Abstract
Comparative analysis of genes, operons and regulatory elements was applied to the lysine biosynthetic pathway in available bacterial genomes. We report identification of a lysine-specific RNA element, named the LYS element, in the regulatory regions of bacterial genes involved in biosynthesis and transport of lysine. Similarly to the previously described RNA regulatory elements for three vitamins (riboflavin, thiamin and cobalamin), purine and methionine regulons, this regulatory RNA structure is highly conserved on the sequence and structural levels. The LYS element includes regions of lysine-constitutive mutations previously identified in Escherichia coli and Bacillus subtilis. A possible mechanism of the lysine-specific riboswitch is similar to the previously defined mechanisms for the other metabolite-specific riboswitches and involves either transcriptional or translational attenuation in various groups of bacteria. Identification of LYS elements in Gram-negative γ-proteobacteria, Gram-positive bacteria from the Bacillus/Clostridium group, and Thermotogales resulted in description of the previously uncharacterized lysine regulon in these bacterial species. Positional analysis of LYS elements led to identification of a number of new candidate lysine transporters, namely LysW, YvsH and LysXY. Finally, the most likely candidates for genes of lysine biosynthesis missing in Gram- positive bacteria were identified using the genome context analysis.
INTRODUCTION
Amino acid lysine is produced from aspartate through the diaminopimelate (DAP) pathway in most bacteria and higher plants (1). In fungi, in thermophilic bacterium Thermus thermophilus, and in several archaeal species, lysine is synthesized by a completely different pathway called the α-aminoadipate pathway (2). In bacteria, DAP is not only a direct precursor of lysine, but it is also an important constituent of the cell wall peptidoglycan (3). The DAP pathway is of special interest for pharmacology, since the absence of DAP in mammalian cells allows for the use of the DAP biosynthetic genes as a bacteria-specific drug target (4).
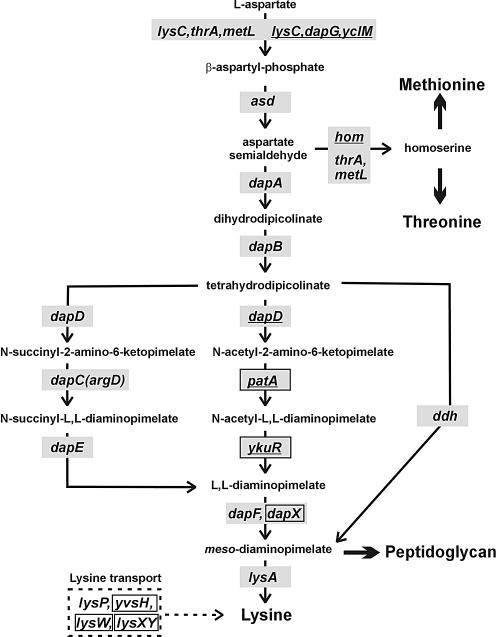
The first two stages of the DAP pathway catalyzed by aspartokinase and aspartate semialdehyde dehydrogenase are common for the biosynthesis of amino acids of the aspartate family, namely lysine, threonine and methionine (Fig. 1). Escherichia coli has two bifunctional aspartokinase/homoserine dehydrogenases, ThrA and MetL, and one monofunctional aspartokinase LysC, which are involved in the threonine, methionine and lysine synthesis, respectively. Transcription of the aspartokinase genes in E.coli is regulated by concentrations of the corresponding amino acids. In addition, ThrA and LysC are feedback inhibited by threonine and lysine, respectively (1). In contrast, Bacillus subtilis has three monofunctional aspartokinases: DAP-inhibited DapG; lysine-inhibited LysC; and aspartokinase-III, that is inhibited by simultaneous addition of threonine and lysine and encoded by the yclM gene (5,6). Single aspartate semialdehyde dehydrogenase serves for the synthesis of the amino acids of the aspartate family both in E.coli and B.subtilis. The next intermediate of the lysine pathway, tetrahydrodipicolinate, is synthesized from aspartate semialdehyde by the products of the dapA and dapB genes in both species (Fig. 1). However, three different types of the meso-DAP synthesis from tetrahydrodipicolinate are known in bacteria. Escherichia coli realizes this synthesis in four steps and uses succinylated DAP derivatives, whereas B.subtilis uses acetylated derivatives for the DAP synthesis (7). The third pathway, observed in Bacillus sphaericus, mediates the same conversion in one step and uses a unique DAP dehydrogenase. Finally, DAP decarboxylase LysA mediates the last step of the lysine synthesis and is common for all studied bacterial species. Escherichia coli is able to uptake lysine by two distinct transport systems, one of which is specific for lysine and the other is inhibited by arginine or ornithine (8). No other lysine transporters were identified in bacteria.
Figure 1.
The DAP pathway of the lysine biosynthesis and lysine transport in bacteria. The B.subtilis gene names are underlined. Genes predicted in this study are boxed.
Although most genes of the lysine synthesis in E.coli are repressed by lysine, little is known about the mechanisms of their regulation. In E.coli, the lysA gene is positively regulated by a transcriptional regulator encoded by the divergently transcribed gene lysR (9). Mutations in the leader regions of the lysC genes release the lysine repression both in E.coli and B.subtilis (10,11). In spite of divergence between these species, the comparison of the lysC leader sequences showed a number of highly conserved regions, and lysine-constitutive mutations were mapped to these regions (11). Based on the identification of a short transcript corresponding to the B.subtilis lysC leader region, it has been proposed that lysine exerts its control by inducing premature transcription termination by an unknown mechanism (12).
