Summary
Most of the 131 cells that die during the development of a C. elegans hermaphrodite do so ~30 min after being generated. Furthermore, in these cells, the pro-caspase proCED-3 is inherited from progenitors and the transcriptional upregulation of the BH3-only gene egl-1 is thought to be sufficient for apoptosis induction. In contrast, the four CEM neurons, which die in hermaphrodites, but not males, die ~150 min after being generated. We found that in the CEMs, the transcriptional activation of both the egl-1 and ced-3 gene is necessary for apoptosis induction. In addition, we show that the Bar homeodomain transcription factor CEH-30 represses egl-1 and ced-3 transcription in the CEMs, thereby permitting their survival. Furthermore, we identified three genes, unc-86, lrs-1 and unc-132, which encode a POU homeodomain transcription factor, a leucyl-tRNA synthetase and a novel protein with limited sequence similarity to the mammalian proto-oncoprotein and kinase PIM-1, respectively, that promote the expression of the ceh-30 gene in the CEMs. Based on these results, we propose that egl-1 and ced-3 transcription are co-regulated in the CEMs to compensate for limiting proCED-3 levels, which most probably are a result of proCED-3 turn over. Similar co-regulatory mechanisms for BH3-only proteins and pro-caspases may function in higher organisms to allow efficient apoptosis induction during development. Finally, we present evidence that the timing of the death of the CEMs is controlled by TRA-1 Gli, the terminal global regulator of somatic sexual fate in C. elegans.
Introduction
The elimination of unwanted cells by apoptosis is critical for animal development (1). A central enzymatic machinery involved in the induction of apoptosis has been defined, and found to be conserved in animals as diverse as the nematode Caenorhabditis elegans and humans (2–5). It is generally believed that most if not all cells in developing animals contain this machinery in an inactive state. Furthermore, BH3-only proteins, which function as receivers of apoptotic stimuli, have emerged as key activators of this central cell-death machinery in both C. elegans and vertebrates (6, 7). However, many questions about apoptosis induction remain to be answered. For example, it is unclear how animals ensure the presence of the central cell death machinery in most if not all cells during development, which is a prerequisite for BH3-only-dependent apoptosis induction.
Due to its invariant pattern of apoptotic cell death during development, C. elegans has become an important model for apoptosis studies. The central cell death machinery in C. elegans is composed of the anti-apoptotic BCL-2-like protein CED-9, the APAF-1-like caspase activator CED-4 and the pro-caspase proCED-3 (4, 5, 8). At least during mid-embryogenesis when the majority of cell deaths occurs, most if not all cells in developing embryos contain CED-4 as well as proCED-3, which once matured is sufficient for apoptosis induction (9–11). However, these cells also contain the CED-9 protein, which through its direct interaction with CED-4, blocks the ability of CED-4 to mediate proCED-3 maturation (9, 10). As a result, apoptosis induction is prevented. During the development of a C. elegans hermaphrodite, exactly 131 cells are programmed to die (12). The majority of these cells die ~30 min after being generated. Furthermore, in these cells, the transcriptional activation of the BH3-only gene egl-1 is thought to be sufficient for the activation of the central cell death machinery and, hence, apoptosis induction (13–16). Specifically, the BH3-only protein EGL-1 can bind to CED-9 thereby causing CED-4 release from CED-9 and CED-4-dependent proCED-3 maturation. Interestingly, recent data suggest that the ced-3 gene is not transcriptionally active in most of the 131 cells that are programmed to die during development (10). This observation suggests that proCED-3 protein present in these cells is derived from their progenitors. It also suggests that once activated, the amount of proCED-3 protein inherited from progenitors is, in general, sufficient for apoptosis induction.
The four male-specific cephalic companion neurons (CEMs) are generated ~320 min after the first cell division. In males, the CEMs survive and differentiate into ciliated sensory neurons implicated in mating behavior (12, 17, 18). In hermaphrodites, however, the CEMs undergo apoptosis ~150 min after being generated (~470 min after the first cell division). As is the case for most of the cells that are programmed to die during development, the death of the CEMs is dependent on the genes egl-1, ced-4 and ced-3 (13, 19). However, the death of the CEMs is also dependent on the gene tra-1, which encodes the terminal, global regulator of somatic sexual fate and which acts to promote female development (20). Specifically, the TRA-1 protein, a GLI-like transcription factor, causes the CEMs in hermaphrodites to undergo apoptosis by directly repressing the transcription of the gene ceh-30, which encodes a Bar homeodomain transcription factor that acts to block the death of the CEMs in males (21, 22).
