Abstract
Background
Cerebrospinal fluid (CSF) and structural magnetic resonance imaging (MRI) show patterns of change in Alzheimer’s disease (AD) that precede dementia. The Alzheimer’s Disease Neuroimaging Initiative (ADNI) studied normal controls (NC), subjects with mild cognitive impairment (MCI) and AD to identify patterns of biomarkers to aid in early diagnosis and effective treatment of AD.
Methods
222 NC underwent baseline MRI and clinical examination at baseline and at least one follow-up. 112 also provided CSF at baseline. Unsupervised clustering based on initial CSF and MRI measures was used to identify clusters of participants with similar profiles. Repeated measures regression modeling assessed the relationship of individual measures, and of cluster membership, to cognitive change over three years.
Results
Most individuals showed little cognitive change. Individual biomarkers had limited predictive value for cognitive decline, but membership in the cluster with the most extreme profile was associated with more rapid decline in ADAS-COG.
Conclusions
Subtypes among NC based on multiple biomarkers may represent the earliest stages of subclinical cognitive decline and AD.
Keywords: Alzheimer’s disease, Dementia, Early diagnosis, Cerebrospinal fluid, Tau protein, Amyloid beta-protein, Structural magnetic resonance imaging, Hippocampal volume, Cognition, Clustering, Normal controls
1. Introduction
Alzheimer’s disease (AD) is a neurocognitive disorder currently estimated to affect some five million people in the United States and more than 25 million worldwide (Evans et al., 1990, Hebert et al., 2003, Brookmeyer et al., 2007, Harvey et al. 2003). Mild cognitive impairment (MCI) has gained recognition as an intermediate clinical category between normal cognitive function and AD, with a greatly increased risk of onset of AD (Bennett 2002, Petersen 2009). AD is characterized not only by cognitive decline, but also by underlying neurobiological changes that likely precede the diagnosis of AD by a considerable period, during MCI and possibly even earlier, before measurable clinical impairment. The process is hypothesized to begin with amyloid deposition, followed by cortical atrophy and decreased metabolism, with effects only gradually becoming apparent in decreased cognitive performance and function (Jack et al. 2010). Identification of early markers of disease would be of great interest to facilitate early diagnosis, improved clinical trials for prevention by targeting individuals at greatest risk, and, ultimately, effective treatment before widespread irreversible neurodegeneration (Clark 2008).
A large number of potential markers have been proposed, including volumetric measures based on MRI, CSF biomarkers, FDG PET and others (Hampel 2007, Shaw 2007). Cortical atrophy, for example, is evident on structural magnetic resonance imaging (MRI) not only in AD but also to some extent in people with mild cognitive impairment (MCI) (Nestor 2008, Morra 2009) and in normal elderly prior to the onset of MCI (Carlson 2008). Between-person differences in cerebrospinal fluid (CSF) protein levels have also been reported to be associated with AD and MCI (Maddalena 2003, Clark 2008, Shaw 2009). White matter hyperintensity (WMH) has been reported to be increased in patients with AD, suggesting that vascular lesions may also play a role in the neurodegenerative process (Barber 1999). Alternatively, WMH may be a vascular pathology contributing to cognitive impairment in an additive or even multiplicative manner. Homocysteine, a risk factor for vascular damage, has also been hypothesized as a possible risk factor for dementia (Smith 2008). Most studies of markers have focused on AD and MCI, as the clinical decline is most evident in these groups and association of candidate markers with clinical benchmarks is more readily established. An earlier biomarker horizon, however, would be of great scientific interest and have substantial clinical relevance.
The Alzheimer’s Disease Neuroimaging Initiative (ADNI), jointly funded by NIH, pharmaceutical partners and the Alzheimer’s Association, is a multi-site research initiative whose aim is to identify biomarkers which would allow the pathological changes of AD to be diagnosed earlier, well before the clinical criteria for dementia are met, and to be tracked more precisely. The goal is to provide earlier diagnosis and better assessment of disease progression and response to therapy. The ADNI participants included normal controls (NC) with detailed standardized assessment of many potential candidate biomarkers and longitudinal follow-up of cognitive outcomes for up to 3 years. We examined a set of imaging and cerebrospinal fluid measures previously proposed in the literature as candidate markers for early diagnosis, and assessed their distribution in normal controls and their relationship to cognitive outcomes over the follow-up period. Our hypothesis was that despite cognitive homogeneity at baseline in the NC subjects, there would be underlying biological heterogeneity in candidate markers, reflecting the earliest detectable changes in the brain. These biological differences would be correlated with each other in a structured way, leading to the ability to construct subgroups based on markers alone, and such subgroups would subsequently have different cognitive trajectories.
2. Methods and material
2.1. The Alzheimer’s Disease Neuroimaging Initiative
Data used in the preparation of this article were obtained from the ADNI database (www.loni.ucla.edu\ADNI). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The research plan called for recruiting 800 adults, ages 55 to 90: approximately 200 healthy elderly controls, 400 subjects with mild cognitive impairment (MCI), and 200 subjects with mild but probable AD. Subjects are followed longitudinally for up to three years, with MRI scans, complete cognitive testing, and blood/urine samples collected at six-month intervals, depending on baseline diagnosis. In addition, subsets of the subjects undergo FDG-PET scans and cerebral spinal fluid (CSF) collection and testing (Mueller 2005a, 2005b).
2.2. Subjects
The individuals studied were recruited between August 17, 2005 and September 4, 2007 as ADNI participants and were identified at baseline clinical evaluation as cognitively normal. NC, MCI and AD participants were frequency-matched by age-group to a common target age profile. NC participants underwent cognitive testing and clinical examination by a physician at baseline and every six months for the first year and then annually for the next two years. MRI scans (1.5 Tesla) were performed in each subject (http://www.loni.ucla.edu/ADNI/Research/Cores/index.shtml) at baseline, repeated at 6, 12, 24 and 36 months. Approximately half the participants also provided CSF at the baseline and m12 visits. Additional details are given in (Petersen 2010). This study was approved by the Institutional Review Boards of all of the participating institutions. Informed written consent was obtained from all participants at each site. A detailed description of the study design and inclusion criteria is available at http://clinicaltrials.gov/show/NCT00106899. Data used in this analysis were downloaded from the ADNI database (www.loni.ucla.edu/ADNI) on September 27, 2009. Analysis focused on NC but the MCI and AD group were described at baseline for comparison purposes.