Recent studies with vitamin-specific regulation in bacteria revealed a number of metabolite-binding RNA domains which are involved in transcriptional and translational regulation of key genes of the vitamin biosynthetic pathways (13–16). In addition, the purine- and methionine-specific riboswitches (the G-box and S-box, respectively) were recently characterized in B.subtilis (17–19). Moreover, a similar mechanism of regulation was suggested for the lysine biosynthetic gene lysC in B.subtilis (19).
Current availability of many complete genomes gives an opportunity to compare genes encoding one metabolic pathway and their regulation in a variety of bacteria. The comparative analysis is a powerful approach for prediction of the conserved RNA secondary structures and detection of novel regulatory RNA elements upstream of co-regulated genes in bacterial genomes (20). In particular, highly conserved RFN, THI and B12 elements were identified in various bacteria upstream of genes involved in the biosynthesis of riboflavin, thiamin and cobalamin, respectively (21–24). In such studies, analysis of complementary substitutions in aligned sequences is used to construct a single conserved structure of an RNA regulatory element.
Here we applied the comparative genomics techniques to the analysis of bacterial lysine metabolism and transport genes. Identification of an RNA element in the regulatory regions of lysine biosynthetic and transport genes in bacteria allowed us to describe the previously uncharacterized lysine regulon. We named this new regulatory RNA structure the LYS element. It is highly conserved on the sequence and structural levels and includes known regions of lysine-constitutive mutations from E.coli and B.subtilis. By analogy to the previously described metabolite-specific RNA elements, we propose a possible mechanism of the lysine regulation mediated by the LYS element. Using a combination of the analysis of regulatory elements and the genome context analysis, several new LYS-element-regulated transporters, which are possibly specific for lysine, were predicted in various bacteria. We also identified a number of new candidate enzymes from the lysine biosynthetic pathway.
MATERIALS AND METHODS
Complete and partial sequences of eubacterial genomes were downloaded from GenBank (25). Preliminary sequence data were obtained also from the WWW sites of The Institute for Genomic Research (http://www.tigr.org), University of Oklahoma’s Advanced Center for Genome Technology (http://www.genome.ou.edu), the Sanger Centre (http://www.sanger.ac.uk), the DOE Joint Genome Institute (http://www.jgi.doe.gov) and the ERGO Database, Integrated Genomics, Inc (26).
The conserved secondary structure of the LYS element was derived using the RNAMultAln program (A.Mironov, unpublished results). This program simultaneously creates a multiple alignment and a conserved secondary structure for a set of RNA sequences. The RNA-PATTERN program (27) was used to search for new LYS elements in bacterial genomes. The input RNA pattern described the RNA secondary structure and the sequence consensus motifs. The RNA secondary structure was described as a set of the following parameters: the number of helices, the length of each helix, the loop lengths and the description of the topology of helix pairs. Additional RNA secondary structures, in particular anti-terminators and anti-sequestors, were predicted using Zuker’s algorithm of free energy minimization (28) implemented in the Mfold program (http://bioinfo.math.rpi.edu/~mfold/rna).
Protein similarity search was done using the Smith–Waterman algorithm implemented in the GenomeExplorer program (29). Orthologous proteins were initially defined by the best bidirectional hit criterion (30) and if necessary confirmed by construction of phylogenetic trees for the corresponding protein families. The phylogenetic trees of the LYS elements and lysine biosynthesis and transport proteins were constructed by the maximum likelihood method implemented in PHYLIP (31). The regulatory ACT domains of aspartokinases, which are specific for amino acid biosynthetic pathways, were not used for construction of the phylogenetic tree. Multiple sequence alignments were done using CLUSTALX (32). Transmembrane segments were predicted using TMpred (http://www.ch.embnet.org/software/TMPREDform.html).
RESULTS
Conserved structure of the LYS element
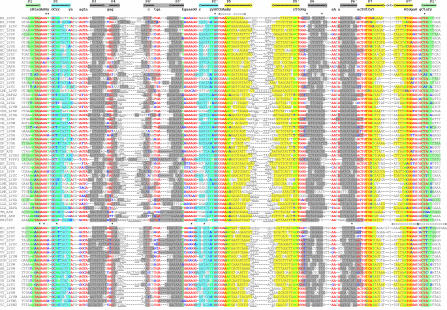
The mRNAs of the lysC genes for lysine-inhibited aspartokinases have extensive 5′-untranslated leaders in E.coli and B.subtilis, containing a stretch of highly conserved regions. In both species, mutations leading to lysine-constitutive expression of the lysC genes have been localized within these conserved regions (10,11) and existence of a riboswitch in this region has been proposed (19). Thus we started with identification of the lysC orthologs in related bacteria. The upstream regions of the lysC orthologs were aligned by the RNAMultAln program and a conserved RNA secondary structure was identified. This RNA structure, named the LYS element, consists of a number of helices and conserved sequence motifs and includes the lysC regulatory regions previously detected in E.coli and B.subtilis. Then, we constructed the pattern of the LYS element and scanned available genomic sequences using the RNA-PATTERN program. As a result, we found 71 LYS elements in 37 bacterial genomes. Multiple alignment of these elements is shown in Figure 2. The LYS element is widely distributed in Gram-positive bacteria from the Bacillus/Clostridium group and Gram-negative γ-proteobacteria, but it has not been observed in most other taxonomic groups of Eubacteria, the exception being two Thermotogales and Fusobacterium nucleatum.
