA3 by the exosome is responsible for the low steady-state levels of 27SA3 observed in both mutants and wild-type strains (Fig. 4D). Figure 4E shows a modest reduction in all 27S rRNA species in mutants relative to the wild type. Oligonucleotide 6, used in Figure 4E, hybridizes to sequences in ITS2. In mutant strains the predominant species recognized by this oligonucleotide is 27SBS, whereas in wild-type cells this oligonucleotide detects relatively high levels of both 27SA2 and 27SBS. The global decrease in 27S rRNAs in mutants is likely to be the result of observed coupling between A2 cleavage and cleavages at A1 and A0 sites within the 5′ external transcribed spacer (ETS). Reduced cleavages at each of these sites gives rise to the increase in 35S rRNA in mutants relative to the wild type (Fig. 4B–E). Thus, flux through the entire rRNA processing pathway is affected in the RPS0B and RPS21A mutants, resulting in lower levels of all 27S rRNA species.
Although flux through the entire rRNA processing pathway is affected in the RPS0B and RPS21A mutants, polysome profiles show a preferential reduction in 40S subunits relative to 60S subunits. This observation is routinely observed in mutants defective in A0, A1 and A2 cleavages, since 35S precursors cleaved directly at the A3 site can still give rise to 5.8S and 25S rRNA. 18S rRNA precursors generated by cleavage at A3, on the other hand, are inefficiently processed to 18S rRNA, accounting for the preferential reduction in 40S subunits in these mutants. Mutants defective in A0, A1 and A2 cleavages have decreased levels of both 20S and 18S rRNA precursors (28–30). 20S rRNA is the immediate precursor to 18S rRNA in the normal processing pathway (Fig. 3). The 20S precursor has a mature 5′ end and is extended through the mature 3′ end of 18S rRNA to the A2 cleavage site. Figure 4F shows that rather than a decrease in 20S rRNA as would be expected in strains defective in A0, A1 and A2 cleavages, there is an increase in 20S rRNA in the RPS0B and RPS21A mutants. These data indicate that in addition to affecting cleavage at A2, the RPS0B and RPS21A mutants may also affect cleavage at site D, which gives rise to the mature 3′ end of 18S rRNA.
Since the 20S and 21S rRNA species are not resolved under the conditions used in the experiments described above, and oligonucleotide 3 recognizes both 20S and 21S rRNAs, it is possible that increased signal in the 20/21S region observed in Figure 4F could be the result of an increase in 21S rRNA rather than 20S rRNA. If so, the data would indicate that it is A2 cleavage alone, rather than cleavage at D and A2, that is affected by the Rps0 and Rps21 proteins. To facilitate the comparison between 20/21S and 21S signals in Figure 4B and F, respectively, signals in each panel were normalized through the 32/35S species, which hybridize with both oligonucleotides 3 and 4. The normalized data derived after phosphorimage analysis is shown below each lane in Figure 4B and F. It is apparent from this analysis that the 20/21S signals in Figure 4F are considerably greater than the 21S signals in Figure 4B. This difference is a reflection of the relative rates of A2 versus A3 cleavage of the 32S rRNA under normal conditions, which gives rise to high steady-state levels of 20S relative to 21S intermediates in the rRNA processing pathway (31). Further analysis reveals that signals derived from Figure 4B corresponding to 21S rRNA alone could account for at most 30–40% of increase in the combined 20S/21S hybridization signal observed in mutants (lanes 2–4) relative to the wild type (lane 1) in Figure 4F.
Support for the conclusion that mutations in the RPS0 and RPS21 genes increase both 20S and 21S rRNA levels comes from the observation that mutations in another ribosomal protein gene, RPS18B, increase 21S rRNA without increasing 20S rRNA. A diploid strain heterozygous at the RPS18B and RPS21B loci was created to compare rRNA processing in RPS18B mutants with a representative mutant where both 20S and 21S rRNAs increase. This diploid was sporulated and the resulting tetrads dissected. One of the tetrads exhibiting a tetratype segregation pattern was used for northern analysis. Figure 5A shows that the 21S rRNA precursor is increased in strains harboring disrupted alleles of either RPS18B or RPS21B. In contrast, the increase in 20S/21S hybridization signal observed in strains with the disrupted RPS21B allele is not observed in the strain containing the RPS18B disruption alone (Fig. 5B). Similar results were observed for the RPS18A gene (data not shown). These data indicate that in contrast to results obtained for the RPS0 and RPS21 genes where cleavages at both sites A2 and D are affected, disruption of RPS18B affects cleavage at the A2 site with little or no influence on processing at site D.
