Abstract
The genome of the sole remaining unsequenced member of species A, human adenovirus type 18 (HAdV-A18), has been sequenced and analyzed. Members of species A are implicated as gastrointestinal pathogens and were shown to be tumorigenic in rodents. These whole genome and in silico proteome data are important as references for re-examining and integrating earlier work and observations based on lower resolution techniques, such as restriction enzyme digestion patterns, particularly for hypotheses based on pre-genomics data. Additionally, the genome of HAdV-A18 will also serve as reference for current studies examining the molecular evolution and origins of human and simian adenoviruses, particularly genome recombination studies. Applications of this virus as a potential vector for gene delivery protocols may be practical as data accumulates on this and other adenovirus genomes.
Keywords: human adenovirus, molecular evolution, species A, oncogenesis
Introduction
Human adenoviruses (HAdV) are partitioned into seven species, based on characteristics including biochemical and genome properties and homologies, tissue tropism and immunochemical examination of serum neutralizing and hemagglutinating epitopes that are found on the virion capsid proteins. Species A contains three members, HAdV-A12, -A18 and -A31, that generated earlier interest due to their oncogenic capabilities, with HAdV-A12 and -A18 shown to cause tumors in newborn hamsters (Huebner et al., 1962; Trentin et al., 1962) and mice (Yabe et al., 1964). The precise basis for this oncogenesis remains unknown; however, the HAdV-A12 E1A proteins have been shown to repress class I major histocompatibility complex expression in transformed rodent cells, and these E1A proteins modulate class I gene expression by similar mechanims in both transformed rodent and human cells (Vasavada et al., 1986). Having the complete genomes of HAdV-A18 and A31 (Hofmayer et al., 2009) allows additional detailed analyses by researchers.
HAdV-A18 was originally isolated from an anal swab of a child with Niemann-Pick syndrome and characterized in the 1950s (Rowe et al., 1956; Rowe et al., 1958). Its role in human health is not well-characterized however another species A member, HAdV-A31, isolated originally from the stool of a healthy child (Pereira et al., 1965), has been shown to be causative agents in gastrointestinal disease with the identification of subsequent field isolates (Adrian and Wigand, 1989; Adrian et al., 1987; Brown et al., 1984; Hammond et al., 1985; Kidd et al., 1982). Several other HAdV species are associated with gasteroenteritis and are isolated from stool samples as well. Among these, the members of species F (de Jong et al., 1983; Uhnoo et al., 1984) are of particular importance because they are the genomes most closely similar to those from species A, as reported in the phylogenetic and percent identity data of this report. The association of HAdV-A31 with gastroenteritis, its genome identity with HAdV-A18 and the close relationship between the HAdV-F and HAdV-A species all suggest a potential pathogenic role as a gastroenteritis agent for HAdV-A18.
Reports of HAdV coinfections (Vora et al., 2006), coupled with the isolation of HAdV mixtures and HAdV types (De Jong et al., 1999) from immunocompromised individuals, suggest the possibility of the host acting as a bioreactor, and this provides a molecular pathway for HAdV type evolution and pathogen emergence by recombination. Novel types of HAdV, isolated as emergent pathogens causing highly contagious outbreaks, have reported recently. These have been characterized using a genomics and bioinformatics approach to take advantage of the high-resolution methodologies (Ishiko and Aoki, 2009; Walsh et al., 2009; Yang et al., 2009). The pathogens include a respiratory, HAdV-B55 (Yang et al., 2009), three ocular pathogens, HAdV-D53 and D54, and a reanalysis of HAdV-D22 (Ishiko and Aoki, 2009; Robinson et al., 2009; Walsh et al., 2009). Reexamination of several prototype strains of HAdV respiratory pathogens archived at the American Type Culture Collection (ATCC; Manassas, VA) by genome sequencing and bioinformatic analysis show a role of recombination in their genesis (HAdV-E4, manuscript in preparation; HAdV-C6, manuscript in preparation; HAdV-B16, manuscript in preparation). However, currently there are limited numbers of whole HAdV genome sequences available in GenBank, handicapping a full comprehensive genomic analysis, particularly of recombination events. In several recombination analysis profiles of HAdV-A18, a suggestion of recombination is present but the data are inconclusive perhaps lacking the appropriate genome sequence[s] for comparison. An example of this problem is demonstrated in the recently characterized emergent keratoconjuntivitis pathogen, HAdV-D53, in which a span of recombinant sequences showed similarity to an unsequenced and/or unknown HAdV genome. This reflects also the recent applications of these tools to the HAdV genomes, as well as the limited number of genomes archived.
To support and enhance continuing computational and comparative studies on the molecular evolution and pathoepidemiology of HAdV, the genome and analysis of HAdV-A18 is presented. Re-examining the oncogenesis data reported for species A genomes in the context of these newer genome data will allow further understanding of the biology of these rarely reported HAdVs.
Materials and Methods
HAdV-A18 was obtained from the collection at The Viral and Rickettsial Disease Laboratory (VRDL; California Department of Public Health), archived as TC-81190. The passage history of this stock is as follows: five times through HeLa cells and eight time through KB cells at the National Institutes of Health, where the virus was first cultured; and followed by passage through HeLa cells twice and human fetal diploid kidney cells four times at VRDL.
Preparation of HAdV-A18 for genome sequencing
For DNA production and collection, the HAdV-A18 stock was passed once onto monolayers of A549cells in 25 cm2 flasks to verify cytopathic effect, and then subsequently amplified in monolayers ofA549 cells in 75 cm2 flasks for intracellular viral DNA extraction, using a protocol described by Kajon and modified from Shinagawa (Kajon and Erdman, 2007). Cells collected from this passage were concentrated two-fold, and viral DNA was extracted and further processed using a MagnaNA Pure LC DNA Isolation Kit I (Roche, Inc.; Indianapolis, IN), in accordance to the manufacturers’ instructions.
Amplification and sequencing of the HAdV-A18 genome
To amplify regions of HAdV-A18, primers based on the conserved adenovirus sequences of HAdV-A12 were designed to flank the known sequences. All amplicons were sequenced using a primer-walking strategy and the Sanger sequencing chemistry with ladders resolved on an ABI3730x.
Sequence analysis and genome annotation
DNA sequence data were parsed and assembled using SeqMan software (Lasergene 8, Madison, WI). In toto, the genome assembly contained 1718 high-quality reads with an average length of 300 bps. Nucleotide coverage for both strands of the genome was 8-fold.