2.3. Measures
Biomarker summary measures for cluster analysis were selected using a list initially specified by researchers from the ADNI Imaging and Biomarker Cores at the time of grant submission, for core hypothesis tests. MRI summary measures were calculated by the Anders Dale Lab at UC San Diego and normalized by their measure of intra-cranial volume (ICV) (Fennema-Notestine 2009). Summary measures used were cerebral volume, hippocampal volume, entorhinal cortex thickness, and ventricle volume. White matter hyperintensities (WMH) were detected by Charles DeCarli and the Imaging of Dementia and Aging lab at UC Davis based on coregistered T1-, T2-, and PD-weighted images using an automated protocol described previously (Schwarz 2009). Total WMH volume was used as the primary summary; it was also standardized by ICV. CSF samples were obtained by the individual centers, then banked and batch-processed using a standardized protocol, under the direction of Drs. Leslie Shaw and John Trojanowski of the ADNI Biomarker Core at the University of Pennsylvania School of Medicine (Shaw 2008). CSF measures at baseline included Aβ1–42, total tau protein (t-tau), tau protein phosphorylated at the 181 threonine position (P-tau181), and the ratios t-tau/Aβ1–42 and P-tau181/Aβ1–42 as previously described (Shaw, et al 2009). Apolipoprotein E genotype was determined and serum homocysteine assayed using a validated enzyme immunoassay methodology by the ADNI Biomarker Core. Two cognitive performance tests were considered for measurement of longitudinal change: the Rey Auditory-Verbal Learning Test (RAVLT) total of five trials, and the Alzheimer’s Disease Assessment Scale-cognitive subsection (ADAS-cog), selected because some NC showed modest changes in these scales. Higher values of RAVLT and lower values of ADAS-cog correspond to better performance.
2.4 Statistical analysis
All demographic, clinical and marker data were first summarized descriptively (means, standard deviations, graphical summaries). Quantitative marker data (Imaging, CSF and serum) were standardized by subtracting the NC mean and dividing by the standard deviation (SD) before inclusion in regression models and clustering, to facilitate comparisons across markers and analyses. As a comparison for the utility of clusters as predictors, univariate and multivariate mixed models for longitudinal data were used to model changes in RAVLT and ADAS-cog over time using candidate biomarkers as predictors (Laird 1982). The annualized rate of change was the model-estimated slope, in units of cognitive test score change per year. Main effects of each predictor estimated the difference in cognitive score at baseline, and interaction terms between predictor and time estimated the effect of a one-unit increase in predictor on the annualized rate of change in cognitive test score. Random effects were included for slope and intercept, and an unstructured correlation matrix was specified for errors. All models also controlled for education effects on cognitive test score and change. Variables in multivariate models were removed step-wise based on the P-value of the variable’s slope component. Univariate P values were reported without adjustment for multiple comparisons, because the goal was not variable selection, but a reference comparison for the utility of any specific, individual biomarker compared to cluster membership.
Risk groups (clusters) based on baseline levels of CSF and serum biomarkers and MRI summaries were created by unsupervised hierarchical clustering, without reference to baseline cognitive test scores or to longitudinal trajectories using the function hclust in R. In this agglomerative approach, each individual begins as a cluster of one person. Clusters are then iteratively combined based on dissimilarity. The calculation of dissimilarity relies on maximizing or minimizing an objective function and a metric for distance between individuals. Many options exist for objective functions and distance metrics; we explored seven different objective function methods (Wards’ method, single linkage, complete linkage, average, Mcquitty’s method, median and centroid [REFS to be added]) and six different distance metrics (Euclidean, supremum norm, Manhattan, Canberra, binary, and Minkowski). Some combinations of these have a tendency to produce clusters containing a very small number of individuals, essentially isolating individual outliers. These methods are not useful in this context because the resulting sample sizes are too small to allow for further exploration. We restricted consideration to methods that had at least 5 individuals per cluster. Our primary approach used Ward’s method, which is defined as minimal increase in the error sum of squares after combining clusters (Ward 1963). Error sum of squares was calculated as the sum of the squared Euclidean distances of each cluster member to the cluster center. Similarity across clustering methods was assessed by Rand’s statistic, a measure of agreement between two partitions, with 1 representing perfect agreement and 0 corresponding to equal amounts of concordance and discordance (Rand 1971). We also examined descriptively the degree of agreement between methods on individuals belonging to the group with “unusual” profiles compared to the typical NC. The choice of the number of clusters (k) was based on a combination of observed visual separation, plots of the log within-cluster dissimilarity against k, and restrictions due to sample size. We chose k = 3 based on these considerations. Although we are forming subgroups, we hypothesize that these subgroups represent a categorization of an unobserved continuum. Thus, for a larger sample size, a reasonable choice for k may be well beyond 3, Secondary analyses examined whether the effect of cluster membership could be accounted for by ApoE genotype or by age. Preliminary analyses examined conversions from NC to MCI but found too few conversions to allow formal statistical analysis. Both regression models and clustering algorithms used a complete-case approach for biomarkers, excluding any individual missing one or more of the biomarkers in that analysis, and regression models treated missing cognitive scores at scheduled visits as missing at random. Statistical analyses were carried out using the R package (R 2009) and SAS/STAT® software (SAS Institute 2004).
3. Results
3.1. Normal control participant characteristics
Baseline MRI were available for 222 NC and CSF biomarker data for 112 NC. However, not all image summary measures were available for all images (range: 192–222). In comparison with ADNI MCI and AD participants, the NC were similar in age and education but had fewer males and fewer participants with one or more ApoE4 alleles (Table 1). As expected, given ADNI’s design to yield three clearly differentiated groups, the NC had substantially higher RAVLT scores and lower (better) ADAS-cog scores at baseline; only about 5% of the NC group had scores as bad as the average MCI participant at baseline. Imaging and CSF biomarkers, however, showed substantial overlap between groups; the 25% of the NC group with the worst baseline measurement for almost all biomarkers was at or beyond the baseline median of the MCI group (Petersen 2010). Means on summaries of MRI showed smaller regional and global volumes, thinner entorhinal cortex, and larger ventricles for AD than MCI, and for MCI than NC. However, the MCI mean was typically just one standard deviation worse than the NC mean. CSF biomarker mean levels for participants who had baseline lumbar puncture were consistent with previously reported patterns with higher tau protein and lower amyloid beta associated with greater cognitive impairment as captured by clinical diagnosis. Considerable variability within the diagnostic groups led to substantial overlap across the groups.
Table 1.