Figure 5.
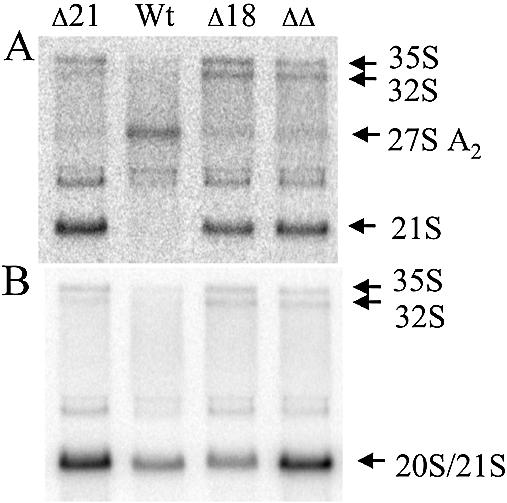
Northern blot analysis of rRNA processing in strains with mutant and wild-type alleles of RPS18B and RPS21B. Total RNA was prepared from strains derived from a tetrad exhibiting a tetratype segregation pattern after sporulation and dissection of a diploid strain heterozygous for RPS18B/rps18B::kanMX4 and RPS21B/rps21B::kanMX4. RNA was fractionated by electrophoresis on a 1.5% agarose–formaldehyde gel, transferred to a Zeta-probe (Bio-Rad), and hybridized with oligonucleotides shown in Figure 3A. (A) Oligonucleotide 4; (B) oligonucleotide 3.
Cleavage at site D is the only rRNA processing step that occurs in the cytoplasm (26). Therefore, the increase in 20S rRNA observed in RPS0 and RPS21 mutants could be the consequence of inefficient transport of 40S subunit precursors from the nucleolus to the cytoplasm, or it could represent a direct effect on D site cleavage. To distinguish between these possibilities, we compared the distribution of rRNA precursors in total and cytoplasmic extracts of wild-type and mutant strains. Cytoplasmic extracts with minimal nuclear contamination were prepared using the procedure of Udem and Warner (26). Figure 6A shows that, as expected, mature 18S rRNA is found in both total and cytoplasmic extracts from wild-type and mutant strains. In contrast, 21S rRNA is under- represented in cytoplasmic extracts relative to 18S rRNA, consistent with a predominantly nucleolar localization (Fig. 6B). The signal in the 20S/21S region in Figure 6A and C that is found in roughly equal proportions to 18S rRNA in total and cytoplasmic RNAs is therefore primarily derived from the 20S rRNA precursor. The 20S to 18S ratio is greater in the mutant strains relative to the wild type in both total and cytoplasmic extracts (Fig. 6). The decrease in this ratio in cytoplasmic relative to total RNA extracts reflects the additional contribution of the predominantly nucleolar 21S rRNA to the total RNA. These data demonstrate that the bulk of the 20S rRNA in mutant and wild-type strains is localized to the cytoplasm and as such, the increased 20S rRNA observed in the RPS0B and RPS21A mutants is a result of inefficient cleavage at site D as opposed to a block in nucleolus to cytoplasm transport.
Figure 6.