The annotated sequence of HAdV-A18 has been deposited in GenBank and is accessioned as GU191019. For the computational analyses, the following HAdV genomes were used: HAdV-A18 (reported here), HAdV-A12 (AC_000005), HAdV-A31 (Heim, personal communication; AM749299), HAdV-B3 (AY599834), HAdV-B7 (AY594255), HAdV-B11 (AY163756), HAdV-B14 (AY803294), HAdV-C2 (AC_000007), HAdV-C5 (AC_000008), HAdV-D9 (AJ854486), HAdV-D53 (FJ169625), HAdV-E4 (AY594253), SAdV-E25 (AC_000011), HAdV-F40 (NC_001454), HAdV-F41 (DQ315364), SAdV-G1 (NC_006879) and HAdV-G52 (DQ923122).
Bioinformatic analyses were performed using protocols and net-accessible software tools similar to those described in earlier publications (Walsh et al., 2009; Seto et al., 2009): 1) MAFFT (http://www.ebi.ac.uk/Tools/mafft/) (Katoh et al., 2002) was used to compute alignments of nucleotide sequences; 2) nucleotide percent identities were calculated using the UCSF Chimera package (Pettersen et al., 2004); 3) Artemis (Rutherford et al., 2000) was used to view, annotate and compare genomes; 4) percent identity values for annotated proteins were calculated using a BioJava (Holland et al., 2008) implementation of a Needleman-Wunsch algorithm; 5) whole genomes were aligned using MEGA (Tamura et al., 2007) 6) phylogenetic bootstrapped neighbor-joining trees (1000 replicates) were constructed using MEGA4 (Tamura et al., 2007); 7) recombination analysis was performed using SimPlot (Lole et al., 1999) and 8) in silico proteome analyses were performed using an in-house beta software tool to calculate percent identities and applying a virtual 2D gel electrophoresis analysis to compare protein charge and size (JVirGel software; http://www.jvirgel.de) (Hiller et al., 2006).
Results and Discussion
Genome
There are three members of the HAdV-A species, originally differentiated by serum neutralization and clustered with respect to biological and genome-derived similarities. All are now sequenced, annotated and analyzed. SmaI in silico restriction enzyme digest patterns are consistent with previously reported data, with additional small sized bands visible (data not shown) (Wadell et al., 1980). The genome of HAdV-A18 has a length of 34,177 nucleotides compared to 34,125 for HAdV-A12 and 33,763 for HAdV-A31. The GC content, of 46.5%, is consistent with species A, even though the three genomes in this species provide a small sample size for such an analysis. Percent GC is a metric for species identification. This and the percent of identity of HAdV-A18 relative to representative HAdVs are noted in Table 1.
Table 1.
GC content and percent identity of HAdV-A18, relative to representative HAdVs
| HAdV | Acc. No. | % Identity | % GC |
|---|---|---|---|
| HAdV-A18 | GU191019 | 100.0 | 46.5 |
| HAdV-A12 | AC_000005 | 83.0 | 46.5 |
| HAdV-A31 | In press | 80.5 | 46.4 |
| HAdV-B3 | AY599834 | 60.3 | 51.1 |
| HAdV-B7 | AY594255 | 60.5 | 51.0 |
| HAdV-B11 | AY163756 | 60.6 | 48.9 |
| HAdV-B14 | AY803294 | 60.6 | 48.1 |
| HAdV-C2 | AC_000007 | 58.7 | 55.2 |
| HAdV-C5 | AC_000008 | 58.7 | 55.2 |
| HAdV-D9 | AJ854486 | 57.0 | 57.1 |
| HAdV-D53 | FJ169625 | 56.6 | 56.2 |
| HAdV-E4 | AY594253 | 58.3 | 57.7 |
| SAdV-E25 | AC_000011 | 58.5 | 59.8 |
| HAdV-F40 | NC_001454 | 61.1 | 51.2 |
| HAdV-F41 | DQ315364 | 61.1 | 51.0 |
| SAdV-G1 | NC_006879 | 58.8 | 55.2 |
| HAdV-G52 | DQ923122 | 58.4 | 55.1 |
An overview of other HAdV species, particularly those with more members, validates grouping by this metric (data not shown). Species A genomes have been noted as being the most diverse within species groupings than the other species. Still, HAdV-A18 shows the highest genome nucleotide identity to the other HAdV-A members (83% to HAdV-A12 and 80.5% to HAdV-A31) than to other HAdV prototypes. For example, lower levels of genome identities are observed in comparison with genomes from other species with HAdV-F40 and F41 having the next highest values of identity to HAdV-A18 (61.1%).
Species A members have been implicated in tumor formation in rodents, initially observed for HAdV-A12 (Huebner et al., 1962). It has also been observed that transformation of cells in vitro by HAdV-A12 requires both E1A and E1B genes based on restriction enzyme digestion products (Graham et al., 1974). These low resolution studies though “state-of-the-art at the time, i.e., using restriction enzyme fragments, can be reinterpreted given the whole genome data. HAdV-A12 was one of the first HAdV genomes sequenced, archived as X73487 (Sprengel et al., 1994). Its genome has been re-annotated as AC_000006, and has been useful as a reference genome for on-going HAdV research. The two remaining species A members were recently sequenced and shown to contain sequences in common with those from HAdV-A12, but are still distinctly different. Significantly, the sequences that constitute the “transforming segment”, originally defined as a restriction enzyme fragment comprising the left 7.2% of the genome, have a high level of similarity with each other, ca. 80%. This similarity is not present in other HAdV, for example in neither HAdV-B7 nor in HAdV-C2 and C5 (Fujinaga et al., 1977; Sawada et al., 1979), which were discussed in the literature as not hybridizing to the HAdV-A12 counterpart; these show less than 61% identity (61%, 58.66% and 59.8%, respectively).
Analysis of Selected Genes
Selected and presumably important and differentiating nucleotide coding sequences from HAdV-A18 were chosen for further analysis and comparison to homologs from other HAdV species from the preliminary computational analysis of all coding sequences. These selected coding sequences include E1A, DNA polymerase (Pol), penton, hexon, fiber and E4 34 kDa. The penton, hexon and fiber genes of HAdV are of particular importance because they are outer coat proteins which play a significant role in the tropism of the virus, as well as in the definition and identification of these prototypes based on serum neutralization and hemagglutination epitopes. E1A, Pol and E4 34 kDa were added to this analysis to provide a more complete picture of HAdV-A18, across the genome from its proximal todistal ends. The Pol sequence is important as a conserved essential gene, while the E1A protein has been implicated as a determinant of the tumorigenic properties of these HAdVs. Whole genome sequences are included in the analysis as a further reference.