Figure 2.
Alignment of LYS-element sequences. The first column contains the genome abbreviations (listed in Table 1) and gene names. The complementary stems of the RNA secondary structure are shown by arrows in the upper line. Base-paired positions are highlighted in matching colors. Conserved positions are set in red; degenerate conserved positions in green; non-conserved positions in black; non-consensus nucleotides in conserved positions in blue. The lengths of the additional stem–loops are given. Asterisk and delta/D symbols indicate operator consitutive mutations and deletions/duplication in the lysC genes of E.coli and B.subtilis (10,11).
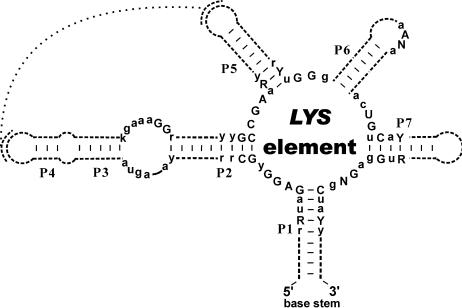
Similarly to the RFN, THI and B12 elements, the LYS element has a set of helices closed by a single base stem, and regions of high sequence conservation that are distributed along the entire element (Fig. 3). The conserved core of the LYS element consists of seven helices (P1 to P7) and single-stranded regions. Existence of the conserved helices is confirmed by compensatory substitutions in base-paired positions. Numerous highly conserved sequence boxes are distributed along the LYS element, mostly concentrated within internal loop regions. We have observed that among conserved nucleotides of the internal loop regions of the LYS element, purines are prevalent over pyrimidines. Interestingly, AG-rich conserved regions were also observed in internal loops of the RFN, THI and B12 elements. The loop in the seventh helix mostly consists of three to five nucleotides, but in some cases it is extended up to 70 nt. In such cases, an additional stem–loop is formed. The end loops of helices P4 and P5 contain stretches of complementary nucleotides that are not conserved on the sequence level but exist in most LYS elements. This possible pseudoknot interaction could be required for the formation of stable tertiary structure of the LYS element.
Figure 3.
Conserved structure of the LYS element. Capitals indicate invariant positions. Lower case letters indicate strongly conserved positions. Degenerate positions: R = A or G; Y = C or U; K = G or U; N = any nucleotide. Conserved helices are numbered P1 to P7. Stem–loops of variable and constant lengths are shown by broken and sold lines, respectively. Possible tertiary interaction between the end loops of helices P4 and P5 is shown.
The detailed functional, positional and phylogenetic analysis of lysine-related genes and the LYS elements is given below.
LYS-element-regulated genes
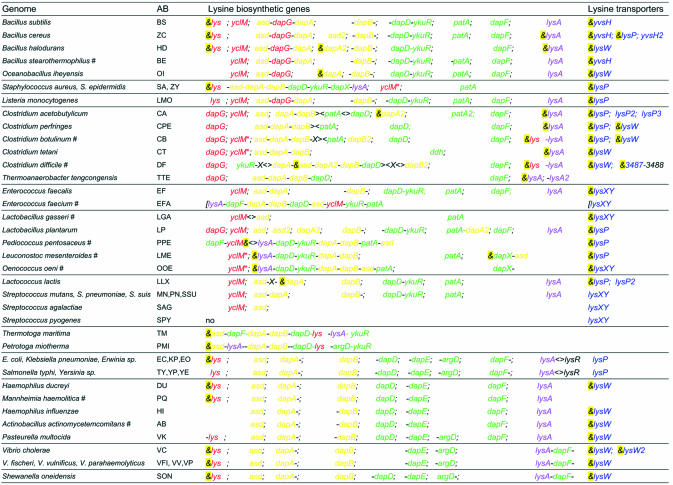
Lysine biosynthesis genes. In E.coli, known lysine biosynthesis (LBS) genes are scattered along the chromosome. Orthologs of these genes were identified in all available bacterial genomes containing LYS elements (Table 1). In all γ-proteobacteria, including enterobacteria, Pasteurellaceae, Vibrionaceae and Shewanella oneidensis, the LBS genes also stand separately. At that, only the lysine-inhibited aspartokinases lysC were found under candidate regulation of the LYS elements (analyzed in detail below). In E.coli, expression of the lysA gene encoding the last stage of the LBS pathway is regulated by the transcriptional regulator LysR reacting to the intracellular concentration of DAP, which acts as an inducer, and lysine, which acts as a corepressor (9). Conservation of the divergent arrangement of the lysA and lysR genes yields possible conservation of the LysR-mediated regulation of lysA in all enterobacteria.
Table 1. The operon structures of the lysine biosynthesis and transport genes in bacteria from the Bacillus/Clostridium group and γ-proteobacteria.
Genes forming one candidate operon (with spacer <100 bp) are separated by ‘–’. The aspartokinase and diaminopimelate decarboxylase genes are shown in red and magenta, respectively. Other genes of the first and second parts of the LBS pathway are shown in orange and green, respectively. Genes encoding transport proteins are in blue. Non-LBS genes are shown as X. Predicted LYS elements are denoted by ‘&’ and highlighted by yellow. Contig ends are marked by square brackets. Genome abbreviations are given in column ‘AB’ with unfinished genomes marked by ‘#’. Additional genome abbreviations are: Yersinia pestis (YP), Yersinia enterocolitica (YE), Erwinia carotovora (EO).