Demographic, clinical, imaging and biomarker characteristics of ADNI participants at baseline, comparing Normal Control (NC), Mild Cognitive Impairment (MCI), and Alzheimer’s Disease (AD) participants. All participants with baseline data were included, but not all participants had data on each measure.
| Variables | NC | MCI | AD | P value |
|---|---|---|---|---|
| Demographic | Mean (SD) | Mean (SD) | Mean (SD) | |
| N1 | 222 | 382 | 181 | |
| Age | 76.0 (5.0) | 74.8 (7.4) | 75.1 (7.6) | 0.12 |
| Percent male | 52% | 64.00% | 53.00% | 0.005 |
| Education (years) | 16.1 (2.9) | 15.6 (3.1) | 14.8 (3.1) | < 0.001 |
| Percent Apoe4+ | 27% | 54% | 67% | < 0.001 |
| Clinical | ||||
| RAVLT score | 43.5 (8.9) | 30.8 (9.1) | 23.3 (7.5) | < 0.001 |
| ADAS-cog score | 6.2 (2.9) | 11.6 (4.5) | 18.5 (6.4) | < 0.001 |
| Imaging | ||||
| Cerebral volume2 | 0.685 (0.025) |
0.671 (0.027) |
0.659 (0.026) |
< 0.001 |
| Ventricle volume2 | 0.026 (0.012) |
0.031 (0.014) |
0.036 (0.016) |
< 0.001 |
| Hippocampal volume2 | 0.0050 (0.0005) |
0.0044 (0.0007) |
0.0040 (0.0007) |
< 0.001 |
| Entorhinal cortex thickness |
6.50 (0.60) | 5.85 (0.93) | 5.14 (0.88) | < 0.001 |
| White matter hyperintensity |
2.65 (2.48) | 2.63 (2.47) | 3.92 (7.32) | < 0.001 |
| Serum Biomarker | ||||
| Homocysteine3 | 9.93 (2.88) | 10.6 (2.90) | 10.6 (3.16) | 0.02 |
| CSF biomarkers | ||||
| CSF sample size | 112 | 189 | 98 | |
| Aβ1–424 | 206 (55) | 164 (55) | 143 (41) | < 0.001 |
| t-tau4 | 69 (28) | 102 (54) | 119 (56) | < 0.001 |
| P-tau181 4 | 25 (14) | 35 (17) | 41 (19) | < 0.001 |
| t-tau/Aβ1–42 | 0.4 (0.25) | 0.7 (0.56) | 0.9 (0.48) | < 0.001 |
| P-tau181/Aβ1–42 | 0.1 (0.12) | 0.3 (0.17) | 0.3 (0.18) | < 0.001 |
Sample sizes are based on subjects with at least one clinical follow-up assessment. Means and standard deviations are based on participants with data at baseline, so calculations for imaging and serum biomarker measures may be based on fewer subjects.
Presented as fraction of ICV.
Presented as µmol/L
Presented in pg/mL
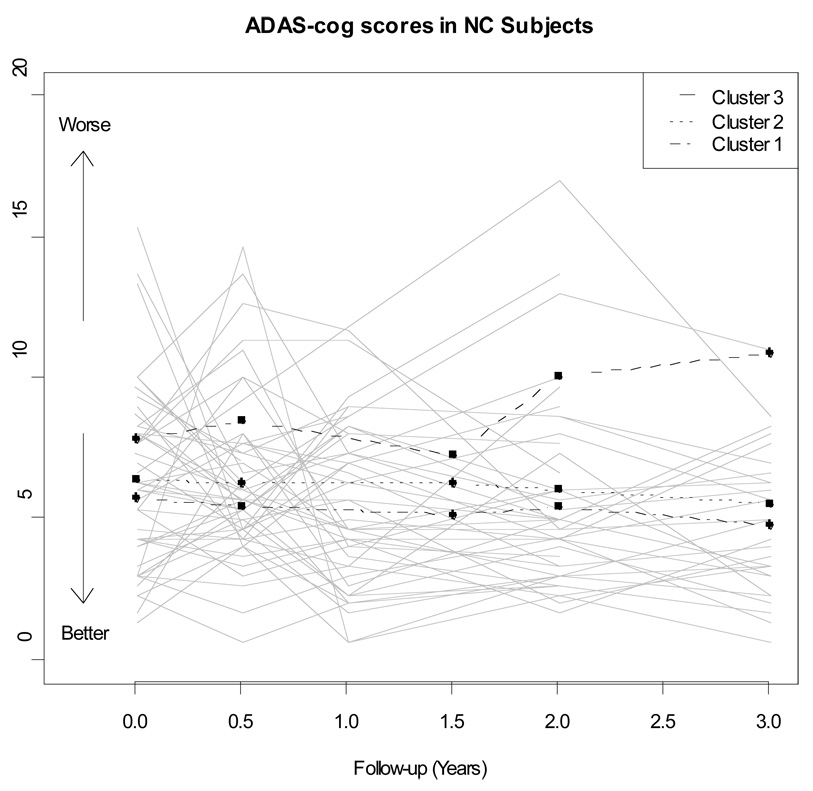
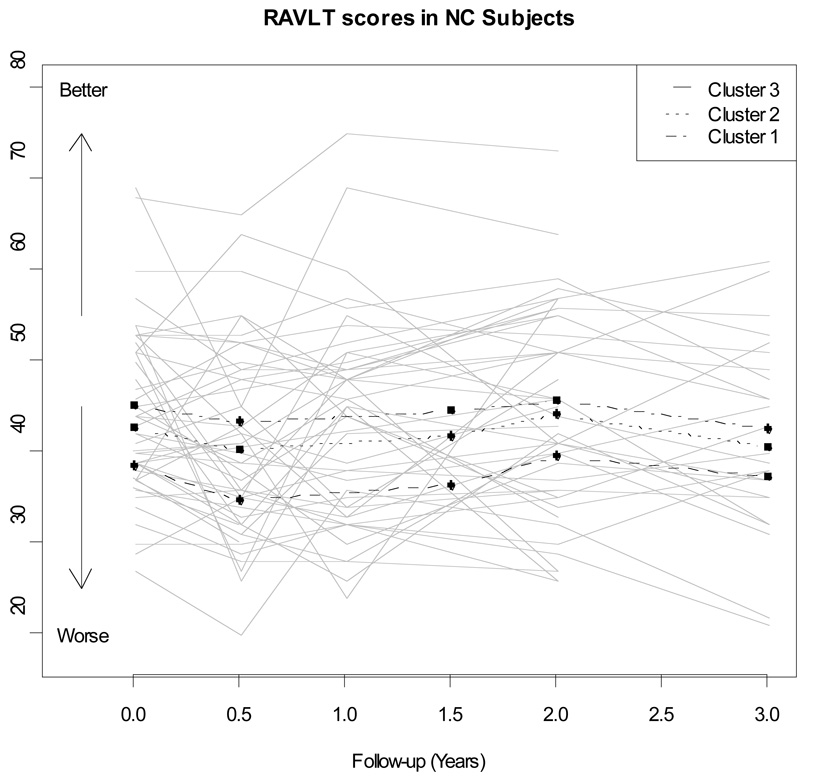
The NC showed little change in clinical status during the first 36 months of follow-up. Only 10 converted to MCI during this period, and 1 of these 10 subsequently to AD. These NC had a mean of 4.4 (SD=0.7) observations per person of ADAS-cog and a mean of 4.3 (0.8) observations of RAVLT. For most participants, there was little change either in ADAS-cog score (Figure 1) or in RAVLT performance (Figure 2), as would be expected in normal controls. Random effects models for RAVLT and ADAS-cog showed significant between-person variance in initial cognitive test scores (random intercept) and rate of change (random slope), indicating unexplained heterogeneity in trajectories and supporting exploration of baseline imaging summaries and fluid biomarkers as possible predictors of early cognitive decline.