Alignments and Percent Identities
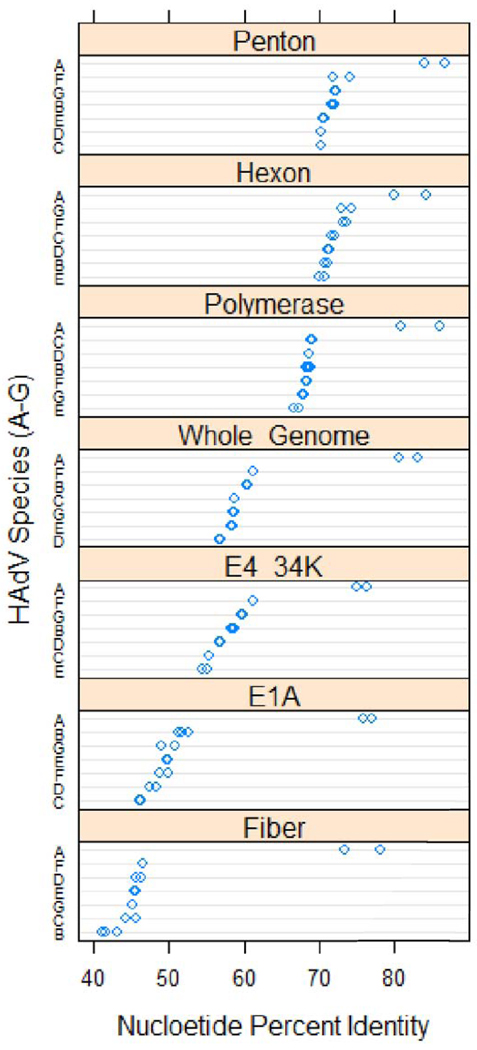
Multiple sequence alignments provide a more detailed understanding of the genes relative to other HAdVs. The nucleotide coding sequences of HAdV-A18 selected genes were analyzed with respect to percent identities and are presented in comparison with homologs found in other HAdV [Figure 1].
Figure 1.
Dotplot Analysis of Percent Identity of Selected Genes. The percent identities of selected genes of HAdV-A18, to homologs in other species, are presented in the form of a dotplot. Two members of each species (A, B1, B2, C, D, E, F and G) were chosen for the analysis to provide a range and completeness. The resulting values were grouped by gene, and sorted in descending order according to the median percent identity values of the genes. Values in each gene panel are grouped by species and sorted according to descending median percent identity. The chart was constructed using the R statistical computing environment (http://www.R-project.org/)
The percent identity values for the selected coding sequences follow a pattern similar to that of the identity values noted (Table 1) for the HAdV whole genome. HAdV-A18 shows the highest identity relationship to the other HAdV-A serotypes, with lower identity values observed for other species. HAdV-A18 shows the highest identity to HAdV-A12 in most of the coding sequences presented. However, the hexon and E4 34kDa genes of HAdV-A18 are more similar to homologs in HAdV-A31. This type of pattern suggests the possibility of a recombination event in one or both of these genes. The genome of HAdV-A18 was examined for recombination events (data described later in this report) and none were found.
Phylogenetic Relationships of Selected Genes
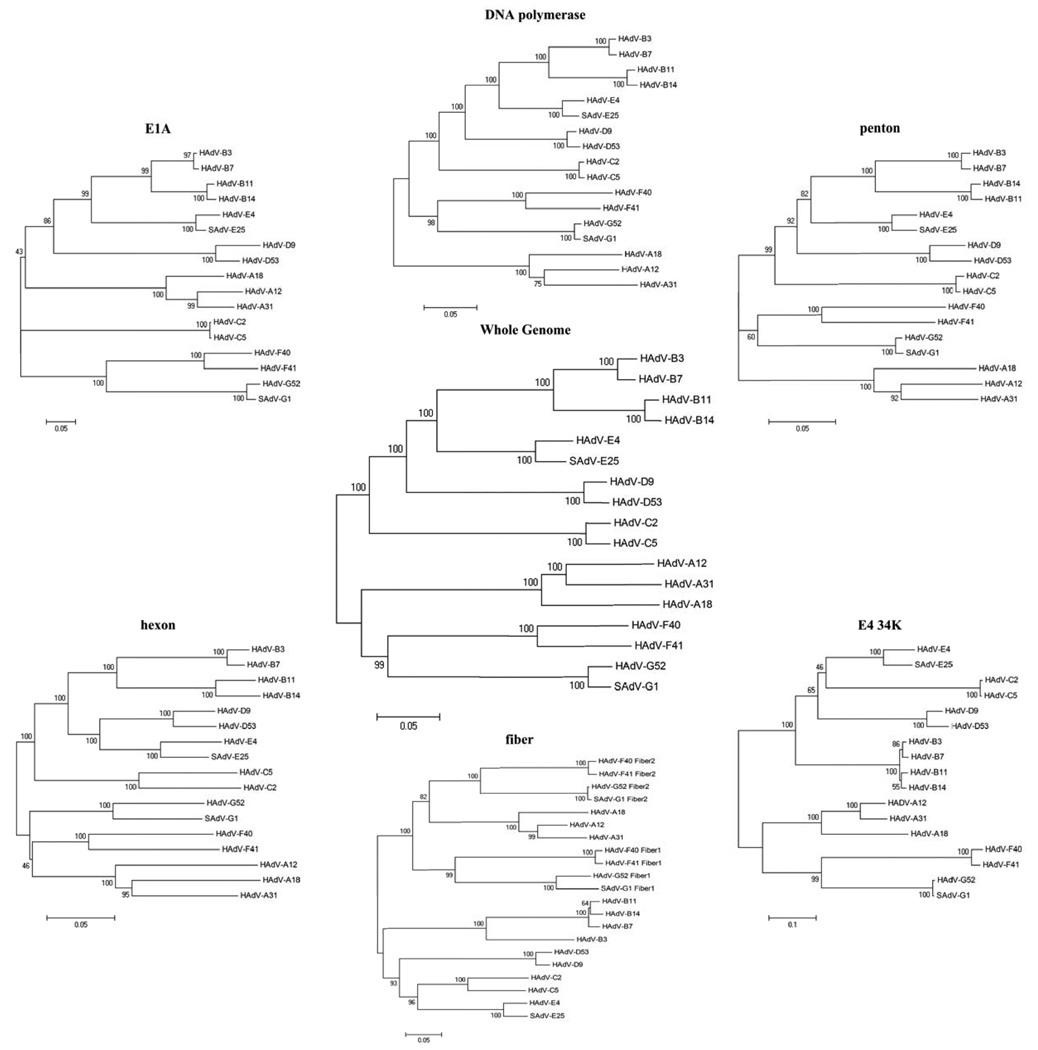
Phylogenetic trees were used to explore the relationship of the coding sequences of HAdV-A18 to homologs in other HAdV species [Figure 2]. Members of the same species form distinct clades in all of the trees. In every tree except the hexon, members of the HAdV-A species subclade in the same pattern with HAdV-A12 and -A31 pairing together, and HAdV-A18 pairing alone within the species A clade. However, in the hexon tree HAdV-A31 subclades with HAdV-A18 rather than HAdV-A12. This change in pattern among the phylogentic trees is suggestive of a possible recombination event between the hexon sequences of HAdV-A18 and -A31.