How the CEH-30 protein acts on the central cell death machinery to block the death of the CEMs has so far been unclear. We now present evidence that CEH-30 blocks the death of the CEMs by repressing the transcription of both the egl-1 and ced-3 gene. Furthermore, we identified three genes, unc-86, lrs-1, and unc-132, which encode a POU homeodomain transcription factor, a leucyl-tRNA synthetase and a novel protein with limited sequence similarity to the mammalian proto-oncoprotein and kinase PIM-1, respectively, that promote ceh-30 expression in the CEMs thereby antagonizing tra-1 function. Our data also demonstrate that in contrast to most cells that are programmed to die during development, in the CEMs, the transcriptional upregulation of both the BH3-only gene egl-1 and the caspase gene ced-3 is required for apoptosis induction. We propose that coupling the transcriptional upregulation of a key activator of apoptosis induction (egl-1) and a component of the central, enzymatic cell death machinery (ced-3) ensures efficient apoptosis induction in the CEMs.
Results
bc151, bc155, and bc159 cause differentiated CEMs to be absent from masculinized hermaphrodites or males
An altered-function mutation of the C. elegans gene sel-10, n1077, which encodes an F-box protein that promotes female development, causes weak masculinization of the hermaphrodite soma (23–25). For example, in wild-type hermaphrodites, the four CEMs undergo apoptosis (12, 17). In contrast, in sel-10(n1077) hermaphrodites, the CEMs inappropriately survive. The CEMs in sel-10(n1077) hermaphrodites have the characteristic CEM morphology when observed by differential interference contrast microscopy (DIC) in larvae of the fourth larval stage (L4 larvae), and express CEM-specific markers in adults, such as the genes pkd-2 (Ppkd-2gfp) and lov-1 (Plov-1gfp) (see below) (26). These observations indicate that the CEMs in sel-10(n1077) hermaphrodites are correctly specified and fully differentiated.
To identify genes required for the survival of the CEMs, we sought mutations that cause the CEMs to be absent from sel-10(n1077) hermaphrodites based on the expression of pkd-2. Using this criterion, we screened 8,500 mutagenized, haploid genomes and identified 49 mutants. Among these mutants, 21 still had CEMs based on DIC and therefore most likely harbor mutations that affect pkd-2 expression. In addition, in 20 mutants, all aspects of the weak masculinization caused by sel-10(n1077) were suppressed, indicating that they most likely carry feminizing mutations or loss-of-function mutations in the sel-10 gene. Of the remaining eight mutations, one behaved non-Mendelian, one could not be outcrossed, and three caused the CEMs to be absent in masculinized sel-10(n1077) hermaphrodites, but not males. The final three mutations, bc151, bc155, and bc159, were characterized further.
Based on pkd-2 expression in adults, the recessive mutations bc151, bc155, and bc159 cause the majority of CEMs to be absent from masculinized hermaphrodites as well as males (Table 1A; data not shown). The CEMs are also absent in bc151, bc155, or bc159 masculinized hermaphrodites based on their characteristic morphology in L4 larvae (Table 1B) and in bc151, bc155, or bc159 males based on lov-1 expression (Table S1). In addition, all three mutations cause a reduced brood size and an uncoordinated or ‘Unc’ phenotype (Table S2; data not shown). None of the mutations affects viability at least in the sel-10(n1077) background (Table S3). Based on these observations, we conclude that bc151, bc155, and bc159 cause differentiated CEMs to be absent in masculinized hermaphrodites and males.
Table 1.
The CEMs are absent in masculinized hermaphrodites or males homozygous for unc-86(bc151), lrs-1(bc155) or unc-132(bc159).
| A. Presence of differentiated CEMs in adults | ||||||||
|---|---|---|---|---|---|---|---|---|
| % GFP+ CEMs (n) | ||||||||
| Genotype | +/+ | sel-10(n1077) | ||||||
| +/+ | ced-3(n717) | |||||||
| XX | XOb | XX | XX | |||||
| +/+ | 0 | (many) | 100 | (many) | 100 | (many) | 100 | (many) |
| egl-1(n1084 n3082) | 68 | (356) | ND | ND | ND | |||
| ced-3(n717) | 78 | (232) | ND | ND | ND | |||
| unc-86(bc151)a | ND | 25 | (80) | 12 | (178) | 8 | (92) | |
| lrs-1(bc155)a | ND | 18 | (76) | 10 | (224) | 7 | (80) | |
| unc-132(bc159) | ND | 0 | (68) | 0 | (232) | 0 | (96) | |
| B. Presence of CEMs in L4 larvae | ||||||
|---|---|---|---|---|---|---|
| % CEMs (n) | ||||||
| Genotype | +/+ | sel-10(n1077) | ced-3(n717); sel-10(n1077) | |||
| XX | XX | XX | ||||
| +/+ | 0 | (36) | 100 | (40) | 100 | (36) |
| egl-1(n1084 n3082) | 96 | (80) | ND | ND | ||
| ced-3(n717) | 98 | (128) | ND | ND | ||
| unc-86(bc151)a | ND | 6 | (32) | 4 | (24) | |
| lrs-1(bc155)a | ND | 7 | (48) | 7 | (48) | |
| unc-132(bc159) | ND | 0 | (20) | 2 | (44) | |
The presence of differentiated CEMs was analyzed in adults using the Ppkd-2 gfp reporter as described in Materials and Methods. All strains were homozygous for the Ppkd-2 gfp integration bcIs9.