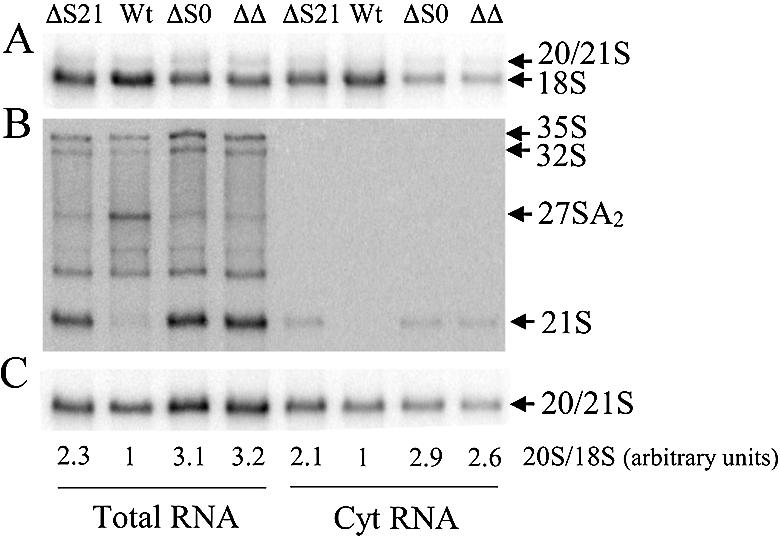
Northern blot analysis of rRNA species present in total and cytoplasmic extracts from strains with mutant and wild-type alleles of RPS21A and RPS0B. Total RNA was prepared using procedures and strains described in Figure 4. Cytoplasmic RNAs were prepared as described in Materials and Methods. RNA was fractionated by electrophoresis on a 1.5% agarose– formaldehyde gel, transferred to a Zeta-probe (Bio-Rad), and hybridized with oligonucleotides shown in Figure 3A. (A) Oligonucleotide 2; (B) oligonucleotide 4; (C) oligonucleotide 3. Relevant phenotypes listed above each lane are designated: wt, RPS21A RPS0B; ΔS21, rps21A::kanMX6 RPS0B; ΔS0, RPS21A rps0B::HIS3; ΔΔ, rps21A::kanMX6 rps0B::HIS3. 18S and 20/21S signals in (A) and (C), respectively, were subjected to phosphorimage analysis. The values below each lane represent an average of 20S/18S ratios from two independent experiments normalized against the wild-type value for either total or cytoplasmic extracts.
Previous results have shown that the 20S rRNA that accumulates in RPS0-disrupted strains is under-represented in polysomes relative to 18S rRNA, indicating 40S precursors containing 20S rRNA have reduced or no translational activity compared with mature 40S subunits (2). To address the translational activity of 40S subunit precursors that accumulate in RPS21-disrupted strains, extracts from an RPS21B-disrupted strain were fractionated on sucrose gradients and analyzed by northern blotting. Hybridization of RNAs from gradient fractions with oligonucleotide 2 shows that 18S rRNA is found in 40S, 80S and polysome fractions with the bulk of the 18S rRNA signal in polysomes (Fig. 7, top). Also evident in Figure 7, top, is a signal just above 18S rRNA that is most abundant in fraction 4 on the leading edge of the 40S subunit peak. This signal is derived from immature 40S subunits containing 18S rRNA precursors. Hybridization with oligonucleotide 3 shows that the signal for 20S/21S rRNAs is most abundant in fractions 4 and 5 with relatively low amounts in 80S and polysome fractions (Fig. 7, middle). Figure 7, bottom, shows that a small amount of the 20S/21S signal in Figure 7, middle, is derived from 21S rRNA, with the bulk of this signal in Figure 7, middle, representing 20S rRNA. Whether the small amount of 21S rRNA-containing precursor is in the cytoplasm or instead, localized to the nucleolus, is unknown since other nucleolar precursors 27SA2, 32S and 35S rRNA precursors are also detected in the gradient. The fact that 20S and 21S rRNAs are under-represented in the 80S peak and polysomes relative to 18S rRNA indicates that immature subunits containing these rRNA precursors have little or no translational activity. Thus, the Rps21 proteins play a role similar to the Rps0 proteins in defining the level of active 40S subunits within yeast cells.
DISCUSSION
The results reported here indicate that the yeast Rps0 and Rps21 proteins have overlapping functions in the maturation of the 3′ end of 18S rRNA. The effects of RPS21 disruptions on the protein synthetic apparatus in yeast are similar to those reported previously for the RPS0 genes (1). In each case, the amount of 40S subunits decreases, larger polysomes are reduced, and the level of free 60S subunits increases. The reduced amounts of 40S subunits in strains disrupted in either of the RPS0 or RPS21 genes result from defects in rRNA processing and 40S subunit maturation. Specifically, these mutant strains are defective in processing of rRNA precursors at the A2 and D cleavage sites involved in forming the mature 3′ end of 18S rRNA.