Figure 2.
Phylogenetic Relationships. A neighbor-joining algorithm was used to determine the phylogenetic relationships of the E1A, DNA polymerase, penton, hexon, fiber and E4 34kDa nucleotide coding sequences. For reference, a whole genome analysis is presented. Numbers next to the branches of the trees indicate the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates).
Hexon Recombination Analysis
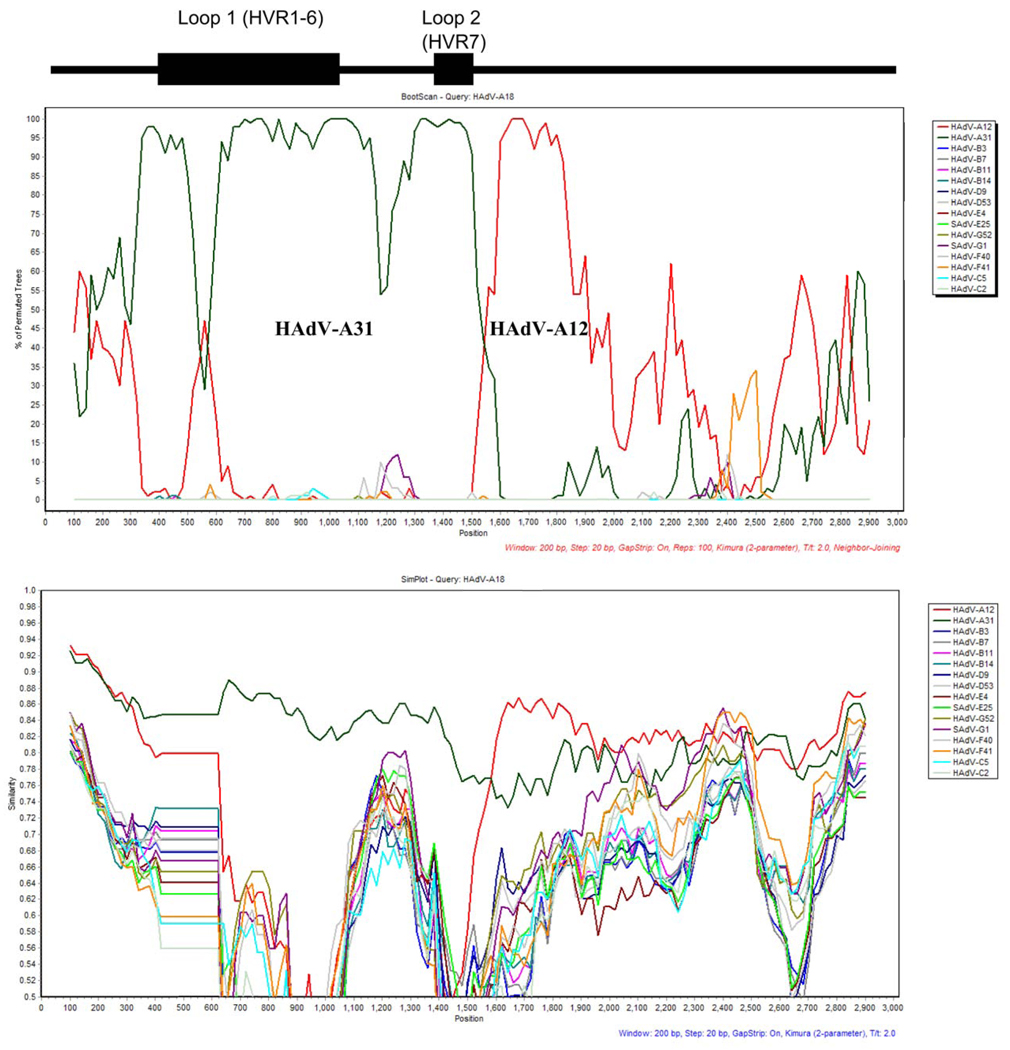
Patterns observed in percent identity values and phylogenetic trees for the coding sequences of HAdV-A18 suggest a possible recombination event with the hexon of HAdV-A31. To explore this observation further, analyses with Bootscan and Simplot (Lole et al., 1999) software were conducted with the hexon of HAdV-A18 [Figure 3]. Bootscan data show a possible recombination event. However, the Bootscan results are not as clear-cut and conclusive as those used to support previously reported recombination events (Robinson et al., 2009; Walsh et al., 2009). For example, in the previous cases demonstrating recombination, peaks in the Bootscans showed long stretches of values at 100%, indicating a strong close phylogentic relationship between the recombinant sequences. The HAdV-A18 Bootscan shows no such pattern, which suggests a weaker phylogentic relationship or perhaps a more ancient one with genetic drift contributing and complicating this result. However, the Bootscan analysis does show two distinct sets of peaks that indicate that the first half of the hexon of HAdV-A18 is closely related to HAdV-A31 while the second half is more related to HAdV-A12. This pattern is common among the HAdV hexons that have been shown to contain a recombination event. In contrast, Simplot and percent identity data reveal that there is only 84.5% identity between the hexons of HAdV-A18 and -A31. Recombinations in other HAdV genomes from the literature have shown percent identity values of greater than 95% (Walsh et al., 2009). Therefore, the sum of the data from these analyses, e.g., Simpots, Bootscans, phylogenetic trees and percent identity values, suggests that if there is a recombination event between the hexons of HAdV-A18 and A31, it is either due to an ancient event or it is to another ‘yet-to-be-sequenced’ HAdV genome.
Figure 3.
Hexon recombination analysis. Bootscan (top panel) and simplot (bottom panel) recombination results are shown. The positions of the putative serotyping epitopes and the two hypervariable loops, of the hexon, are noted above the graphs as reference. The parameters are listed in red or blue lettering under each.
Other Sequences
VA RNAs
The HAdV genomes contain coding sequences for one or two “virus associated RNA” (VA RNA) sequences. These are transcribed by the host RNA polymerase III (Gonçalves and de Vries, 2006) and are noted to play a role in the inhibition of the host anti-viral functions (Mathews and Shenk, 1991). The number of VA RNAs varies according to species, with the genomes of HAdV species B2, C, D and E encoding for two VA RNAs (VA RNA I and VA RNA II) and the members of species A, B1, F and G encoding for a single VA RNA (VA RNA I).