Indicates that the strains were homozygous for dpy-17(e164) III.
Indicates that the strains were homozygous for him-5(e1490) V.
The presence of CEMs was scored at the L4 stage using DIC as described in Materials and Methods. All strains were homozygous for the Ppkd-2 gfp integration bcIs9.
Indicates that the strains were homozygous for dpy-17(e164) III.
bc151 is a loss-of-function mutation of unc-86 POU
unc-86, which encodes a POU transcription factor, has previously been shown to be required for the presence of differentiated CEMs in males (21, 27, 28). We found that bc151 animals carry two mutations in the unc-86 gene: first, a missense mutation leading to an aspartic acid (GAU) -to-asparagine (AAU) substitution at position 203 of the protein sequence of the UNC-86.b protein (generated by transcript C30A5.7b); second, a nonsense mutation, changing codon 220 of the C30A5.7b coding sequence (CAA) to a STOP codon (UAA) (Figure 1A). Both unc-86 transcripts (C30A5.7a, C30A5.7b) are predicted to be affected by the two mutations. Since loss-of-function mutations of unc-86 have previously been shown to cause differentiated CEMs to be absent in males, we conclude that bc151 is a new loss-of-function mutation of the unc-86 gene.
Fig. 1. Cloning of unc-86(bc151), lrs-1(bc155) and unc-132(bc159).
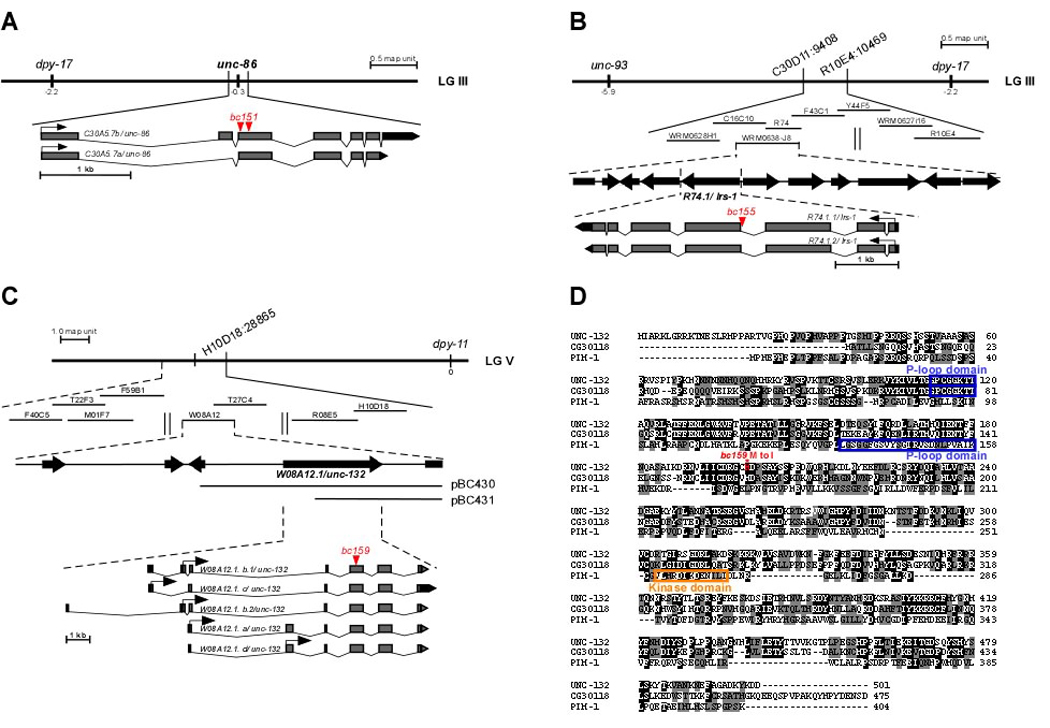
(A) The gene dpy-17 used for mapping bc151 is indicated on the genetic map (LG III). The two mutations identified in the C30A5.7/unc-86 gene in bc151 animals are indicated in red. (B) Genes and SNPs used for mapping bc155 on LG III are indicated on the genetic map. Cosmids and fosmids tested for bc155 rescue are shown below the genetic map. The bc155 mutation in R74.1/lrs-1 is indicated in red. (C) dpy-11 and H10D18:28865 used for mapping bc159 on LG V are indicated on the genetic map. Cosmids tested for bc159 rescue are shown below. pBC430 and pBC431 represent rescuing subclones of cosmid W08A12. The bc159 mutation in W08A12.1/unc-132 is indicated in red. (D) Alignment of the protein sequences of UNC-132 (isoform a), D. melanogaster CG30118 and human PIM-1. The alignment was done using the EMBOSS algorithm. Identical amino acids have a black background. Amino acids with similar biochemical properties have a gray background. The blue boxes indicate the P-loop domains and the orange box the kinase domain. The amino acid that is changed as a result bc159 (M200I) is indicated by a red asterisk.