Figure 1.
Longitudinal trajectories of ADAS-COG scores for a random sample of 50 ADNI normal controls. The average longitudinal trajectory for each of the three clusters is also presented.
Figure 2.
Longitudinal trajectories of RAVLT scores for a random sample of 50 ADNI normal controls. The average longitudinal trajectory for each of the three clusters is also presented.
3.2 Imaging and CSF measures as predictors of cognitive decline
We fitted mixed models for possible predictors of cognitive decline, separately with RAVLT and ADAS-cog trajectories as the outcome. Each imaging or fluid biomarker predictor, standardized as described above was first considered individually in a model adjusted for education, and the association of a one standard deviation (SD) higher level of the biomarker at baseline with cognitive score at baseline and with annualized rate of change in cognitive score was estimated using models with random effects for intercept and slope (Table 2). Baseline test scores were significantly correlated in univariate models with baseline levels of many MRI and CSF markers. Higher (worse) initial ADAS-cog scores were associated with lower cortical volume at baseline as measured by hippocampal volume (0.41 points better for each SD larger, P=0.02), entorhinal cortex (0.7 points better for every SD thicker, P<0.001), and ventricular volume (0.52 points worse for every SD larger, P=0.003). CSF biomarker differences at baseline were also associated with worse initial performance on the ADAS-cog; a one SD lower CSF Aβ1–42 (P=0.017) predicted about a half-point worse ADAS-cog, as did a one SD higher P-tau181/Aβ1–42 ratio (P=0.027). Higher (better) initial RAVLT scores were associated with larger cortical volume, whether measured by a one SD increase in cerebral volume (1.34 point higher score, P=0.026), hippocampal volume (1.62 point better, P=0.007), or entorhinal thickness (2.20 points better, P<0.001). A one SD increase in ventricular volume was associated with initial RAVLT scores almost two points worse (P=0.001). Higher homocysteine levels at baseline were also associated with significantly worse initial RAVLT score (1.85 points worse for a one SD higher level, P=0.001.) A one SD higher P-tau181/Aβ1–42 ratio was also associated with a 1.5 point lower baseline RAVLT score (P=0.041).
Table 2.
Univariate estimates from mixed models (random slope and intercept) of association of one-standard-deviation higher level in baseline imaging, serum, or CSF marker with cognitive test outcome at baseline and with annualized rate of change in cognitive test score. All imaging summaries were normalized to total intracranial volume before standardizing by baseline mean and standard deviation of normal controls. Predictors significant at 0.05 level are indicated with bold face.
| Predictor | Outcome: ADAS-cog score | Outcome: RAVLT score | ||||
|---|---|---|---|---|---|---|
| Estimate | SE | p-value | Estimate | SE | p-value | |
| Effects on baseline score | ||||||
| Male | −1.01 | 0.35 | 0.004 | 5.92 | 1.15 | <0.001 |
| ApoE4+ | 0.60 | 0.38 | 0.11 | 1.01 | 1.32 | 0.44 |
| Homocysteine | 0.28 | 0.17 | 0.10 | −1.85 | 0.58 | 0.001 |
| Cerebral vol | −0.31 | 0.17 | 0.07 | 1.34 | 0.60 | 0.026 |
| Hipp vol | −0.41 | 0.17 | 0.02 | 1.62 | 0.60 | 0.007 |
| Vent vol | 0.52 | 0.17 | 0.003 | −1.96 | 0.59 | 0.001 |
| Entorhin thick | −0.71 | 0.17 | <0.001 | 2.20 | 0.60 | <0.001 |
| White matter | 0.09 | 0.18 | 0.63 | −0.06 | 0.65 | 0.93 |
| t-tau | 0.20 | 0.22 | 0.36 | −0.27 | −0.77 | 0.73 |
| P-tau181 | 0.30 | 0.22 | 0.18 | −1.21 | 0.77 | 0.11 |
| Aβ1–42 | −0.52 | 0.22 | 0.017 | 0.60 | 0.76 | 0.43 |
| t-tau/Aβ1–42 | 0.36 | 0.22 | 0.10 | −0.73 | 0.76 | 0.33 |
| P-tau181/Aβ1–42 | 0.49 | 0.22 | 0.027 | −1.57 | 0.76 | 0.041 |
| Effects on annual change | ||||||
| Male | 0.05 | 0.16 | 0.76 | 0.47 | 0.39 | 0.23 |
| ApoE4+ | 0.29 | 0.18 | 0.10 | 0.21 | 0.44 | 0.63 |
| Homocysteine | 0.16 | 0.08 | 0.047 | 0.10 | 0.20 | 0.64 |
| Cerebral vol | −0.09 | 0.08 | 0.28 | 0.14 | 0.20 | 0.50 |
| Hipp vol | −0.16 | 0.08 | 0.046 | 0.17 | 0.20 | 0.38 |
| Vent vol | 0.08 | 0.08 | 0.31 | 0.01 | 0.20 | 0.97 |
| Entorhin thick | −0.05 | 0.08 | 0.55 | −0.06 | 0.20 | 0.76 |
| White matter | −0.01 | 0.08 | 0.90 | −0.20 | 0.19 | 0.29 |
| t-tau | 0.02 | 0.11 | 0.90 | 0.05 | 0.25 | 0.83 |
| P-tau181 | 0.21 | 0.11 | 0.048 | 0.08 | 0.24 | 0.73 |
| Aβ1–42 | −0.09 | 0.11 | 0.42 | −0.27 | 0.25 | 0.29 |
| t-tau/Aβ1–42 | 0.08 | 0.12 | 0.50 | 0.26 | 0.27 | 0.33 |
| P-tau181/Aβ1–42 | 0.23 | 0.11 | 0.036 | 0.16 | 0.24 | 0.52 |
Baseline imaging and biomarker scores, though, had little predictive impact on ADAS-cog or RAVLT change, assuming cognitive test score changes are linear over the period of observation. A one SD higher homocysteine level was associated with a 0.16 point per year faster worsening in ADAS-cog performance (P=0.047), while a one SD smaller hippocampus was associated with about the same magnitude of difference in rate of ADAS-cog decline (P=0.046). A one SD higher P-tau181 or P-tau181/Aβ1–42 ratio was associated with about a 0.2 point faster annual worsening of ADAS-cog. These effects are modest; it would take about 5 years at this rate to account for a single additional point worse on the ADAS-cog. Adjusting P-values in Table 2 for multiple comparisons, as would be done for selection of best predictors, would further reduce the evidence for utility of individual biomarkers as predictors of cognitive baseline or decline.
In multivariate models adjusted for education, no individual baseline measure of cortical volume or CSF remained a significant predictor of cognitive decline, either for the ADAS-cog or the RAVLT.