In Gram-positive bacteria from the Bacillus/Clostridium group, the LBS genes were found either as single genes or within a LBS gene cluster, potentially forming an operon (Table 1). For instance, the asd-dapG-dapA gene cluster is conserved in bacilli and Listeria species, while the conserved asd-dapA-dapB gene cluster was found in clostridia. Single lysC, lysA and dapA genes in the Bacillus/Clostridium group are preceded by LYS elements in most cases. Complete LBS gene clusters containing all or almost all LBS genes were detected in Staphylococcus aureus, Enterococcus faecium, Pediococcus pentosaceus, Leuconostoc mesenteroides and Oenococcus oeni, where they are mostly regulated by candidate LYS elements. In addition, potential LBS operons of two thermophilic bacteria, Thermotoga maritima and Petrotoga miotherma, and the potential lysC-lysA operons of two clostridia species are also preceded by LYS elements. Among bacteria from the Bacillus/Clostridium group, the LBS genes are completely absent only in the genomes of two streptococci and Lactobacillus gasseri, arguing for existence of lysine-specific transport systems in these bacteria (see below). Finally, DAP dehydrogenase encoded by the ddh gene complements the absence of four DAP synthesis genes in Clostridium tetani.
Both E.coli and B.subtilis genomes encode three aspartokinase isozymes, required for different biosynthetic pathways starting from aspartate. To identify the aspartokinase genes that are specific for the lysine biosynthesis, similarity search for the lysC homologs was complemented by positional and phylogenetic analysis. The phylogenetic tree of bacterial aspartokinases consists of at least five main branches, corresponding to the lysine-inhibited aspartokinases LysC from Gram- and Gram-negative species, the methionine- and threonine-regulated aspartokinases MetL and ThrA from proteobacteria, and the DAP- and threonine/lysine-inhibited enzymes from the Bacillus/Clostridium group, DapG and YclM, respectively (see Supplementary Material Fig. 1).
In contrast to E.coli, most of the dap genes were not characterized experimentally in B.subtilis. Using a similarity search, we identified orthologs of the E.coli dapB, dapD and dapF genes in bacteria from the Bacillus/Clostridium group, but we did not find the dapE and dapC counterparts. Positional analysis of the LBS genes allowed us to identify the most likely candidates for the missing DAP/lysine biosynthetic genes in bacteria from the Bacillus/Clostridium group. As result, we tentatively assigned the previously missing in B.subtilis N-acetyl-diaminopimelate deacetylase function to the ykuR gene, whose product is a member of the N-acyl-l-amino acid amidohydrolase family. In most bacteria from the Bacillus/Clostridium group, as well as in Thermotogales, this gene is possibly co-transcribed with the ykuQ gene, which is an ortholog of the E.coli dapD gene. Moreover, in some cases the ykuR gene belongs to a long LYS-element-regulated LBS operon. Based on similar reasoning, the acetyl-diaminopimelate aminotransferase function was assigned to the B.subtilis patA gene, whose product belongs to the class I pyridoxal-phosphate-dependent aminotransferases. Orthologs of the patA gene were identified in most bacteria from the Bacillus/Clostridium group, and they are co-localized with the LBS genes in some cases. The observed involvement of unrelated genes for the DAP synthesis in Gram-negative and Gram-positive bacteria is consistent with a key difference in their LBS pathways: E.coli and B.subtilis use succinylated and acetylated DAP derivatives, respectively (Fig. 1).
Orthologs of the dapF gene encoding the DAP epimerase were not found in S.aureus, O.oeni, L.mesenteroides and in streptococci. However, the LBS operon of S.aureus contains a hypothetical gene, named herein dapX, whose product is similar to the alanine racemase and other epimerases. In addition, the dapX-asd cluster is preceded by a LYS element in O.oeni, and the single dapX gene was also detected in L.mesenteroides. We conclude that dapX is the most suitable candidate for a non-orthologous displacement of the dapF gene.
Lysine transporters. Orthologs of the lysine-specific permease lysP from E.coli were identified in other Gram-negative bacteria, including enterobacteria, pseudomonads and γ-proteobacteria, as well as in several Gram-positive species, including clostridia, lactobacilli, listeria, S.aureus and B.cereus. In contrast to Gram-negative bacteria, the lysP genes from Gram-positive bacteria have candidate LYS elements in their regulatory regions. At that, there are two and three lysP paralogs in the genomes of Lactococcus lactis and Clostridium acetobutylicum, respectively, but in each genome only one of them has an upstream LYS element. The lysP orthologs never form potential operons with other genes. The absence of lysP in many complete bacterial genomes, including several species without lysine biosynthetic genes, hints to the existence of previously unknown transport systems for lysine. The search for LYS elements in bacterial genomes allowed us to identify a number of new candidate lysine transporters.
The LYS-regulated gene yvsH (the B.subtilis gene name) was found in three Bacillus species (Table 1). YvsH has 11 predicted transmembrane segments and is similar to the arginine:ornithine antiporter ArcD from Pseudomonas aeruginosa and the lysine permease LysI from Corynebacterium glutamicum. All these proteins belong to the basic amino acid/polyamine antiporter APA family. Based on identification of highly-specific LYS elements upstream of the yvsH orthologs, we tentatively predict that YvsH is involved in the lysine transport in the above bacilli. Interestingly, B.cereus has two yvsH paralogs (yvsH1 and yvsH2), with and without an upstream LYS element, respectively. The upstream region of yvsH2 contains a candidate transcriptional attenuator that includes a leader peptide region with a run of histidine regulatory codons (A.Vitreschak, unpublished results). Thus YvsH2 is possibly involved in the histidine transport. The predicted specificity of these identified transporters from the APA family is consistent with experimental data for the homologous HisJ and LAO transporters, which both bind histidine, arginine, lysine and ornithine, albeit with different affinities towards these ligands (33).