bc155 is a loss-of-function mutation of the gene lrs-1 leucyl-tRNA synthetase
We found that bc155 is a mutation in the gene lrs-1, which encodes a leucyl-tRNA synthetase, a Class I tRNA synthetase (Figure 1B; data not shown) (29). bc155 is a G-to-A transition that changes the 3’ splice acceptor site of intron 3 from AG to AA, which results in reduced levels of fully spliced lrs-1 mRNA (Figure 1B; data not shown). Both lrs-1 transcripts (R74.1.1, R74.1.2) are predicted to be affected by the bc155 mutation. Reducing lrs-1 function by RNAi results in the absence of differentiated CEMs in masculinized hermaphrodites (Table 2A). Based on these findings, we conclude that bc155 is a loss-of-function mutation of the gene lrs-1. Furthermore, we found that reducing the function by RNAi of genes encoding other Class I tRNA synthetases (isoleucyl-tRNA synthetase [irs-2], valyl-tRNA synthetase [vrs-2]) or Class II tRNA synthetases (histidyl-tRNA synthetase [hrs-1], tryptophanyl-tRNA synthetase [wrs-2], threonyl-tRNA synthetase [trs-1]) can result in the absence of at least 20% of differentiated CEMs in masculinized hermaphrodites (Table 2A). Hence, the presence of differentiated CEMs in masculinized hermaphrodites is sensitive to changes in the activity level of the translational machinery.
Table 2.
The inactivation by RNAi of lrs-1 and other genes encoding tRNA synthetases as well as unc-132 by RNAi results in the absence of differentiated CEMs in masculinized hermaphrodites.
| A. Inactivation by RNAi of lrs-1 and other genes encoding tRNA synthetases | ||||
|---|---|---|---|---|
| Genotype | tRNA synthetase affected |
%GFP+ CEMs (n) | Other RNAi phenotypes | |
| sel-10(n1077) | ||||
| XX | ||||
| +/+ | NA | 100 | (many) | NA |
| Class I tRNA synthetases | ||||
| lrs-1(RNAi) | Leu | 62 | (60) | larval arrest, lethality |
| irs-2(RNAi) | Ile | 63 | (248) | none detected |
| irs-1(RNAi) | Ile | 83 | (12) | larval arrest, lethality |
| vrs-2(RNAi) | Val | 60 | (32) | larval arrest, lethality |
| vrs-1(RNAi) | Val | 90 | (124) | none detected |
| mrs-1(RNAi) | Met | 89 | (132) | none detected |
| Class II tRNA synthetases | ||||
| hrs-1(RNAi) | His | 71 | (24) | larval arrest, lethality |
| srs-1(RNAi) | Ser | 95 | (112) | none detected |
| yrs-1(RNAi) | Tyr | 88 | (68) | none detected |
| ers-1(RNAi) | Glu | 85 | (100) | none detected |
| wrs-2(RNAi) | Trp | 75 | (124) | none detected |
| drs-1(RNAi) | Asp | 92 | (24) | larval arrest, lethality |
| prs-1(RNAi) | Pro | 97 | (116) | none detected |
| ars-1(RNAi) | Ala | 88 | (124) | none detected |
| trs-1(RNAi) | Thr | 74 | (72) | none detected |
| nrs-1(RNAi) | Asn | 86 | (28) | none detected |
| frs-1(RNAi) | Phe | 100 | (36) | larval arrest, lethality |
| crs-1(RNAi) | Cys | 91 | (108) | larval arrest, lethality |
| krs-1(RNAi) | Lys | 100 | (32) | larval arrest, lethality |
| B. Inactivation of unc-132 by RNAi | ||
|---|---|---|
| Genotype | % GFP+ CEMs (n) | |
| sel-10(n1077) | ||
| +/+ | 99 | (200) |
| unc-132(RNAi ; W08A12.1b cDNA) | 49 | (216) |
| unc-132(RNAi ; W08A12.1c cDNA) | 64 | (172) |
The presence of differentiated CEMs was analyzed in adults using the Ppkd-2gfp reporter as described in Materials and Methods. All strains were homozygous for the Ppkd-2gfp integration bcIs9. RNAi was performed by feeding as described in Materials and Methods.