These analyses found little evidence of associations between either the individual standardized imaging and biomarker measures of this study or weighted sums of the measures and subsequent cognitive decline in NC, over a period of up to 3 years. These results served as a comparison for analyses which focused on an effort to identify individuals with a distinctive profile of biomarkers that separated them out from typical NC, without reference to their cognitive outcomes, and then to assess the behavior of cognitive trajectories for these subgroups.
3.3. Cluster analysis of NC using baseline imaging and biomarker measures
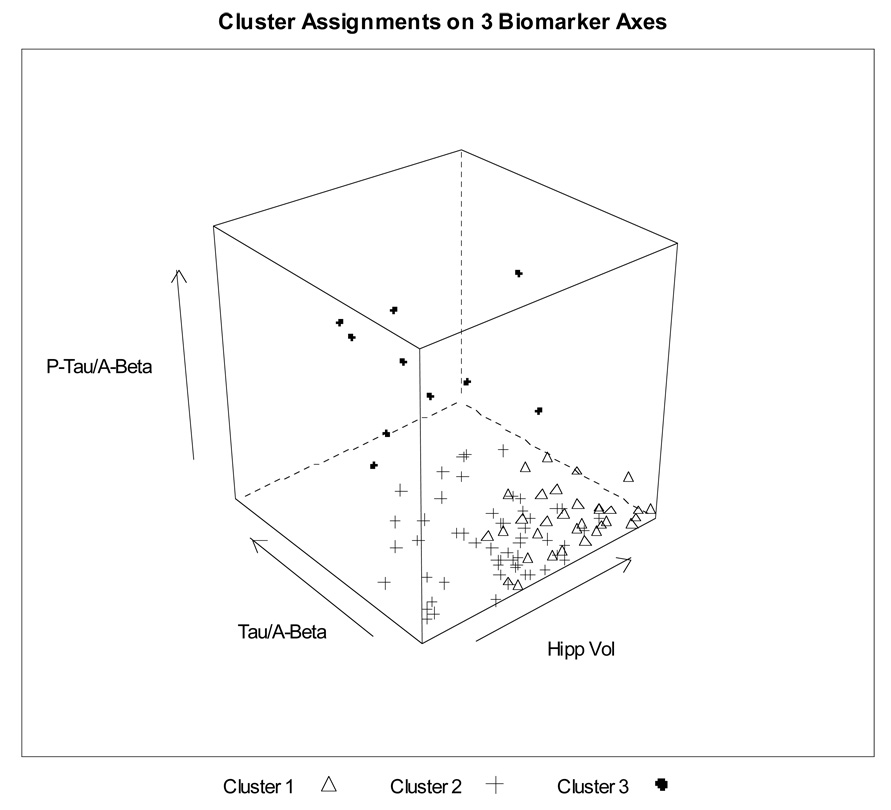
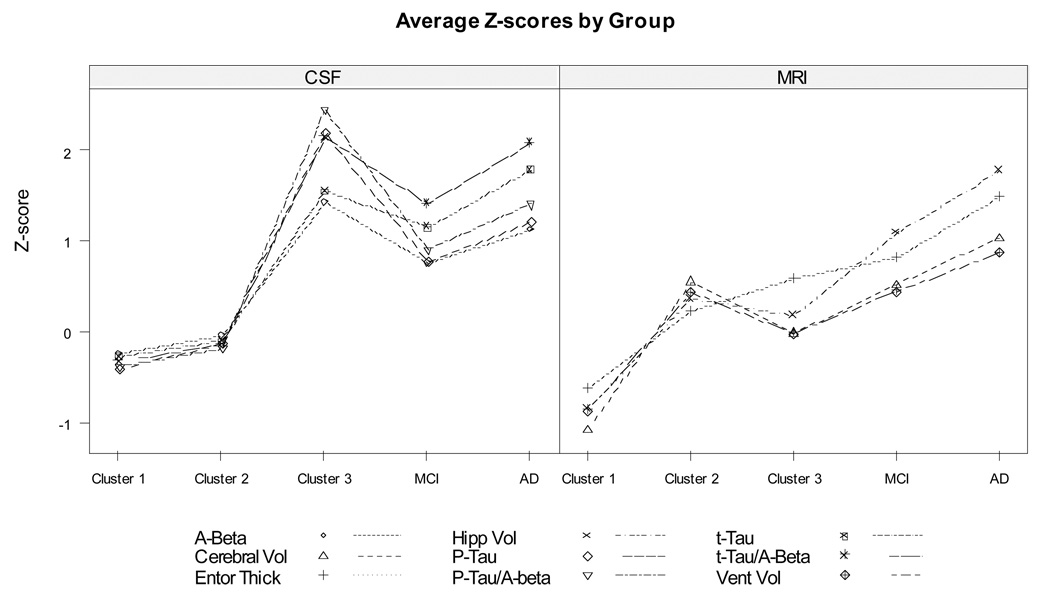
Cluster analysis of the NC using the eleven imaging and biomarker measures (96 complete cases) identified three distinct clusters, of size 32, 54 and 10. The first cluster was relatively compact, while the third cluster was well separated in multidimensional space even when individual measurements might fall in the normal range (Figure 3, projection from 11 dimensions to 3, projection chosen to separate cluster centers for best visualization of cluster locations). The second cluster was intermediate, but closer to the first, “healthier” cluster. We calculated the center (11-dimensional point representing the average across all people in the cluster for each marker) for each of the three clusters and for the MCI and AD groups (Table 3). Note that the MCI and AD data were not used to develop the clusters among the NC, but are included for comparison. Because all of the markers were standardized using the NC mean and standard deviation, components of the centers are in units of NC SD. The center for Cluster 3, the atypical group, is 1.5–2.5 SD away from the normal mean for all of the baseline CSF biomarkers. The average person in this cluster has a smaller cortical volume than the compact, “healthier” Cluster 1 but not as small as a typical individual in Cluster 2. Overall, Cluster 3 looks very close to the AD group for CSF and closer to MCI than to normal for the MRI variables (Figure 4). We also calculated Euclidean distances for the centers of each of the three clusters to the MCI and AD groups. Cluster 3, the “atypical” cluster, was closer to the MCI and AD than to either of the other two NC clusters (2.6 and 2.8 SD versus 4.8 and 5.6 SD). Cluster 2 was equidistant from Cluster 1, the “typical” normal group, and MCI (2.7SD). Cluster 1 was furthest from AD (6.1 SD). The MCI and AD groups were closer together than any other two groups (1.7 SD).
Figure 3.
Three-dimensional representation of cluster assignments for 96 ADNI normal controls, based on unsupervised clustering using eleven baseline MRI and CSF markers and serum homocysteine, without use of clinical data to define clusters. This representation is shown on three axes of the original 11-dimensional data space, chosen to maximize separation of cluster centers for clearest visualization on paper of the cluster locations.
Table 3.