The Rps0 and Rps21 proteins could exert their effects on processing the 3′ end of 18S rRNA by direct interactions with rRNA precursors that facilitate cleavage at the D and A2 sites in ITS1, or through more general effects on small subunit structure. The observation that disruption of another small subunit ribosomal protein gene, RPS18B, affects processing at the A2 site without affecting cleavage at site D indicates that the influence of small subunit proteins on cleavages at these sites should be considered separately.
Analysis of structure of small ribosomal subunits suggests that the influence of the Rps0 and Rps21 proteins on cleavage at site D may be more direct, whereas their influence on cleavage at site A2 may have more to do with more general aspects of subunit structure. Cryo-EM structures of the yeast 40S subunit place the Rps0 proteins on the solvent surface near the mRNA exit channel (32). This region contains the 3′ end of the 18S rRNA. However, these are relatively low-resolution structures that do not provide detail regarding the relationship between the Rps0 proteins and the 3′ end of 18S rRNA. The structure of the Thermus thermophilus 30S ribosomal subunit, on the other hand, has been solved at 3 Å resolution (33). This subunit contains ribosomal protein S2, the bacterial homolog of the Rps0 proteins. S2 is located on the solvent surface of the bacterial 30S subunit, at the base of head, near the bottom of the cleft in a position analogous to that of the Rps0 proteins on the yeast 40S subunit (Fig. 8). The 3′ end of 16S rRNA enters the cleft from the subunit interface in a 5′–3′ orientation, and terminates within the cleft before exiting to the solvent surface (33,34). The bacterial S2 protein does not appear to be close enough to make contact with the 3′ end of mature 16S rRNA, but is positioned in such a manner that it could make contact with 3′ extended precursors. If a similar relationship were to occur in the yeast 40S subunit, the Rps0 proteins would be in a position to interact with sequences in ITS1 and promote cleavage at the D site.
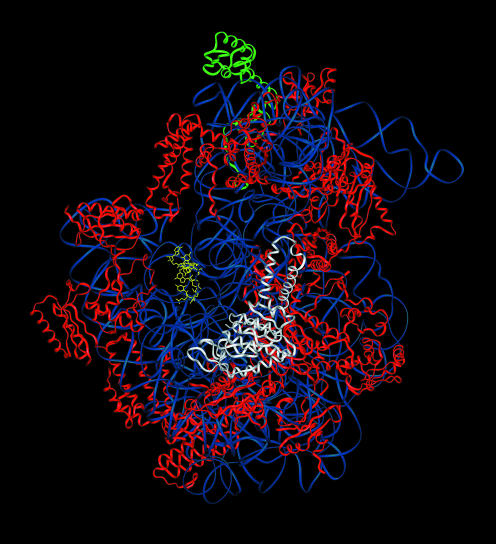
Figure 8.
The structure and location of ribosomal protein S2 relative to the 3′ end of 16S rRNA in T.thermophilus 30S subunits. The coordinates of T.thermophilus 30S subunits are those of Wimberly et al. (33) obtained from the Brookhaven Protein Database (accession no. 1FJF). The bulk of the 16S rRNA backbone structure is shown in blue with the 3′ end shown in yellow. A ribbon diagram of the S2 protein is shown in white and the S13 protein in green. Other ribosomal proteins are shown in red.
The Rps18B protein, on the other hand, is homologous to ribosomal protein S13, which is located on the subunit interface of the T.thermophilus 30S subunit at the top of the head (Fig. 8). This location places S13 on the surface of the small subunit opposite that of the 3′ end of small subunit rRNA. Thus, the effect of Rps18B on cleavage at site A2 is likely to be indirect and related to some more general aspect of small subunit structure required for efficient processing at this site within the nucleolus. The effect of the Rps0 and Rps21 proteins on cleavage at A2 site indicates that they too influence aspects of small subunit structure within the nucleolus that are necessary for efficient processing at this site.