bc159 is a loss-of-function mutation of the PIM-1-like gene unc-132
bc159 is a mutation in a previously uncharacterized gene, W08A12.1, which generates five transcripts. Transcript W08A12.1.a encodes a 501 amino acid protein of unknown function with a P-loop domain and sequence similarity to an uncharacterized protein of Drosophila melanogaster, CG30118, and the human proto-oncoprotein and kinase PIM-1 (Figure 1C; see below) (30, 31). As mentioned above, apart from causing differentiated CEMs to be absent in masculinized hermaphrodites and males, bc159 also causes an Unc phenotype. For this reason, we named W08A12.1 ‘unc-132’. The bc159 mutation affects the second exon that is common to all five unc-132 transcripts. Specifically, bc159 is a missense mutation that leads to a methionine (AUG) -to-isoleucine (AUA) substitution at position 200 of the protein sequence of the UNC-132.a protein (generated by transcript W08A12.1.a). Since the reduction of unc-132 function by RNAi causes differentiated CEMs to be absent in a sel-10(n1077) background (Table 2B), we conclude that bc159 is a loss-of-function mutation of the unc-132 gene.
unc-132 is orthologous to the gene CG30118 of D. melanogaster (30). Over its entire length, the UNC-132 protein is 38.2% identical and 53.5% similar to the CG30118 protein (Figure 1D). The similarities between UNC-132 and CG30118 are mainly found in the central (57.5% identity, 76.4% similarity) and C-terminal (42.3% identity, 59.6% similarity) regions of the two proteins. In addition, both proteins contain a P-loop domain. Among mammalian proteins, UNC-132 is most similar to the human kinase PIM-1 (12.7% identical and 22.0% similar over its entire length); however the two proteins are not orthologous (31). The similarity between UNC-132 and PIM-1 is mainly restricted to their N-terminal (29.9% identical, 46.8% similar) regions. As is the case for UNC-132 and CG30118, PIM-1 has a P-loop domain. However, in contrast to UNC-132 and CG30118, PIM-1 also contains a kinase domain.
The loss of unc-86, lrs-1 or unc-132 function causes a defect in CEM specification
To determine whether the CEMs inappropriately undergo apoptosis in masculinized hermaphrodites lacking unc-86, lrs-1 or unc-132 function, we tested whether a mutation that blocks apoptotic cell death in general (ced-3(n717)) can suppress the CEM phenotype observed in these mutant backgrounds (19). We found that ced-3(n717) failed to suppress the lack of differentiated CEMs in these animals, which indicates that the loss of unc-86, lrs-1 or unc-132 function does not cause the CEMs to inappropriately die (Table 1A, B). This notion is supported by the following observation. In wild-type hermaphrodites, cells with a refractile, corpse-like morphology indicative of apoptotic cells are detected at the position at which the CEMs are located ~470 min after the first cell division, which is the time at which the CEMs die in these animals (Table S4). In contrast, in masculinized hermaphrodites, in which the CEMs survive, such cells are not detected. Similarly, in masculinized hermaphrodites lacking unc-86, lrs-1 or unc-132 function, cells with a refractile, corpse-like morphology are rarely detected at this position (Table S4).
To determine whether the CEMs are generated, we analyzed the fate of their sisters, the dorsal URAs in the case of the dorsal CEMs and the amphid socket cells in the case of the ventral CEMs (12). The rationale for these analyses was that the presence of properly differentiated sisters would suggest that the cell division that generates the CEMs and their sisters had occurred appropriately. Using markers specifically expressed in differentiated URAs (Pglr-4glr-4∷gfp and Pcfi-1gfp) or amphid socket cells (Punc-53gfp) (32–34), we found that the loss of lrs-1 or unc-132 function does not affect the presence of differentiated dorsal URAs or amphid socket cells (Table 3). Based on these results we conclude that the CEMs are generated in animals lacking lrs-1 or unc-132 function. In the case of unc-86, we found that animals lacking unc-86 function had amphid socket cells, but lacked differentiated dorsal URAs (Table 3). However, as shown below, we can detect CEMs in unc-86 mutant embryos using a transcriptional egl-1 reporter. Therefore, we conclude that the CEMs are also generated in animals lacking unc-86 function. Based on these observations we propose that the loss of unc-86, lrs-1 or unc-132 function causes a defect in the specification (rather than generation or survival) of the CEMs.
Table 3.