Marker values at center of NC clusters, compared to center of MCI and AD groups. All values are in SD units away from the overall NC mean.
| Variable name | Location of center of group, in SD from NC mean | ||||
|---|---|---|---|---|---|
| NC Cluster 1 | NC Cluster 2 | NC Cluster 3 | MCI | AD | |
| Cerebral vol | 1.08 | −0.57 | 0.00 | −0.54 | −1.05 |
| Hippo vol | 0.84 | −0.38 | −0.19 | −1.11 | −1.79 |
| Vent vol | −0.86 | 0.45 | 0.00 | 0.46 | 0.88 |
| Entor thick | 0.62 | −0.24 | −0.60 | −0.83 | −1.50 |
| WMH | −0.24 | 0.12 | −0.27 | −0.02 | 0.50 |
| Homocysteine | −0.44 | 0.23 | 0.10 | 0.23 | 0.23 |
| t-tau | −0.26 | −0.09 | 1.56 | 1.17 | 1.80 |
| Ab1–42 | 0.22 | 0.03 | −1.44 | −0.77 | −1.15 |
| P-tau181 | −0.39 | −0.12 | 2.19 | 0.79 | 1.22 |
| t-tau/Ab1–42 | −0.32 | −0.13 | 2.15 | 1.43 | 2.10 |
| P-tau181/Ab1–42 | −0.37 | −0.17 | 2.45 | 0.92 | 1.42 |
Figure 4.
Difference for each biomarker of cluster means from the mean for all ADNI normal controls, standardized by normal control standard deviation, compared to distances for ADNI MCI and AD groups. Sign of standardized differences was reversed for all variables except Aβ1–42 and ventricular volume so that high values (top of figure) represent worse biomarker measurements.
Alternative clustering techniques generally led to similar cluster assignments, particularly with regard to the atypical cluster (Cluster 3). Among methods that did not result in clusters with too few individuals, the results were fairly consistent with the results from Ward’s method, as measured by Rand’s statistic (average 0.64, range = [0.45, 1]) and by the tendency to locate the same atypical cluster (over half kept at least 9 of the 10 members of the atypical cluster together, most of these locating exactly the same cluster of 10). Using just MRI summary measures and omitting CSF biomarkers approximately doubled the available sample size but led to less clear separation between clusters, consistent with hypotheses that changes in amyloid levels precede volumetric change (Petersen 2009, Jack 2010).
We fitted mixed effects models to assess the predictive value of cluster membership at baseline on subsequent cognitive change (Table 4). Members of Cluster 3, the profile farthest from a typical NC, had baseline scores 2 points worse on average than the typical individuals in Cluster 1 for the ADAS-cog (P=0.021) and 9 points worse on average for the RAVLT (P=0.002). The atypical baseline marker profile of Cluster 3 was also accompanied by a faster rate of worsening on the ADAS-cog (1.25 points worse per year, P<0.001). The rate of decline for RAVLT, while of similar magnitude, was not statistically significant (P=0.17). A permutation test was performed to determine the likelihood of a different clustering with groups of these sizes resulting in an equal or smaller P-value for the cluster variable indicating decline in ADAS-cog (P = 0.0025). Out of 5000 random clusterings, only 21 (less than 1%) resulted in a P-value ≤ 0.0025.
Table 4.
Mixed-model estimates of effects of cluster membership based on baseline imaging and biomarker measures on cognitive test outcome at baseline and on annualized rate of change. Predictors significant at 0.05 level are indicated with bold face. Cluster 3 is the most atypical and Cluster 2 closer to the “typical” Normal Control.
| Variable | ADAS-cog | RAVLT | ||||
|---|---|---|---|---|---|---|
| Estimate | SE | p-value | Estimate | SE | p-value | |
| Effects at baseline | ||||||
| Cluster 3 | 1.96 | 0.85 | 0.021 | −9.08 | 2.84 | 0.002 |
| Cluster 2 | 0.85 | 0.52 | 0.10 | −3.45 | 1.73 | 0.048 |
| Effects on annual change | ||||||
| Cluster 3 | 1.25 | 0.37 | <0.001 | 1.18 | 0.87 | 0.17 |
| Cluster 2 | 0.03 | 0.22 | 0.88 | 0.32 | 0.51 | 0.54 |
Further characterization of the three clusters found that 50% of the members of Cluster 3 had at least one ApoE4 allele, while 25% and 22% of the members of Clusters 1 and 2 were E4+. However, the association of cluster membership with worse baseline performance and more rapid deterioration of ADAS-cog performance was not accounted for by ApoE genotype or by participant age; results were not materially changed by including in the model the effects of these factors on baseline cognition and rate of change. Cluster size and composition were sensitive at the margins to choice of measures included and algorithms used but were overall quite robust.
Among the 96 people assigned to clusters, only 7 people converted to MCI, 2 in Cluster 3 (20%), 4 in Cluster 2 ( 8%) and 1 in Cluster 1 ( 3%). These numbers, while too small for statistical analysis, indicate the importance of additional follow-up of the NC.
4. Discussion
Our analyses in a large, well-characterized cohort of normal controls found heterogeneity in imaging summary measures and fluid biomarker summaries, indicating that the biomarker profile could not be encompassed by a single, compact, well-defined set of boundaries. Instead, the patterns were suggestive of several distinct clusters of individuals, even though these individuals were cognitively intact at baseline and showed very little clinical progression over three years of follow-up. Regression models suggested that baseline levels of some measures of brain size and CSF biomarkers are associated with cross-sectional differences in cognitive performance, and that increased P-tau181 and the P-tau181/Aβ1–42 ratio may foreshadow slightly more rapid declines in ADAS-cog. The effects, however, were modest, and not improved in multivariate regression analysis. Cluster analysis identified three distinct groups of individuals, based solely on their baseline imaging and biomarker measures without reference to cognitive status or change. One group in particular, comprising about 10% of the NC group, was well separated from the bulk of the NC and lay closer to the centers of the MCI and AD groups, even when some individual marker measurements might be closer to the center for typical NC. This subgroup had strikingly lower baseline scores on the RAVLT, significantly worse scores on the ADAS-cog, and a significantly more rapid deterioration on the ADAS-cog than the typical NC. Membership in this cluster was associated with annual cognitive decline approximately 5 times as rapid as that predicted for a person one standard deviation worse than average on the strongest individual marker. A second group, centered between the atypical group and the more typical NC group, showed structural MRI profiles closer to the MCI group and had somewhat worse baseline performance but showed little difference in cognitive trajectory compared to typical NC. The division into distinct clusters was robust across several clustering algorithms, with the same people consistently identified as a distinct group whose center placed them closer to the profile of MCI and AD. As a complementary approach, unsupervised regression trees were used to generate a proximity matrix which was then plotted on multidimensional scaling axes and colored by cluster membership. The plot showed almost no mixing of clusters, supporting the clusters identified with agglomerative clustering despite the fact that methodologies have almost nothing in common. This suggests that the clusters are not simply an artifact of the clustering process and may instead represent meaningful structure in the data.