Table 2 lists the lengths and percent identities of VA RNA I of HAdV-A18 relative to its homolog from other HAdVs. VA RNA I of HAdV-A18 has an identical length to its homolog in species A members and shows the highest identity scores to the VA RNA I of HAdV-A12. Among the non-HAdV-A species, HAdV-A18 shows the highest identity to the VA RNA of HAdV-G52 (65.3%).
Table 2.
Comparison of VA-RNA Coding Regions. Percent identities and coding lengths of VA-RNA I from HAdV-A18, relative to counterparts in other species.
| Length of VA-RNA I |
VA-RNA I | Length of VA-RNA II |
VA-RNA II | |
|---|---|---|---|---|
| HAdV-A12 | 141 | 97.2 | - | - |
| HAdV-A31 | 141 | 90.1 | - | - |
| HAdV-B3 | 175 | 52.5 | 177 | 50.4 |
| HAdV-B7 | 175 | 52.5 | 171 | 52.5 |
| HAdV-B11 | 158 | 53.2 | - | - |
| HAdV-B14 | 162 | 51.8 | - | - |
| HAdV-C2 | 160 | 48.9 | 158 | 54.6 |
| HAdV-C5 | 160 | 48.9 | 158 | 54.6 |
| HAdV-D9 | 163 | 58.9 | 154 | 47.5 |
| HAdV-D53 | 163 | 58.2 | 179 | 47.5 |
| HAdV-E4 | 159 | 58.2 | 101 | 33.7 |
| SAdV-E25 | 159 | 56.0 | 160 | 46.8 |
| HAdV-F40 | 171 | 63.1 | - | - |
| HAdV-F41 | 174 | 63.8 | - | - |
| HAdV-G52 | 164 | 65.3 | - | - |
| SAdV-G1 | 164 | 63.8 | - | - |
Inverted Terminal Repeats
Both ends of the HAdV genomes contain “Inverted Terminal Repeat” (ITR) sequences (Dán et al., 2001; Hatfield and Hearing, 1993). The ITRs are conserved and are crucial for the replication of the virus as they contain the sequence motifs for DNA replication (Dán et al., 2001; Hatfield and Hearing, 1993; Temperley and Hay, 1992). Three important components of the ITR are the canonical core origin (Temperley and Hay, 1992), NF I binding site and NF III binding site (Hatfield and Hearing, 1993). The core origin binds a pre-terminal protein (pTP)-Pol heterodimer, and the NF I and NF III binding sites interact with the host transcription factors which are required for adenovirus replication.
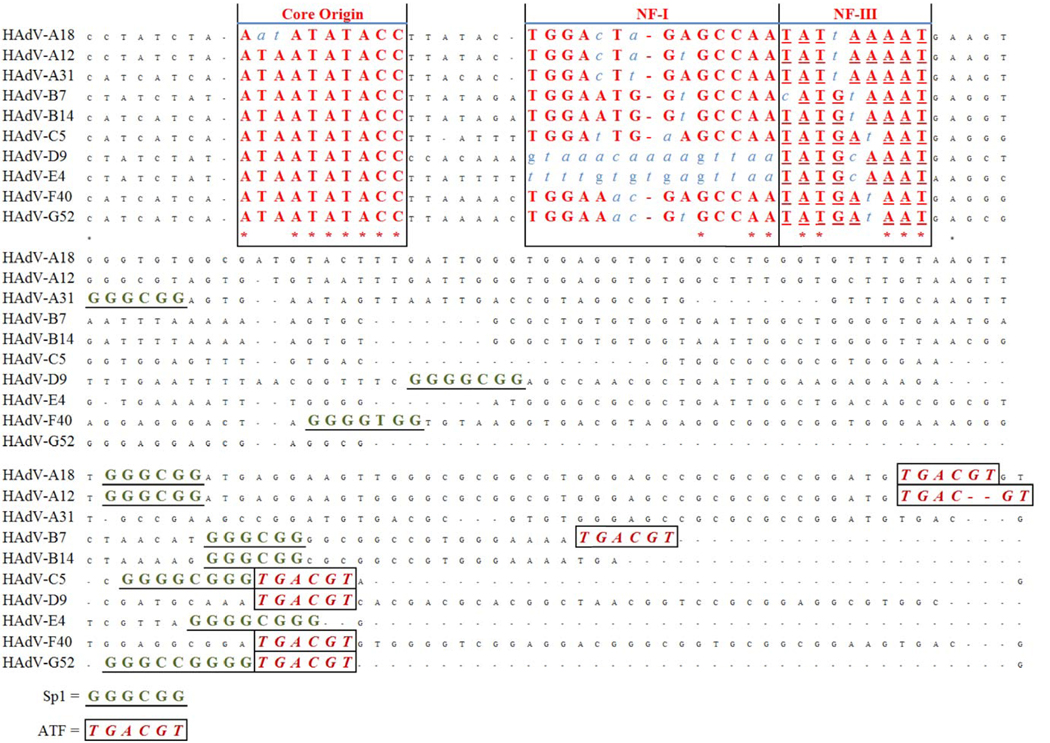
Figure 4 shows a schematic representation of an alignment of the ITRs of several HAdV. The ITR of HAdV-A18 contains all of the requisite components: a core origin and both NF I/ NF III binding sites. However, the core origin of HAdV-A18 contains mutations in its second and third bases, “TA” to “AT”, not found elsewhere to date. This mutation causes the core origin of HAdV-A18 to be unique among sequenced HAdV ITRs, but its significance and effects are undetermined. It has been shown the core origins of non-human adenoviruses are not as well conserved, in contrast to the HAdVs, and observations from the literature suggest that an “AT rich” domain is all that is required for the core origin to be functional (Dán et al., 2001). This indicates that while this mutation is unique among the HAdVs, it may not have noticeable effect on the replication of the virus. Alternatively, it may also indicate a closer relationship with and a more recent origin from the non-human adenoviruses.
Figure 4.
HAdV ITR analysis. An alignment of the ITRs of selected HAdV is presented, with critical sequence motifs marked: core origin, NF I binding site and NF III binding site. Non-conserved nucleotides within the boxes appear as blue italics.
Proteome Analysis
Examining the in silico proteome of HAdV-A18 can provide information on the effects of the differences observed in nucleotide alignments discussed earlier. To this end, the proteins of HAdV-A18 were aligned with their homologs in HAdV-A12 and -A31. Percent identities, which were calculated based on these alignments, are displayed in Figure 5.
Figure 5.