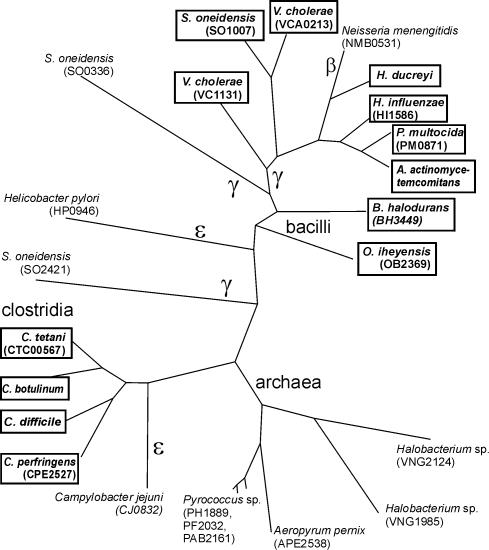
Another new type of candidate lysine transporter, named herein LysW, was identified in microorganisms from various taxonomic divisions, such as γ-, β- and ε-proteobacteria, the Bacillus/Clostridium group and archaea (Fig. 4). Among γ-proteobacteria, only Pasteurellaceae, Vibrio and Shewanella species have the lysW genes that are preceded by an upstream LYS element in all cases, except two additional lysW paralogs in S.oneidensis. Interestingly, the genome of V.cholerae contains two LYS-element-regulated lysW genes. The absence of lysW in the genomes of other enterobacteria is consistent with the presence of the lysP lysine transporter in these genomes. Among Gram-positive bacteria, only two bacilli and four clostridia species have the lysW genes, which are also regulated by candidate LYS elements. The detected LysW proteins contain 11 candidate transmembrane segments and comprise a unique protein family (see the phylogenetic tree in Fig. 4), which is a part of the NhaC Na+:H+ antiporter superfamily.
Figure 4.
Maximum likelihood phylogenetic tree of the LysW family of predicted lysine transporters. Gene identifiers for annotated complete genome sequences are shown in parentheses. Genes predicted to be regulated by LYS elements are boxed and shown in bold.
Candidate components of an ATP-dependent lysine transport system, named lysX-lysY, were identified in Lactobacillus gassei, O.oeni, Enterococcus and Streptococcus species (see, for reference, the EF0247-EF0246 genes in Enterococcus faecalis). The lysX-lysY system complements the absence of the lysW, lysP and yvsH lysine transporters in these Gram-positive bacteria. The N-terminal substrate-binding and C-terminal transmembrane domains of the LysX proteins, as well as the ATP-binding LysY proteins, show significant similarity to the corresponding components of ATP-dependent transport systems for various amino acids, including arginine, histidine and glutamine. We tentatively assigned the lysine specificity for the LysX-LysY transport system based on two following observations. First, the candidate lysX-lysY operons of L.gasseri, O.oeni and E.faecalis have upstream LYS elements. Secondly, the lysX-lysY transport system possibly complements the absence of known lysine biosynthesis and transport genes in the genomes of S.agalactiae, S.pyogenes and L.gasseri (Table 1).
Lysine utilization genes. The genome of T.tengcongensis contains two candidate LYS elements. The first one precedes the lysine biosynthetic operon, whereas the second one is located upstream of a long hypothetical gene cluster beginning with the pspF3 gene. Three genes from this cluster are highly homologous to the kamA, kamD and kamE lysine aminomutase genes, which are involved in the lysine catabolism in Clostridium species (34). Among available bacterial genomes, a similar gene cluster was found only in F.nucleatum, albeit without pspF3 and atoDA orthologs. The only candidate LYS element detected in F.nucleatum is located upstream of the FN1869 gene, the first gene of the kam cluster. In addition, this gene cluster contains an ortholog of the lysine exporter LysE of C.glutamicum, which is induced by an excess of intracellular lysine that might be harmful to the cell (35). Therefore, the LYS elements could be involved not only in regulation of widely distributed lysine biosynthesis and uptake genes, but also in control of relatively rare genes for the lysine catabolism and export. However, unlike the first group of genes that is obviously repressed by lysine, the second group should be activated by lysine. A possible explanation for this discrepancy is given below following description of a suggested mechanism for the LYS-element-mediated gene regulation.