Our findings are consistent with previous work suggesting P-tau181, Aβ1–42, and their ratio as among the measures most closely reflecting early preclinical neurobiological changes [Stomrud et al. 2007, Shaw et al. 2009]. We also found evidence for correlation between brain atrophy measures and cross-sectional measures of cognitive performance, even in this very high-functioning cohort. None of these measures, however, had substantial predictive power for future cognitive decline, either taken individually or as linear combinations. Little previous work has explored the multi-dimensional structure of imaging and CSF biomarkers in clinically normal older people. A recent study (Fagan 2009) assessed correlations between CSF measures and normalized whole brain volume in cognitively normal elderly subjects and found that Abeta, but not Tau or P-tau181, correlated inversely with whole-brain volume in elderly control subjects, while Tau and P-tau181 correlated in very mild and mild AD. Such findings have suggested a hypothetical model of the sequence of biomarker changes in the neurobiological process leading to AD, with amyloid changes occurring very early, followed by tau pathology, volumetric and metabolic decline, and with a very gradual onset of measurable cognitive decline, reaching the diagnosis of dementia only very late in the cascade (Jack, 2010). Our analyses offer additional support for this conceptual model. We identified a subgroup of normal controls, based solely on biomarker profiles, who were notable primarily for amyloid-related CSF abnormalities. Not only were people in this group much closer to the AD group in their CSF profile, but they also had a striking pattern of ADAS-cog decline over 2–3 years, despite being clinically well within the normal range to start. Figure 4 suggests strong similarity to the conceptual model proposed in Jack. Clustering based just on MRI measures failed to isolate such a distinctive subgroup. Our findings are consistent with Cluster 3 representing a group of people who have already progressed so far in amyloid deposition that they are starting to experience the earliest signs of clinical decline, even though their scores remain, for the most part, within the wider normal range and do not yet verge on dementia.
Methods looking at best classification strategies to predict conversion among NC are not yet applicable in ADNI, as there are so few conversions. We are not aware of any work with data mining or other dimension-reduction strategies based on rate of cognitive change as an outcome. Our study represents a first attempt to seek structure based strictly on the imaging and fluid biomarker measures, and to map the observed heterogeneity to distinct subgroups.
One limitation of our approach is that the CSF markers were available only for half the participants, thus reducing the sample size available for clustering. This, in turn, led to a fairly small though stable “atypical” cluster of about 10% of the NC with CSF. The remaining participants split with every clustering algorithm we examined, with some uncertainty in locating the border between a compact, “typical” group and a second group leaning toward the “atypical” group. A larger sample size might have allowed better definition of the margins, as well as the variables most important for identifying the “atypical” cluster. A second limitation is that length of follow-up and the generally healthy nature of this cohort led to only a few conversions to MCI, precluding our assessment of the predictive value of the cluster for change in clinical status. A third limitation is that we used only a fraction of the many potential biomarkers, especially from the imaging summaries, and we included 5 participants with some imaging quality control problems whose effect on our findings appeared minimal in the primary analysis. Our selection was based on previous literature, but may have omitted variables with better prognostic power in the ADNI cohort. A fourth limitation is the approach to standardization of the imaging variables. Dividing by ICV may diminish differences between individuals or groups, although the cluster definitions were more dependent on the CSF biomarkers. Finally, we have not used ADNI’s FDG PET or PiB imaging data because the number of participants with complete data on the MRI, CSF and PET imaging data would be too small for analysis.
Our approach has several notable strengths. First, the ADNI data offer one of the largest, uniformly ascertained databases available, with standardized protocols not only for data collection but also for image processing and biomarker specimen assays.
Second, our approach makes use of 36 months of follow-up with standardized cognitive testing. Third, our clustering strategy was “unsupervised”, that is, based on the imaging and biomarker data without reference to cognitive endpoints or diagnostic categories, so that we address directly the question of whether a strikingly unusual profile on these measures may foreshadow clinical decline, without letting the outcome define the profile of interest. Notably, a recent study on ADNI CSF biomarkers used another unsupervised learning approach that identified nearly the same CSF Aβ cut point as Shaw et al 2009 wherein this cut point was established in subjects with autopsy confirmed diagnoses of AD thereby demonstrating the power of unsupervised analytical approaches (de Meyer et al, 2010). Supervised techniques such as regression trees or discriminant analysis offer a complementary approach, focused on identifying subgroups of clinical interest based on their outcome, but would require cross-validation, potentially problematic in our sample of under 100 NC with little clinical change as yet. In comparison to univariate or multivariate regression models, our approach takes advantage of the correlated relationship among the biomarkers, rather than being limited by this interrelationship. This strength of the clustering approach may account for the improvement in our ability to detect associations with future cognitive change, compared to the more traditional univariate and multivariate regression models.
Our findings indicate that not just individual abnormalities, but a distinctive pattern of imaging and biomarker deviations from typical healthy older adults may be an early warning sign of neurobiological pathology. Additional follow-up would help to establish the prognostic value for longer-term cognitive decline and conversion to MCI and eventually to AD. A larger sample, with CSF on all participants, would help to confirm the cluster pattern for our “atypical” group and better define the boundary between the other two clusters. In addition, the recruitment of early MCI participants, as currently underway in the recently funded ADNI-GO grant, will offer an opportunity to test whether our atypical group have a profile similar to the earliest stages of clinically-identified impairment. Longitudinal analysis of the imaging and biomarker measures can also address whether the distance of the atypical cluster relative to the center for typical NC is increasing over time, and whether the location of the intermediate cluster shifts toward the atypical cluster’s baseline location over time.
5. Conclusions
A pattern of imaging and biomarker deviations can identify a subgroup of normal controls distinct from typical cognitively normal older participants. This subgroup, defined based strictly on imaging and biomarkers, had substantially worse cognitive performance at baseline and a more rapid rate of deterioration on the ADAS-cog than the typical NC. Additional follow-up and analysis is warranted to investigate the long-term clinical implications and to characterize the imaging and biomarker trajectories of subgroups over time.
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co.,Inc., Novartis AG, Pfizer Inc, F. Hoffman-La Roche, Schering-Plough, Synarc, Inc., and Wyeth, as well as non-profit partners the Alzheimer's Association and Alzheimer's Drug Discovery Foundation, with participation from the U.S. Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129, K01 AG030514, and the Dana Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu\ADNI). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators is available at www.loni.ucla.edu\ADNI\Collaboration\ADNI_Manuscript_Citations.pdf.
Conflict of interest
None reported for JN, JB, LB, DH.
Contributor Information
Jasmine Nettiksimmons, Email: janettik@ucdavis.edu.
Danielle Harvey, Email: djharvey@ucdavis.edu.
Laurel Beckett, Email: labeckett@ucdavis.edu.