The fate of the AMso and URAs in unc-86(bc151), lrs-1(bc155) and unc-132(bc159).
| Genotype | % GFP+ AMso (n) | % GFP+ URAs (n) | % GFP+ URAs (n) | |||
|---|---|---|---|---|---|---|
| Punc-53gfp | Pglr-4grl-4∷gfp | Pcfi-1gfp | ||||
| +/+ | 100 | (120) | 95 | (92) | 91 | (124) |
| unc-86(bc151) a | 100 | (100) | 8 | (40) | 0 | (100) |
| lrs-1(bc155) a | 100 | (140) | 93 | (84) | 89 | (60) |
| unc-132(bc159) | 100 | (140) | 77 | (88) | 93 | (60) |
The presence of the AMso and URAs was determined in hermaphrodites using Punc-53gfp(bgEx21, AMso) or Pglr-4grl-4∷gfp (akEx32) and Pcfi-1gfp (nsEx37) (URAs) as described in Materials and Methods. Strains carrying the array akEx32 were homozygous for lin-15(n765ts) X. Strains carrying the array nsEx37 were homozygous for him-5(e1490) V and lin-15(n765ts) X.
Indicates that the strains were homozygous for dpy-17(e164) III.
unc-86, lrs-1 and unc-132 are required for ceh-30 expression in the CEMs in masculinized hermaphrodites
The Bar homeodomain transcription factor CEH-30, whose expression is under the direct control of the terminal, global regulator of somatic sexual fate, TRA-1, acts to block the death of the CEMs (21, 22). Specifically, the loss of ceh-30 function causes the CEMs to inappropriately die in males as well as in masculinized hermaphrodites, indicating that ceh-30 is required for CEM survival (21, 22) (Grote, P. and Conradt, B., unpublished data). Using a translational ceh-30 reporter (Pceh-30ceh-30∷gfp) (22), we determined the expression pattern of the ceh-30 gene in the CEMs in embryos ~450 min after the first cell division, which is just prior to the time at which the CEMs normally die in hermaphrodites (~470 min) (12, 16). The CEMs were identified based on their position using DIC and observed for ~30 min. We found that in wild-type hermaphrodites, the CEMs do not express ceh-30 and adopt a corpse-like morphology (Figure 2A, B, +/+). In contrast, in males (XO +/+) or masculinized hermaphrodites (sel-10(n1077)), the CEMs do express ceh-30 and do not adopt a corpse-like morphology (Figure 2A, B). These observations confirm that ceh-30 expression correlates with CEM survival. These observations also confirm that inappropriately surviving CEMs in masculinized hermaphrodites faithfully recapitulate molecular events that occur in the CEMs in males.
Fig. 2. unc-86, lrs-1 and unc-132 are required for ceh-30 expression in the CEMs in masculinized hermaphrodites.
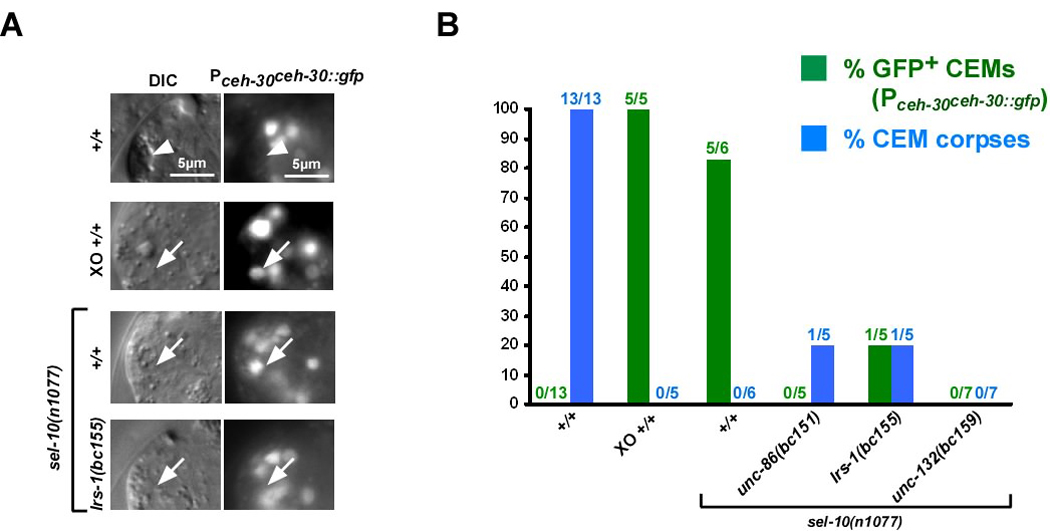
(A) DIC and fluorescence images (Pceh-30ceh-30∷gfp) of CEM corpses (white arrow heads) or CEMs (white arrows) in wild-type hermaphrodites (+/+), males (XO +/+), sel-10(n1077) hermaphrodites or lrs-1(bc155); sel-10(n1077) hermaphrodites. (B) Summary of data obtained on Pceh-30ceh-30∷gfp expression. Green bars indicate the percentage of CEMs that were GFP-positive and green numbers above indicate the fraction of CEMs analyzed that were GFP-positive. Blue bars indicate the percentage of CEMs that acquired a corpse-like morphology and blue numbers above indicate the fraction of CEMs analyzed that had a corpse-like morphology. All strains analyzed were homozygous for unc-76(e911) and carried the extrachromosomal array nEx1171 (Pceh-30ceh-30∷gfp). The wild-type hermaphrodites and males analyzed were homozygous for him-5(e1467ts). The strains unc-86(bc151); sel-10(n1077) and lrs-1(bc155); sel-10(n1077) were also homozygous for dpy-17(e164).