Possible attenuation mechanism for the LYS-mediated regulation
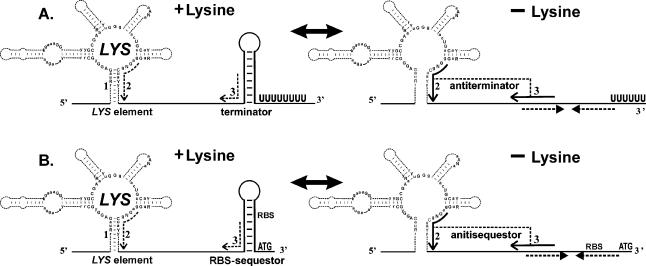
Downstream of all LYS elements, except some elements of T.tengcongenesis and F.nucleatum, there are additional RNA regulatory hairpins (Fig. 5). In Gram-positive bacteria, these additional hairpins are followed by runs of thymidines and therefore are candidate ρ-independent terminators of transcription. In Gram-negative bacteria, the hairpins overlap the ribosome-binding site of the first gene in the LYS-regulated operon and therefore are candidate translational sequestors that prevent ribosome binding to the ribosome-binding site (RBS). Therefore, the regulation of lysine biosynthetic and transport genes is likely to operate mainly at the level of transcription in the former group of bacteria and at the level of translation in the latter group. Previously it has been shown that lysine regulates the expression of the B.subtilis lysC gene by effecting the premature termination of transcription at a ρ-independent terminator site in its leader region (12). In addition, we observed complementary RNA regions that partially overlap both the proposed regulatory hairpin (terminator or sequestor) and one of the conserved helices in the LYS element. Furthermore, these complementary fragments always form the base stem of a new, more stable alternative secondary structure (Fig. 5). We predict that this structure functions as an anti-terminator/anti-sequestor, alternative to both the LYS element and the terminator/sequestor hairpin.
Figure 5.
Conserved RNA elements upstream of the LYS-regulated genes. The P1′ and P7′ stems of the LYS element and proposed regulatory hairpins (terminator in Gram-positive bacteria; RBS-sequestors in proteobacteria) are indicated by a gray background. The main stem of possible anti-terminator/anti-sequestor structure is underlined. Arrows show the complementary stems of RNA secondary structures. RBSs, start codons and poly-T tracts are set in bold.
Genetic control by metabolite-binding mRNAs, or riboswitches, was experimentally confirmed for various regulons specific for vitamins, amino acids and nucleotides, in particular thiamin, riboflavin, vitamin B12, methionine and guanine (13–19). By analogy with the previously suggested riboswitch regulatory mechanisms (22,23), we propose here a lysine-specific riboswitch (Fig. 6). As in previous cases, the model for the lysine regulation is based on competition between alternative RNA secondary structures. In the repressing conditions, the LYS element is stabilized by binding of effector molecules, possibly lysine. In Gram-positive bacteria, it leads to formation of the terminator hairpin and premature termination of transcription. In Gram-negative bacteria, stabilization of the LYS element leads to formation of the RBS-sequestor hairpin which represses initiation of translation. In the absence of lysine, the non-stabilized LYS element is replaced by the more energetically favorable anti- terminator/anti-sequestor structure, thus allowing for the transcriptional readthrough or translation initiation.
Figure 6.
Predicted mechanisms of the LYS-mediated regulation of the lysine biosynthetic/transport genes: (A) transcription attenuation, (B) translation attenuation. Dashed lines show location of complementary regions. Point lines show interactions in derepressed conditions. RBS, the ribosome-binding site; ATG, start-codon; UUUU, poly-U tract in the terminator.
Analysis of the upstream regions of the lysine utilization operons from T.tengcongenesis and F.nucleatum reveals another possible mode of regulation. In this case the LYS element overlaps the predicted terminator hairpin directly, possibly acting as an anti-terminator (Fig. 5). Therefore, formation of these LYS elements in conditions of lysine excess promotes expression of the lysine utilization operons. This mode of regulation contrasts with the above-predicted regulatory mechanism for the lysine biosynthetic genes, which are obviously expressed in the conditions of lysine limitation.
DISCUSSION
The lysine-mediated gene regulation in bacteria appears to operate via a unique RNA structural element. The LYS element is characterized by its compact secondary structure with a number of conserved helices and extended regions of sequence conservation, which could be necessary for specific metabolite binding. It was found in 5′-untranslated regulatory regions of the lysine biosynthetic and transport genes in both Gram-positive and Gram-negative bacteria, as well as in Thermotogales. All previously detected lysine-constitutive mutations in the lysC leader regions of E.coli and B.subtilis hit either highly conserved nucleotides of the LYS elements or one of its paired regions (10,11).
Using multiple alignment of 71 identified LYS element sequences, we constructed the maximum likelihood phylogenetic tree for these RNA elements and compared it with the standard trees for ribosomal proteins (36). The tree of LYS elements has a number of branches that correspond to various taxonomic groups of the Bacillus/Clostridium group, namely Bacillales, Clostridiales and Lactobacillales, as well as a highly diverged branch of the LYS elements from γ-proteobacteria (Supplementary Material Fig. 2). However, these lineage-specific branches contain also a number of gene-specific sub-branches. For instance, the branch of LYS elements from γ-proteobacteria can be divided into two parts corresponding to the lysC and lysW genes, respectively.
On the whole, the possible mechanism of regulation of the lysine biosynthetic and transport genes is similar to the previously proposed mechanisms of regulation of riboflavin, thiamin and vitamin B12 genes (22–24). Similarly to the previously established direct binding of an effector (flavin mononucleotide, thiamin pyrophosphate or adenosylcobalamin) to a highly conserved RNA element (RFN, THI or B12, respectively), we propose that lysine is an effector molecule for the LYS element, acting in a similar manner. In the repressing conditions of lysine excess, an adjacent regulatory hairpin, terminator or RBS-sequestor can fold that leads to the transcriptional or translational repression of the target genes. At low concentration of effector molecules, the unstable RNA element is replaced by an alternative anti-terminator or anti-sequestor RNA conformation allowing for transcription readthrough or translation initiation. Interestingly, the observed phylogenetic distribution of the LYS-element-associated terminators and sequestors in Gram-positive and Gram-negative bacteria, respectively, is similar to the previously observed distribution of the regulatory hairpins in the analyzed vitamin regulons (20).