In contrast to the Rps0 and Rps18 proteins, the Rps21 proteins have not been localized on the yeast 40S subunit, nor do they have a homolog on the bacterial 30S subunit from which their location could be inferred. It is known, however, that human and Drosophila Rps21 proteins physically interact with homologs of the Rps0 proteins (17,20). This observation suggests that the Rps0 and Rps21 proteins may make contact with one another on the 40S subunit. In this context, cryo-EM models of the yeast 40S subunit have a region of electron density just above the Rps0 proteins that is not accounted for in homology models with the bacterial 30S subunit (32). This electron density may be derived from the Rps21 proteins. In the bacterial structure, this region contains the H35–H37 helices of the 3′ major domain of 16S rRNA that interact with an extended coiled-coil motif in the S2 protein. The S2 protein makes relatively few contacts with 16S rRNA, and of these, the strongest interactions are formed between the coiled-coil domain and the H35–H37 helices (35). Intriguingly, the coiled-coil structure present in bacterial S2 proteins is absent in eukaryotic members of this family (36). The absence of this domain would presumably make the eukaryotic S2-like proteins more dependent on protein–protein interactions for their incorporation into 40S subunits. If so, an Rps0/Rps21 interaction could potentially compensate for the protein–RNA interaction lost by the absence of the coiled-coil domain in Rps0 proteins. One explanation for the similar effects of RPS0 and RPS21 deletions on rRNA processing is therefore that the Rps21 proteins are involved in the binding of Rps0 proteins to the 40S ribosomal subunit.
How a severe reduction in the amounts of p40/Rps0 and Rps21 proteins leads to a loss of control of cellular proliferation in Drosophila is unknown but may be relevant to human disease since the human p40 gene is found on the short arm of chromosome 3 in a region frequently subject to loss of heterozygosity in tumors (37). Recent results have provided intriguing links between factors involved in ribosome synthesis and cancer susceptibility syndromes (38). Dyskeratosis congenita is caused by mutations in DSK1, which encodes a pseudouracil synthase required for rRNA processing (39). Similarly, cartilage hair hypoplasia has been shown to result from mutations in the RNA subunit of RNase MRP, an endonuclease involved in rRNA processing (40). Finally, Diamond-Blackfan anemia, which has an associated increase in cancer incidence, has been linked to mutations in the gene encoding ribosomal protein S19 (41,42). Whether the link between disruption of ribosome synthesis and cancer susceptibility is general and potentially includes the hundreds of factors involved in ribosome assembly, processing, and transport or is instead restricted to particular aspects of the overall process of ribosome synthesis and function remains an open question.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by an intramural research incentive grant from the Vice President for Research at the University of Louisville, the University of Louisville Medical School Research Committee and the Kentucky Lung Cancer Research program.
REFERENCES
- 1.Demianova M., Formosa,T.G. and Ellis,S.R. (1996) Yeast proteins related to the p40/laminin receptor precursor are essential components of the 40S ribosomal subunit. J. Biol. Chem., 271, 11383–11391. [DOI] [PubMed] [Google Scholar]
- 2.Ford C.L., Randal-Whitis,L. and Ellis,S.R. (1999) Yeast proteins related to the p40/laminin receptor precursor are required for 20S rRNA processing and the maturation of 40S ribosomal subunits. Cancer Res., 59, 704–710. [PubMed] [Google Scholar]
- 3.Tabb A.L., Utsugi,T., Wooten-Kee,C.R., Sasaki,T., Edling,S.A., Gump,W., Kikuchi,Y. and Ellis,S.R. (2001) Genes encoding ribosomal proteins Rps0A/B of Saccharomyces cerevisiae interact with TOM1 mutants defective in ribosome synthesis. Genetics, 157, 1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auth D. and Brawerman,G. (1992) A 33-kDa polypeptide with homology to the laminin receptor: component of the translational machinery. Proc. Natl Acad. Sci. USA, 89, 4368–4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis S.C., Tzagoloff,A. and Ellis,S.R. (1992) Characterization of a yeast mitochondrial ribosomal protein structurally related to the mammalian 68-kDa high affinity laminin receptor. J. Biol. Chem., 267, 5508–5514. [PubMed] [Google Scholar]