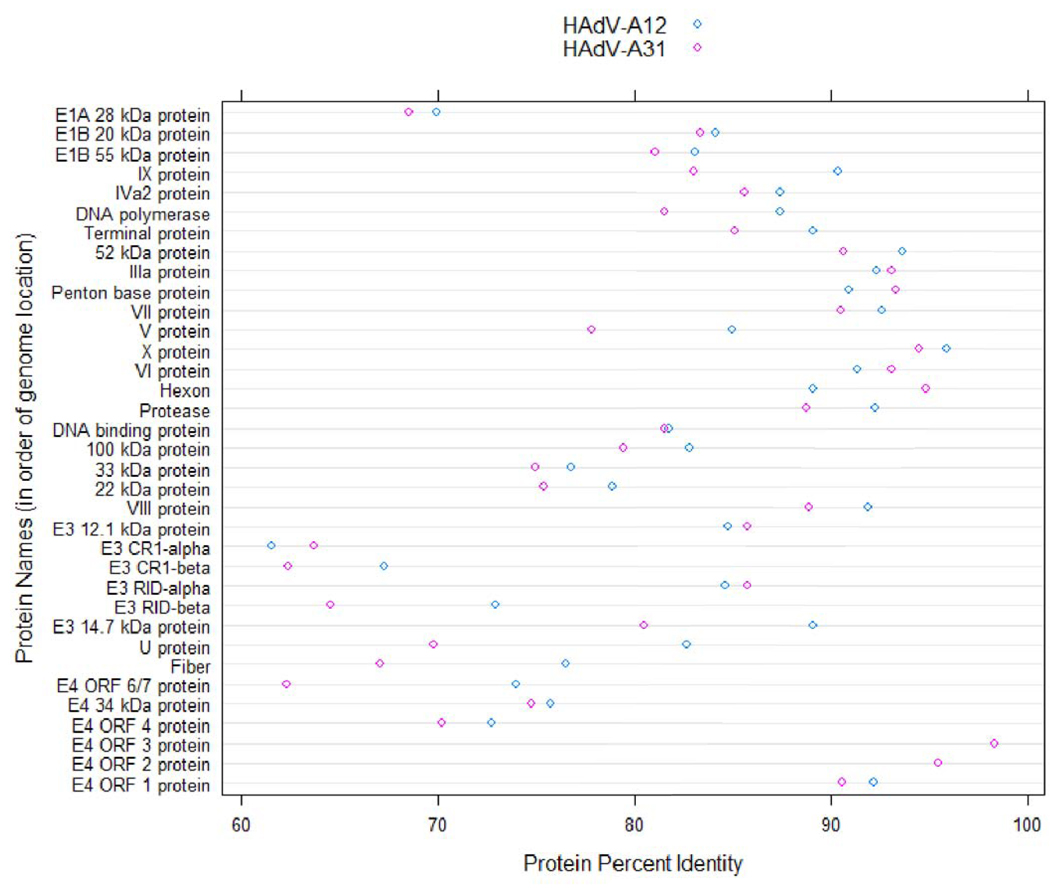
Dotplot of Protein Percent Identities. The percent identities of proteins of HAdV-A18 to homologs in the other members of the HAdV-A species are plotted in a dotplot format. The proteins are sorted according to the location of their nucleotide coding sequence. This chart was constructed using the R statistical computing environment (http://www.R-project.org/).
Protein Percent Identities
The percent identity values show that the proteome of HAdV-A18 is most similar to that of HAdV-A12. Within the two proteomes, the highest identity values are in the E4 ORF 3 protein and the least similar values are in the CR1-α and CR1-β proteins.
Virtual 2D Gel Electrophoresis
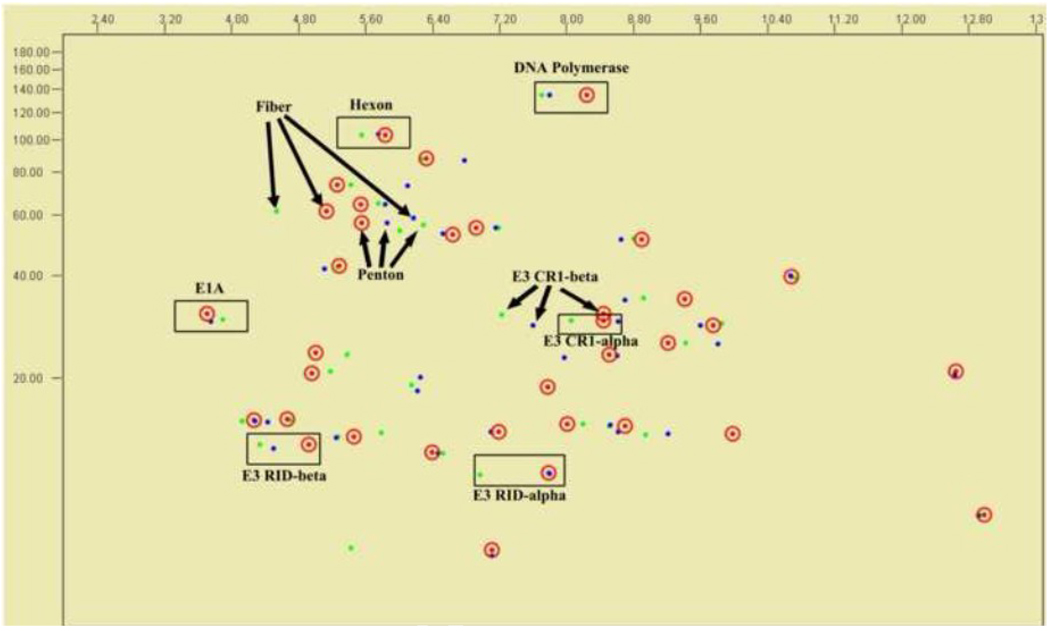
To examine the differences in the proteomes of HAdV-A18, -A31 and –A12, all sequence-based predicted proteins from the genomes were examined via in silico 2D gel electrophoresis, which parses the proteins based on their computed isoelectric point (pI) and molecular weight. None of the homologs from the three genomes shows significant differences in molecular weight. However, some homologs display charge differences. Selected proteins are noted and presented in Figure 6: the coat proteins that are putatively involved in tissue tropism and serve as serum neutralization and hemagglutination epitopes for laboratory serotyping; E1A protein, which is reported in the literature as the transforming or tumorigenic factor; DNA polymerase, a conserved essential protein; and several E3 transcript-derived proteins, which are reported to be involved in host immune system evasion. Of particular interest, the fiber and CR1-β proteins show the greatest discrepancy in charge density and molecular size, which is not unexpected given the low identity values of these proteins to one another. The fiber protein provides recognition of host cell surface proteins and provides entry into the cell, as a cell tropism parameter. CR1-β is important as it is noted to have a role in escaping the host immunosurveillance (Burgert and Blusch, 2000). Interestingly, the RID-alpha proteins of HAdV-A18 and -A31 show very similar charge/mass ratios in spite of the fact that they share only 85.7% identity. The 2D gel supports an earlier report of species A polypeptide analysis that the patterns are “clearly distinct from each other” (Wadell et al., 1980). The hexon protein analysis, both 2D gel and phylogeny, also support the past observation that these three viruses cross-react via serum neutralization, while the phylogeny analysis agrees with the observation of similar hemagglutination-inhibition assay results for these species A members; the 2D gel shows physical differences between the three fibers (Wadell et al., 1980).