Using positional analysis of lysine-specific regulatory elements in bacteria, we have identified three new types of candidate lysine transporters, namely LysW, YvsH and LysXY, in addition to the previously known LysP transporter. In some bacteria species, the suggested lysine transporter LysXY complements the absence of lysine biosynthetic genes and other lysine transporters. Noteworthy, the lysine transport systems of all four types has been observed in Gram-positive bacteria from the Bacillus/Clostridium group. At that, the genomes of B.cereus and two Clostridium species encode candidate lysine transporters of two different types. In contrast, most Gram-negative γ-proteobacteria possess only one lysine-specific transporter, LysP or LysW. On the whole, most lysine transport systems, both known and predicted, are preceeded by LYS elements.
Analysis of possible operon structures for the lysine biosynthetic genes shows that the LYS element predominantly regulates either single lysC and lysA genes, or a composite lysine operon. Arrangement of the LBS genes significantly differs in Gram-negative and Gram-positive bacteria: the LBS gene clusters exist only in the latter taxonomic group. Using a combination of positional and phylogenetic analyses applied to numerous aspartokinase genes we associated most of them with a certain metabolic pathway (lysine, thereonine, methionine or DAP). This study demonstrated that the evolutionary history of the aspartokinase genes involves a number of duplication and gene fusion events with consequent specialization to a certain biosynthetic pathway when the feedback regulation of the enzyme is mediated by the pathway’s end-product (Supplementary Material Fig. 1). In the Bacillus/Clostridium group of bacteria we identified two candidate missing genes of the DAP pathway for lysine biosynthesis, analogs of the E.coli dapC and dapE genes. Finally, a non-orthologous gene displacement of the dapF gene for another epimerase gene was observed in some Gram-positive bacteria.
From the practical point of view, this work, in addition to our previous analyses of the vitamin-specific regulons (22–24), demonstrate one more example of the power of comparative genomics for the functional gene annotation. Comparative analysis of pathway-specific regulatory sites in bacterial genomes is very effective in this respect. Combination of genomic techniques allowed us to identify candidates for previously missing lysine biosynthetic and transport genes in a variety of bacterial species.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This study was partially supported by grants CDF RBO-1268 from the Ludwig Institute for Cancer Research and 55000309 from the Howard Hughes Medical Institute.
NOTE ADDED IN PROOF
Recently, it has been demonstrated by in vitro experiment that lysine can directly regulate the B.subtilis lysC gene using termination/anti-termination mechanism (37). At that, lysine itself, but not its direct precursor, diaminopimelate, promotes a structural rearrangement of the lysC leader RNA. Thus this result may be considered as a confirmation of our hypothesis of lysine riboswitch. Grundy et al. also proposed a conserved secondary structure of lysine-regulatory RNA (L-box) using phylogenetic analysis of 22 leader regions (37). The structure of the LYS element derived from comparison of 71 leader sequences in this work is consistent with the L-box structure.
REFERENCES
- 1.Patte J.C. (1994) Biosynthesis of threonine and lysine. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella. Cellular and molecular biology. American Society for Microbiology, Washington, DC, pp. 528–541. [Google Scholar]
- 2.Nishida H., Nishiyama,M., Kobashi,N., Kosuge,T., Hoshino,T. and Yamane,H. (1999) A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: a key to the evolution of amino acid biosynthesis. Genome Res., 9, 1175–1183. [DOI] [PubMed] [Google Scholar]
- 3.Cirillo J.D., Weisbrod,T.R., Banerjee,A., Bloom,B.R. and Jacobs,W.R.,Jr (1994) Genetic determination of the meso-diaminopimelate biosynthetic pathway of mycobacteria. J. Bacteriol., 176, 4424–4429. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Hutton C.A., Southwood,T.J. and Turner,J.J. (2003) Inhibitors of lysine biosynthesis as antibacterial agents. Mini Rev. Med. Chem., 3, 115–127. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J.J., Hu,F.M., Chen,N.Y. and Paulus,H. (1990) Comparison of the three aspartokinase isozymes in Bacillus subtilis Marburg and 168. J. Bacteriol., 172, 701–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobashi N., Nishiyama,M. and Yamane,H. (2001) Characterization of aspartate kinase III of Bacillus subtilis. Biosci. Biotechnol. Biochem., 65, 1391–1394. [DOI] [PubMed] [Google Scholar]
- 7.Velasco A.M., Leguina,J.I. and Lazcano,A. (2002) Molecular evolution of the lysine biosynthetic pathways. J. Mol. Evol., 55, 445–459. [DOI] [PubMed] [Google Scholar]
- 8.Steffes C., Ellis,J., Wu,J. and Rosen,B.P. (1992) The lysP gene encodes the lysine-specific permease. J. Bacteriol., 174, 3242–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stragier P., Richaud,F., Borne,F. and Patte,J.C. (1983) Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. I. Identification of a lysR gene encoding an activator of the lysA gene. J. Mol. Biol., 168, 307–320. [DOI] [PubMed] [Google Scholar]