Next we analyzed the expression of the ceh-30 reporter in the CEMs in masculinized hermaphrodites lacking unc-86, lrs-1 or unc-132 function in which, as described above, the CEMs are generated and survive, but are mis-specified. We found that in these animals, ceh-30 is not expressed in most CEMs (Figure 2A, B). This result was surprising since the loss of ceh-30 function causes the CEMs to inappropriately die in masculinized hermaphrodites. Based on these observations we conclude that unc-86, lrs-1 and unc-132 are required for the expression of ceh-30 in the CEMs in masculinized hermaphrodites (Figure 5). We also conclude that apart from promoting ceh-30 expression, unc-86, lrs-1 and unc-132 have an additional function, which is to promote CEM death in a ceh-30-independent manner (Figure 5).
Fig. 5.
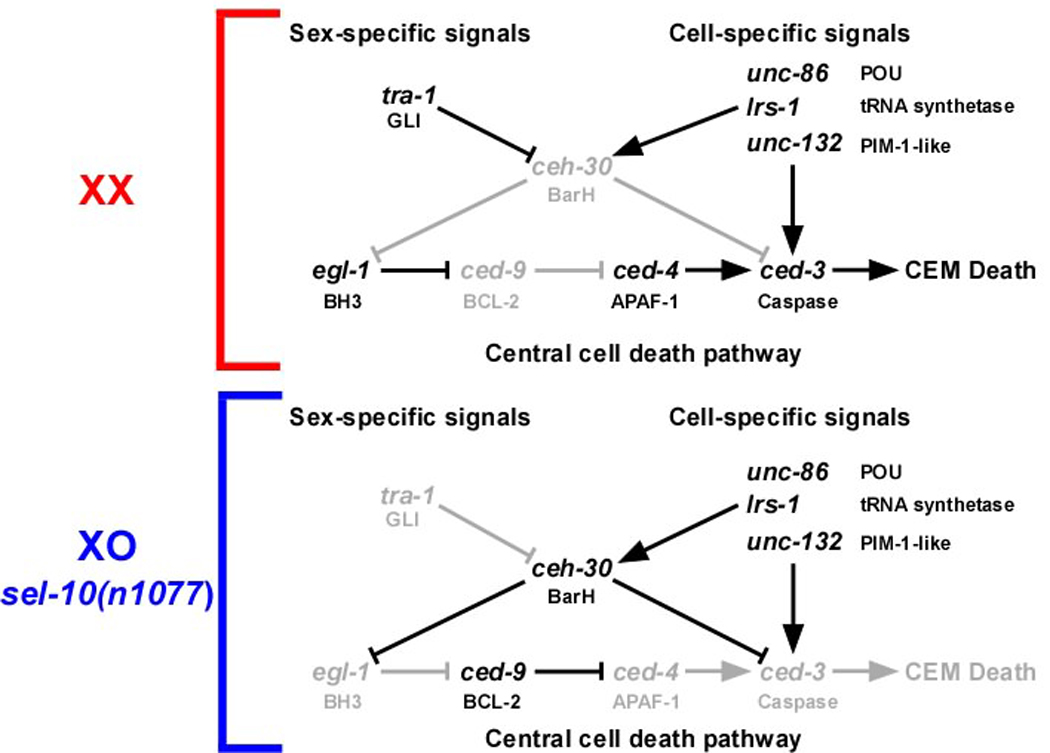
Genetic pathway of the CEM death. Upper panel. In wild-type hermaphrodites (XX), tra-1 is active in the CEMs, thereby blocking ceh-30. Thus the pro-apoptotic genes egl-1 and ced-3 are active, which promotes CEM death. See text for details. Lower panel. In males (XO) or masculinized hermaphrodites (sel-10(n1077)), tra-1 is not active in the CEMs, which results in the activation of ceh-30, and, consequently, the repression of egl-1 and ced-3. Thus, CEM death is inhibited. See text for details.