Figure 6.
Virtual 2D Gel Electrophoresis Analysis. The in silico migration patterns of select proteins of HAdV-A18 (red, circled), HAdV -A12 (green) and HAdV-A31 (blue) are noted, with arrows or boxes clustering the homologs.
Conclusion
This report of the HAdV-A18 genomic sequence data and analysis completes the set of genomes classified within the HAdV-A species. One of the major contributions of this HAdV-A18 genome sequence is that it serves as a reference for further genomic and wet-bench studies, both as a reference for recombination analysis in understanding the phylogeny, molecular evolution and for studies of the origins of HAdVs. It is important also for understanding the putative roles of several HAdV genomes and their genes in reported and potential oncogenesis in rodents; the exact nucleotide sequences will allow additional wet-bench investigations.
Recombination has been implicated as a mechanism in the emergence of HAdVs as pathogens (Walsh et al., 2009). HAdV-A18 was examined for recombination events but no overwhelming evidence of an event was found. However, as these types of in silico analyses are novel, further and additional genome studies are required to define the exact metrics for recombination. The few recent HAdV recombination events reported in the literature to date are very clearly supported by these data (Robinson et al., 2009; Walsh et al., 2009). However, in the case of HAdV-A18, while the Bootscan analysis evidence revealed the strong possibility of a recombination event in the hexon with HAdV-A31, complementary Simplot analysis and percent identity data are not supportive. Therefore, either the hexon of HAdV-A18 has no recombination event or it does have a recombination event that is from an ancient event that has been complicated by genetic drift. As more HAdV recombination events are revealed and studied, the predicament of conflicting results between phylogeney-based methods (Bootscan) and similarity-based methods (Simplot and percent identity data) will likely to become a more prominent problem.
Acknowledgments
The work reported herein was performed with the support of the United States Air Force Surgeon General, Clinical Investigation No. FDG20040024E. This was also supported in part by U.S. Public Health Service NIH grants EY013124 and P30EY013104, Massachusetts Lions Eye Research Fund, Inc, and an unrestricted grant to the Department of Ophthalmology, Harvard Medical School from Research to Prevent Blindness, Inc. DS and MPW were also supported by an USAF contract, FA442709P0262. The views expressed in this material are those of the authors, and do not reflect the official policy or position of the U.S. Government, the Department of Defense, or the Department of the Air Force.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Michael P. Walsh, Email: mwalsh5@gmu.edu.
Jason Seto, Email: jseto@gmu.edu.
Damaris Tirado, Email: Damaris.Tirado@travis.af.mil.
James Chodosh, Email: James_Chodosh@meei.harvard.edu.
Morris S. Jones, Email: Morris.Jones@travis.af.mil.
References
- Adrian T, Wigand R. Genome type analysis of adenovirus 31, a potential causative agent of infants' enteritis. Arch. Virol. 1989;105(1–2):81–87. doi: 10.1007/BF01311118. [DOI] [PubMed] [Google Scholar]
- Adrian T, Wigand R, Richter J. Gastroenteritis in infants, associated with a genome type of adenovirus 31 and with combined rotavirus and adenovirus 31 infection. Eur. J. Pediatr. 1987;146(1):38–40. doi: 10.1007/BF00647280. [DOI] [PubMed] [Google Scholar]
- Brown M, Petric M, Middleton PJ. Diagnosis of fastidious enteric adenoviruses 40 and 41 in stool specimens. J. Clin. Microbiol. 1984;20(3):334–338. doi: 10.1128/jcm.20.3.334-338.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgert H, Blusch JH. Immunomodulatory Functions Encoded by the E3 Transcription Unit of Adenoviruses. Virus Genes. 2000;21(1):13–25. [PubMed] [Google Scholar]
- Dán Á, Élő P, Harrach B, Zádori Z, Benkő M. Four New Inverted Terminal Repeat Sequences from Bovine Adenoviruses Reveal Striking Differences in the Length and Content of the ITRs. Virus Genes. 2001;22(2):175–179. doi: 10.1023/a:1008125324346. [DOI] [PubMed] [Google Scholar]
- De Jong JC, Wermenbol AG, Verweij-Uijterwaal MW, Slaterus KW, Wertheim-Van Dillen P, Van Doornum GJ, Khoo SH, Hierholzer JC. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 1999;37(12):3940–3945. doi: 10.1128/jcm.37.12.3940-3945.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Ojima S, Yano S, Shinagawa M. The Transforming DNA Sequence Common to Highly Oncogenic Human Adenoviruses, Type 12, 18 and 31. Proceedings of the Japan Academy. Ser. B: Physical and Biological Sciences. 1977;53(4):152–155. [Google Scholar]
- Gonçalves MAFV, de Vries AAF. Adenovirus: from foe to friend. Rev. Med. Virol. 2006;16(3):167–186. doi: 10.1002/rmv.494. [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ, Heijneker HL. Size and location of the transforming region in human adenovirus type 5 DNA. Nature. 1974;251(5477):687–691. doi: 10.1038/251687a0. [DOI] [PubMed] [Google Scholar]
- Hammond GW, Mauthe G, Joshua J, Hannan CK. Examination of uncommon clinical isolates of human adenoviruses by restriction endonuclease analysis. J. Clin. Microbiol. 1985;21(4):611–616. doi: 10.1128/jcm.21.4.611-616.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield L, Hearing P. The NFIII/OCT-1 binding site stimulates adenovirus DNA replication in vivo and is functionally redundant with adjacent sequences. J. Virol. 1993;67(7):3931–3939. doi: 10.1128/jvi.67.7.3931-3939.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller K, Grote A, Maneck M, Munch R, Jahn D. JVirGel 2.0: computational prediction of proteomes separated via two-dimensional gel electrophoresis under consideration of membrane and secreted proteins. Bioinformatics. 2006;22(19):2441–2443. doi: 10.1093/bioinformatics/btl409. [DOI] [PubMed] [Google Scholar]