- 10.Lu Y., Shevtchenko,T.N. and Paulus,H. (1992) Fine-structure mapping of cis-acting control sites in the lysC operon of Bacillus subtilis.FEMS Microbiol. Lett., 71, 23–27. [DOI] [PubMed] [Google Scholar]
- 11.Patte J.C., Akrim,M. and Mejean,V. (1998) The leader sequence of the Escherichia coli lysC gene is involved in the regulation of LysC synthesis. FEMS Microbiol. Lett., 169, 165–170. [DOI] [PubMed] [Google Scholar]
- 12.Kochhar S. and Paulus,H. (1996) Lysine-induced premature transcription termination in the lysC operon of Bacillus subtilis. Microbiology, 142, 1635–1639. [DOI] [PubMed] [Google Scholar]
- 13.Nahvi A., Sudarsan,N., Ebert,M.S., Zou,X., Brown,K.L. and Breaker,R.R. (2002) Genetic control by a metabolite binding mRNA. Chem. Biol., 9, 1043–1049. [DOI] [PubMed] [Google Scholar]
- 14.Winkler W., Nahvi,A. and Breaker,R.R. (2002) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature, 419, 952–956. [DOI] [PubMed] [Google Scholar]
- 15.Winkler W., Cohen-Chalamish,S. and Breaker,R.R. (2002) An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA, 99, 15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mironov A.S., Gusarov,I., Rafikov,R., Lopez,L.E., Shatalin,K., Kreneva,R.A., Perumov,D.A. and Nudler,E. (2002) Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell, 111, 747–756. [DOI] [PubMed] [Google Scholar]
- 17.McDaniel B.A., Grundy,F.J., Artsimovitch,I. and Henkin,T.M. (2003) Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl Acad. Sci. USA, 100, 3083–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epshtein V., Mironov,A.S. and Nudler,E. (2003) The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl Acad. Sci. USA, 100, 5052–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandal M., Boese,B., Barrick,J.E., Winkler,W.C. and Breaker,R.R. (2003) Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell, 113, 577–586. [DOI] [PubMed] [Google Scholar]
- 20.Vitreschak A.G., Rodionov,D.A., Mironov,A.A and Gelfand,M.S. (2003) Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet., in press. [DOI] [PubMed] [Google Scholar]
- 21.Gelfand M.S., Mironov,A.A., Jomantas,J., Kozlov,Y.I. and Perumov,D.A. (1999) A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet., 15, 439–442. [DOI] [PubMed] [Google Scholar]
- 22.Vitreschak A.G., Rodionov,D.A., Mironov,A.A. and Gelfand,M.S. (2002) Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res., 30, 3141–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodionov D.A., Vitreschak,A.G., Mironov,A.A. and Gelfand,M.S. (2002) Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem., 277, 48949–48959. [DOI] [PubMed] [Google Scholar]
- 24.Rodionov D.A., Vitreschak,A.G., Mironov,A.A. and Gelfand,M.S. (2003) Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem., 278, 41148–41159. [DOI] [PubMed] [Google Scholar]
- 25.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.I. (2003) GenBank. Nucleic Acids Res., 31, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overbeek R., Larsen,N., Walunas,T., D’Souza,M., Pusch,G., Selkov,E.Jr., Liolios,K., Joukov,V., Kaznadzey,D., Anderson,I., Bhattacharyya,A., Burd,H., Gardner,W., Hanke,P., Kapatral,V., Mikhailova,N., Vasieva,O., Osterman,A., Vonstein,V., Fonstein,M., Ivanova,N. and Kyrpides,N. (2003) The ERGO genome analysis and discovery system. Nucleic Acids Res., 31, 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vitreschak A.G., Mironov,A.A. and Gelfand,M.S. (2001) Proceedings of the third International Conference ‘Complex Systems: Control and Modeling Problems’. Samara, Russia, September 4–9, 2001, The Institute of Control of Complex Systems, Samara, Russia, 623–625.
- 28.Lyngso R.B., Zuker,M. and Pedersen,C.N. (1999) Fast evaluation of internal loops in RNA secondary structure prediction. Bioinformatics, 15, 440–445. [DOI] [PubMed] [Google Scholar]
- 29.Mironov A.A., Vinokurova,N.P. and Gelfand,M.S. (2000) GenomeExplorer: software for analysis of complete bacterial genomes. Mol. Biol., 34, 222–231. [Google Scholar]
- 30.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res., 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felsenstein J. (1981) Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol., 17, 368–376. [DOI] [PubMed] [Google Scholar]
- 32.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreimer D.I., Malak,H., Lakowicz,J.R., Trakhanov,S., Villar,E. and Shnyrov,V.L. (2000) Thermodynamics and dynamics of histidine-binding protein, the water-soluble receptor of histidine permease. Implications for the transport of high and low affinity ligands. Eur. J. Biochem., 267, 4242–4252. [DOI] [PubMed] [Google Scholar]
- 34.Chang C.H. and Frey,P.A. (2000) Cloning, sequencing, heterologous expression, purification and characterization of adenosylcobalamin-dependent D-lysine 5, 6-aminomutase from Clostridium sticklandii. J. Biol. Chem., 275, 106–114. [DOI] [PubMed] [Google Scholar]
- 35.Bellmann A., Vrljic,M., Patek,M., Sahm,H., Kramer,R. and Eggeling,L. (2001) Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology, 147, 1765–1774. [DOI] [PubMed] [Google Scholar]
- 36.Wolf Y.I., Rogozin,I.B., Grishin,N.V., Tatusov,R.L. and Koonin,E.V. (2001) Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol. Biol., 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grundy F.J., Lehman,S.C. and Henkin,T.M. (2003) The L box regulon: Lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl Acad. Sci. USA, 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.