- 6.Ouzonis C., Kyrpides,N. and Sander,C. (1995) Novel protein families in archaean genomes. Nucleic Acids Res., 23, 565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mafune K., Ravikumar,T.S., Wong,J.M., Yow,H., Chen,L.B. and Steele,G.D. (1990) Expression of a MR 32,000 laminin-binding protein messenger RNA in human carcinoma correlates with disease progression. Cancer Res., 50, 3888–3891. [PubMed] [Google Scholar]
- 8.D'Errico A., Garbisa,S., Liotta,L.A., Castronovo,V., Stetler-Stevenson,W.G. and Grigioni,W.F. (1991) Augmentation of type IV collagenase, laminin receptor and ki67 proliferation antigen associated with human colon, gastric and breast carcinoma progression. Mod. Pathol., 4, 239–246. [PubMed] [Google Scholar]
- 9.Castronovo V. (1993) Laminin receptors and laminin binding proteins during tumor invasion and metastasis. Invasion Metastasis, 13, 1–30. [PubMed] [Google Scholar]
- 10.Vacca A., Ribatti,D., Roncali,L., Lospalluti,M., Serio,G., Carrel,S. and Dammacco,F. (1993) Melanocyte tumor progression is associated with changes in angiogenesis and expression of the 67-kilodalton laminin receptor. Cancer, 72, 455–461. [DOI] [PubMed] [Google Scholar]
- 11.Pei D.P., Han,Y., Narayan,D., Herz,D. and Ravikumar,T.S. (1996) Expression of 32-kDa laminin-binding protein mRNA in colon cancer tissues. J. Surg. Res., 61, 120–126. [DOI] [PubMed] [Google Scholar]
- 12.Halatsch M.E., Hirsch-Ernst,K.I., Kahl.,G.F. and Weinel,R.J. (1997) Increased expression of alpha6-integrin receptors and of mRNA encoding the putative 37 kDa laminin receptor precursor in pancreatic carcinoma. Cancer Lett., 118, 7–11. [DOI] [PubMed] [Google Scholar]
- 13.Daheron L., Salmeron,S., Patri,S., Brizard,A., Guilhot,F., Chomel,J.C. and Kitzis,A. (1998) Identification of several genes differentially expressed during progression of chronic myelogenous leukemia. Leukemia, 12, 326–332. [DOI] [PubMed] [Google Scholar]
- 14.Verlaet M., Deregowski,V., Denis,G., Humblet,C., Stalmans,M.T., Bours,V., Castronovo,V., Boniver,J. and Defresne,M.P. (2001) Genetic imbalances in preleukemic thymuses. Biochem. Biophys. Res. Commun., 283, 12–18. [DOI] [PubMed] [Google Scholar]
- 15.Liotta L. (1986) Tumor invasion and metastasis—role of extracellular matrix. Rhoads Memorial Lecture. Cancer Res., 50, 1479–1483. [PubMed] [Google Scholar]
- 16.Tabb A.L. and Ellis,S.R. (2001) A new perspective on the role of the 37-kDa laminin binding protein in cancer. Recent Res. Dev. Cancer, 3, 257–266. [Google Scholar]
- 17.Torok I., Herrmann-Horle,D., Kiss,I., Tick,G., Speer,G., Schmitt,R. and Mechler,B.M. (1999) Down-regulation of Rps21, a putative translation initiation factor interacting with p40, produces viable minute imagos and larval lethality with overgrown hematopoietic organs and imaginal discs. Mol. Cell. Biol., 19, 2308–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher E.M.C., Beer-Romero,P., Brown,L.G., Ridley,A., McNeil,J.A., Lawrence,J.B., Willard,H.F., Bieber,F.R. and Page,D.C. (1990) Homologous ribosomal protein genes on the X and Y chromosomes: escape from X inactivation and possible implications for Turner syndrome. Cell, 63, 1205–1218. [DOI] [PubMed] [Google Scholar]
- 19.Melnick M.B., Noll,E. and Perrimon,N. (1993) The Drosophila stubarista phenotype is associated with a dosage effect of the putative ribosome-associated protein D-p40 on spineless. Genetics, 135, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato M., Saeki,Y., Tanaka,K. and Kaneda,Y. (1999) Ribosome-associated protein LBP/p40 binds to S21 protein of the 40S ribosome: analysis using a yeast two-hybrid system. Biochem. Biophys. Res. Commun., 256, 385–390. [DOI] [PubMed] [Google Scholar]
- 21.Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 1–21. [DOI] [PubMed] [Google Scholar]