References
- Barber R, Scheltens P, Gholkar A, Ballard C, McKeith I, Ince P, Perry R, O’Brien J. White matter lesions on magnetic resonance imaging in dementia with Lewy bodies, Alzheimer’s disease, vascular dementia, and normal aging. British Medical Journal. 1999;67(1):66–72. doi: 10.1136/jnnp.67.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DA, Wilson RS, Schneider JA, Evans DA, Beckett LA, Aggarwal NT, Barnes LL, Fox JH, Bach J. Natural history of mild cognitive impairment in older persons. Neurology. 2002;59(2):198–205. doi: 10.1212/wnl.59.2.198. 23. [DOI] [PubMed] [Google Scholar]
- Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer’s disease. Alzheimer’s and dementia. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- Carlson NE, Moore MM, Dame A, Howieson D, Silbert LC, Quinn JF, Kaye JA. Trajectories of brain loss in aging and the development of cognitive impairment. Neurology. 2008;70:828–833. doi: 10.1212/01.wnl.0000280577.43413.d9. [DOI] [PubMed] [Google Scholar]
- Clark CM, Davatzikos C, Borthakur A, Newberg A, Leight S, Lee VM, Trojanowski JQ. Biomarkers for early detection of Alzheimer pathology. Neurosignals. 2008;16(1):11–18. doi: 10.1159/000109754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer G, Shapiro F, Vanderstichele H, Vanmechelen E, Engleborghs B, De Deyn P-P, Coart E, Hanson O, Minthon L, Zetterberg H, Blennow K, Shaw LM, Trojanowski JQ Alzheimer’s Disease Neuroimaging Initiative. Diagnosis-independent Alzheimer’s disease biomarker signature in cognitively normal elderly people. Arch. Neurol. 2010 doi: 10.1001/archneurol.2010.179. and the, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DA, et al. Estimated prevalence of Alzheimer’s disease in the United States. The Milbank Quarterly. 1990;68(2):267–289. [PubMed] [Google Scholar]
- Fagan AM, Head D, Shah AR, Marcus D, Mintun M, Morris JC, Holtzman DM. Decreased cerebrospinal fluid Ab42 correlates with brain atrophy in cognitively normal elderly. Ann Neurol. 2009;65:176–183. doi: 10.1002/ana.21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennema-Notestine C, Hagler DJ, Jr, McEvoy LK, Fleisher AS, Wu EH, Karow DS, Dale AM ADNI. Structural MRI biomarkers for preclinical and mild Alzheimer's Disease. Human Brain Mapping. 2009;30:3238–3253. doi: 10.1002/hbm.20744. and the. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel H, Burger KB, Teipel SJ, Bokde ALW, Zetterberg H, Blennow K. Core candidate neurochemical and imaging biomarkers of Alzheimer’s disease. Alzheimer’s & Dementia: The Journal of the Alzheimer’s Association. 2007;4(1):38–48. doi: 10.1016/j.jalz.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Harvey RJ, Skelton-Robinson M, Rossor MN. The prevalence and causes of dementia in people under the age of 65 years. J Neurol Neurosurg Psychiatry. 2003;74:1206–1209. doi: 10.1136/jnnp.74.9.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol. 2003;60:1119–1122. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen P, Weiner MW, Petersen RC, Trojanowski JQ. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- Maddalena A, Papassotiropoulos A, Muller-Tillmanns B, Jung HH, Hegi T, Nitsch RM, Hock C. Biochemical Diagnosis of Alzheimer Disease by Measuring the Cerebrospinal Fluid Ratio of Phosphorylated tau Protein to beta-Amyloid Peptide42. Archives of Neurology. 2003;60(9):1202. doi: 10.1001/archneur.60.9.1202. [DOI] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Toga AW, Jack CR, Schuff N, et al. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer’s disease, mild cognitive impairment, and elderly controls. Neuroimage. 2009;45(1S1):3–15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Tro janowski JQ, Toga AW, Beckett L. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clinics of North America. 2005;15(4):869–877. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimers disease: The Alzheimers Disease Neuroimaging Initiative (ADNI) Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2005;1(1):55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor SM, Rupsingh R, Borrie M, Smith M, Accomazzi V, Wells JL, Fogarty J, Bartha R. Ventricular enlargement as a possible measure of Alzheimer’s disease progression validated using the Alzheimer’s disease neuroimaging initiative database. Brain. 2008;131:2443–2454. doi: 10.1093/brain/awn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Robers RO, Knopman DS, Boeve BF, Geda YE, Ivnik RJ, Smith GE, Jack CR. Mild Cognitive Impairment: Ten years later. Archives of Neurology. 2009;66:1477–1455. doi: 10.1001/archneurol.2009.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, Jack CR, Jr, Jagust WJ, Shaw LM, Toga AW, Trojanowski JQ, Weiner MW Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74:201–209. doi: 10.1212/WNL.0b013e3181cb3e25. and the. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria; 2009. ISBN 3-900051-07-0. [Google Scholar]
- Rand WM. Objective criteria for the evaluation of clustering methods. Journal of the American Statistical Association. 1971;66:846–850. [Google Scholar]
- SAS Institute, Inc. SAS/STATÒ Version 9.0. Cary, NC: SAS Institute, Inc.; 2004. [Google Scholar]
- Schwarz C, Fletcher E, DeCarli C, armichael O. Fully-automated white matter hyperintensity detection with anatomical prior knowledge and without FLAIR. Inf Process Med Imaging. 2009;21:239–251. doi: 10.1007/978-3-642-02498-6_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, Korecka M, Clark CM, Lee VM-Y, Trojanowski JQ. Biomarkers of neurodegeneration for diagnosis and monitoring therapeutics. Nat. Rev. Drug Discovery. 2007;6:295–303. doi: 10.1038/nrd2176. [DOI] [PubMed] [Google Scholar]
- Shaw LM. PENN Biomarker Core of the Alzheimer’s Disease Neuroimaging Initiative. Neurosignals. 2008;16:19–23. doi: 10.1159/000109755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen P, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, Dean R, Siemers E, Potter W, Lee VM-Y, Trojanowski JQ Alzheimer's Disease Neuroimaging Initiative. Cerebrospinal fluid biomarker signature in Alzheimer’s Disease Neuroimaging Initiative subjects. Ann. Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. and the. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD. The worldwide challenge of the dementias: a role for B vitamins and homocysteine? Food and nutrition bulletin. 2008;29(2 Suppl):S143. doi: 10.1177/15648265080292S119. [DOI] [PubMed] [Google Scholar]
- Stomrud E, Hansson O, Blennow K, Minthon L, Londos E. Cerebrospinal fluid biomarkers predict decline in subjective cognitive function over 3 years in healthy elderly. Dementia Geriatr Cogn Disord. 2007;24:118–124. doi: 10.1159/000105017. [DOI] [PubMed] [Google Scholar]
- Ward J. Hierarchical Grouping to optimize an objective function. Journal of American Statistical Association. 1963;58(301):236–244. [Google Scholar]