- Hofmayer S, Madisch I, Darr S, Rehren F, Heim A. Unique sequence features of the Adenovirus 31 complete genomic sequence are conserved in clinical isolates. BMC Genomics. 2009;10(1):557. doi: 10.1186/1471-2164-10-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland RCG, Down TA, Pocock M, Prlić A, Huen D, James K, Foisy S, Dräger A, Yates A, Heuer M, Schreiber MJ. BioJava: an open-source framework for bioinformatics. Bioinformatics. 2008;24(18):2096–2097. doi: 10.1093/bioinformatics/btn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner RJ, Rowe WP, Lane WT. Oncogenic effects in hamsters of human adenovirus types 12 and 18. Proc. Natl. Acad. Sci. U. S. A. 1962;48(12):2051–2058. doi: 10.1073/pnas.48.12.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiko H, Aoki K. Spread of Epidemic Keratoconjunctivitis Due to a Novel Serotype of Human Adenovirus in Japan. J. Clin. Microbiol. 2009;47(8):2678–2679. doi: 10.1128/JCM.r00313-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JC, Wigand R, Kidd AH, Wadell G, Kapsenberg JG, Muzerie CJ, Wermenbol AG, Firtzlaff RG. Candidate adenoviruses 40 and 41: fastidious adenoviruses from human infant stool. J. Med. Virol. 1983;11(3):215–231. doi: 10.1002/jmv.1890110305. [DOI] [PubMed] [Google Scholar]
- Kajon AE, Erdman DD. Assessment of genetic variability among subspecies b1 human adenoviruses for molecular epidemiology studies. Methods Mol. Med. 2007;131:335–355. doi: 10.1007/978-1-59745-277-9_23. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl. Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd AH, Cosgrove BP, Brown RA, Madeley CR. Faecal adenoviruses from Glasgow babies. Studies on culture and identity. J. Hyg. (Lond) 1982;88(3):463–474. doi: 10.1017/s0022172400070327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73(1):152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Shenk T. Adenovirus virus-associated RNA and translation control. J. Virol. 1991;65(11):5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MS, Pereira HG, Clarke SK. Human adenovirus type 31. A new serotype with oncogenic properties. Lancet. 1965;1(7375):21–23. doi: 10.1016/s0140-6736(65)90925-6. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Robinson CM, Rajaiya J, Walsh MP, Seto D, Dyer DW, Jones MS, Chodosh J. Computational analysis of human adenovirus type 22 provides evidence for recombination between human adenoviruses species D in the penton base gene. [Accessed August 11, 2009];J. Virol. 2009 doi: 10.1128/JVI.00786-09. Available at: http://www.ncbi.nlm.nih.gov/pubmed/19553309. [DOI] [PMC free article] [PubMed]
- Rowe WP, Hartley JW, Huebner RJ. Additional serotypes of the APC virus group. Proc. Soc. Exp. Biol. Med. 1956;91(2):260–262. doi: 10.3181/00379727-91-22231. [DOI] [PubMed] [Google Scholar]
- Rowe WP, Hartley JW, Huebner RJ. Serotype composition of the adenovirus group. Proc. Soc. Exp. Biol. Med. 1958;97(2):465–470. doi: 10.3181/00379727-97-23776. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16(10):944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sawada Y, Ojima S, Shimojo H, Shiroki K, Fujinaga K. Transforming DNA sequences in rat cells transformed by DNA fragments of highly oncogenic human adenovirus type 12. J. Virol. 1979;32(2):379–385. doi: 10.1128/jvi.32.2.379-385.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto J, Walsh MP, Mahadevan P, Purkayastha A, Clark JM, Tibbetts C, Seto D. Genomic and bioinformatics analyses of HAdV-14p, reference strain of a re-emerging respiratory pathogen and analysis of B1/B2. Virus Res. 2009;143(1):94–105. doi: 10.1016/j.virusres.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Sprengel J, Schmitz B, Heuss-Neitzel D, Zock C, Doerfler W. Nucleotide sequence of human adenovirus type 12 DNA: comparative functional analysis. J Virol. 1994;68(1):379–389. doi: 10.1128/jvi.68.1.379-389.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Temperley SM, Hay RT. Recognition of the adenovirus type 2 origin of DNA replication by the virally encoded DNA polymerase and preterminal proteins. EMBO. J. 1992;11(2):761–768. doi: 10.1002/j.1460-2075.1992.tb05109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentin JJ, Yabe Y, Taylor G. The Quest for Human Cancer Viruses: A new approach to an old problem reveals cancer induction in hamsters by human adenovirus. Science. 1962;137(3533):835–841. doi: 10.1126/science.137.3533.835. [DOI] [PubMed] [Google Scholar]
- Uhnoo I, Wadell G, Svensson L, Johansson ME. Importance of enteric adenoviruses 40 and 41 in acute gastroenteritis in infants and young children. J. Clin. Microbiol. 1984;20(3):365–372. doi: 10.1128/jcm.20.3.365-372.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasavada R, Eager KB, Barbanti-Brodano G, Caputo A, Ricciardi RP. Adenovirus type 12 early region 1A proteins repress class I HLA expression in transformed human cells. Proc. Natl. Acad. Sci. U. S. A. 1986;83(14):5257–5261. doi: 10.1073/pnas.83.14.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora GJ, Lin B, Gratwick K, Meador C, Hansen C, Tibbetts C, Stenger DA, Irvine M, Seto D, Purkayastha A, Freed NE, Gibson MG, Russell K, Metzgar D. Co-infections of adenovirus species in previously vaccinated patients. Emerging Infect. Dis. 2006;12(6):921–930. doi: 10.3201/eid1206.050245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadell G, Hammarskjold ML, Winberg G, Varsanyi TM, Sundell G. Genetic variability of adenoviruses. Ann. N. Y. Acad. Sci. 1980;354:16–42. doi: 10.1111/j.1749-6632.1980.tb27955.x. [DOI] [PubMed] [Google Scholar]
- Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D, Jones MS. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS. ONE. 2009;4(6):e5635. doi: 10.1371/journal.pone.0005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe Y, Samper L, Bryan E, Taylor G, Trentin JJ. Oncogenic Effect of Human Adenovirus Type 12, in Mice. Science. 1964;143(3601):46–47. doi: 10.1126/science.143.3601.46. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhu Z, Tang L, Wang L, Tan X, Yu P, Zhang Y, Tian X, Wang J, Zhang Y, Li D, Xu W. Genomic analyses of recombinant adenovirus type 11a in China. J. Clin. Microbiol. 2009;47(10):3082–3090. doi: 10.1128/JCM.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]