- 22.Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 23.Wach A. (1996) PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast, 12, 259–265. [DOI] [PubMed] [Google Scholar]
- 24.Baim S.B., Pietras,D.F., Eustice,D.C. and Sherman,F. (1985) A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol. Cell. Biol., 5, 1830–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udem S.A. and Warner,J.R. (1973) The cytoplasmic maturation of a ribosomal ribonucleic acid in yeast. J. Biol. Chem., 248, 1412–1416. [PubMed] [Google Scholar]
- 27.Henry Y., Wood,H., Morrissey,J.P., Petfalski,E., Kearsey,S. and Tollervey,D. (1994) The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J., 13, 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee W.C., Zabetakis,D. and Melese,T. (1992) NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol. Cell. Biol., 12, 3865–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunbar D.A., Wormsley,S., Agentis,T.M. and Baserga,S.J. (1997) Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol., 17, 5803–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venema J., Bousquet-Antonelli,C., Gelugne,J.-P., Caizergues-Ferrer,M. and Tollervey,D. (1997) Rok1p is a putative RNA helicase required for rRNA processing. Mol. Cell. Biol., 17, 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tollervey D. (1987) A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J., 6, 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spahn C.M., Beckmann,R., Enswar,N., Penczek,P.A., Sali,A., Blobel,G. and Frank,J. (2001) Structure of the 80S ribosome from Saccharomyces cerevisiae tRNA–ribosome and subunit–subunit interactions. Cell, 107, 373–386. [DOI] [PubMed] [Google Scholar]
- 33.Wimberly B.T., Brodersen,D.E., Clemons,W.M., Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrisnan,V. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 34.Yusupova G.Z., Yusupov,M.M. Cate,J.H.D. and Noller,H.F. (2001) The path of messenger RNA through the ribosome. Cell, 106, 233–241. [DOI] [PubMed] [Google Scholar]
- 35.Brodersen D.E., Clemons,W.M.,Jr, Carter,A.P., Wimberly,B.T. and Ramakrishnan,V. (2002) Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16S RNA. J. Mol. Biol., 316, 725–768. [DOI] [PubMed] [Google Scholar]
- 36.Kazmin D.A., Chinenov,Y., Larson,E. and Starkey,J.R. (2003) Comparative modeling of the N-terminal domain of the 67kDa laminin-binding protein: implications for putative ribosomal function. Biochem. Biophys. Res. Commun., 300, 161–166. [DOI] [PubMed] [Google Scholar]
- 37.Protopopov A., Kashuba,V., Zabarovska,V.I., Muravenko,O.V., Lerman,M.I., Klein,G. and Zabarovsky,E.R. (2003) An integrated physical map of the 3.5-Mb chromosome 3p21.3 (AP20) region implicated in major human epithelial malignancies. Cancer Res., 63, 404–412. [PubMed] [Google Scholar]
- 38.Ruggero D. and Pandolfi,P.P. (2003) Does the ribosome translate cancer? Nature Rev., 3, 178–192. [DOI] [PubMed] [Google Scholar]
- 39.Ruggero D., Grisendi,S., Piazza,F., Rego,E., Mari,F., Rao,P.H., Cordon-Cardo,C. and Pandolfi,P.P. (2003) Dyskeratosis congenita and cancer in mice deficient in rRNA processing. Science, 299, 259–262. [DOI] [PubMed] [Google Scholar]
- 40.Ridanpaa M., van Eenennaam,H., Pelin,K., Chadwick,R., Johnson,C., Yuan,B., van Venrooij,W., Pruijn,G., Salmela,R., Rockas,S., Makitie,O., Kaitila,I. and de la Chapelle,A. (2001) Mutations in the RNA component of RNAse MRP cuase a pleiotropic human disease, cartilage hair hypoplasia. Cell, 104, 195–203. [DOI] [PubMed] [Google Scholar]
- 41.Draptchinskaia N., Gustavsson,P., Andersson,B., Pettersson,M., Willig,T.N., Dianzani,I., Ball,S., Tchernia,G., Klar,J., Matsson,H., Tentler,D., Mohandas,N., Carlsson,B. and Dahl,N. (1999) The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anemia. Nature Genet., 21, 169–175. [DOI] [PubMed] [Google Scholar]
- 42.Matsson H., Klar,J. Draptchinskaia,N., Gustavsson,P., Carlsson,B., Bowers,D., de Bont,E. and Dahl,N. (1999) Truncating ribosomal protein S19 mutations and variable clinical expression in Diamond-Blackfan anemia. Hum. Genet., 105, 496–500. [DOI] [PubMed] [Google Scholar]
- 43.Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]