The loss of unc-86, lrs-1 or unc-132 function causes de-repression of egl-1 transcription in the CEMs in masculinized hermaphrodites
Using a transcriptional reporter (Pegl-1his-24∷gfp), we analyzed the expression of the egl-1 gene in the CEMs ~450 min after the first cell division (12, 16). We found that in wild-type hermaphrodites, the CEMs transcribe egl-1 and adopt a corpse-like morphology (Figure 3A, B, +/+). In contrast, in masculinized hermaphrodites, the CEMs do not transcribe egl-1 and do not adopt a corpse-like morphology (Figure 3A, B, sel-10(n1077)). As mentioned above, the loss of ceh-30 function causes the CEMs to inappropriately die in masculinized hermaphrodites. We found that the loss of ceh-30 function causes egl-1 to be inappropriately transcribed in the CEMs in masculinized hermaphrodites (Figure 2B, C, ceh-30(bc272); sel-10(n1077)). These results demonstrate that egl-1 transcriptional upregulation in the CEMs correlates with CEM death. In addition, they demonstrate that ceh-30 functions to repress egl-1 transcription in the CEMs in masculinized hermaphrodites (Figure 5).
Fig. 3. egl-1 is transcribed in surviving CEMs in masculinized unc-86(bc151), lrs-1(bc155), or unc-132(bc159) hermaphrodites.
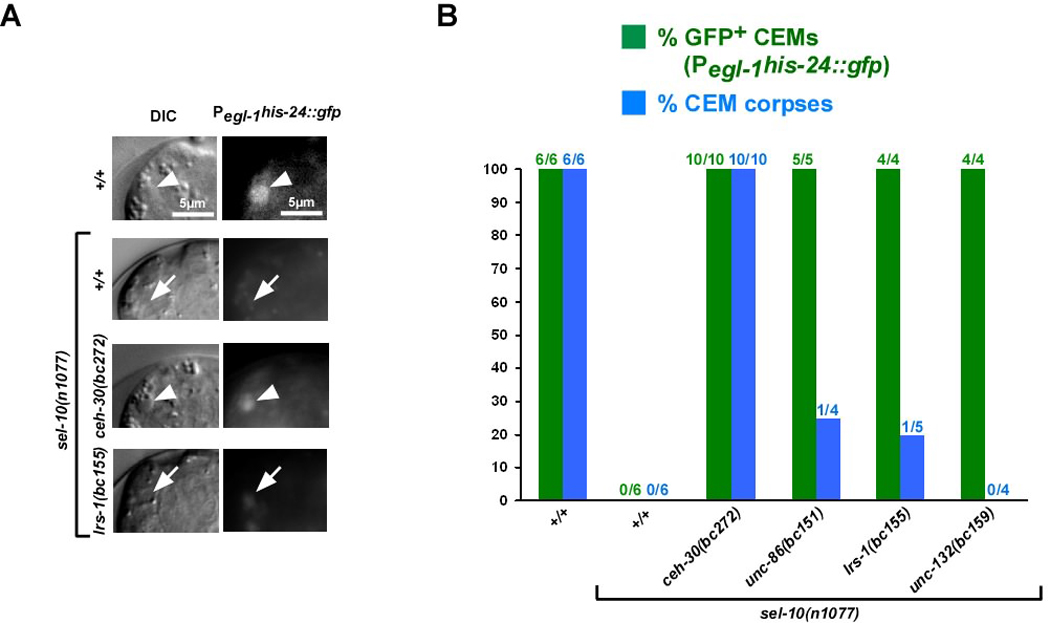
(A) DIC and fluorescence images (Pegl-1his-24∷gfp) of CEMs (white arrows) or CEM corpses (white arrow heads) in wild-type hermaphrodites (+/+), sel-10(n1077), sel-10(n1077); ceh-30(bc272) or lrs-1(bc155); sel-10(n1077) hermaphrodites. (B) Summary of data obtained on Pegl-1his-24∷gfp expression. Green bars indicate the percentage of CEMs that were GFP-positive and blue bars indicate the percentage of CEMs that acquired a corpse-like morphology. All strains analyzed were homozygous for the integrated Pegl-1his-24∷gfp array bcIs37. The strain ceh-30(bc272); sel-10(n1077) was also homozygous for the integrated array bcIs9. The strains unc-86(bc151); sel-10(n1077) and lrs-1(bc155); sel-10(n1077) were homozygous for dpy-17(e164).
To determine why the CEMs in masculinized hermaphrodites lacking unc-86, lrs-1 or unc-132 function, but not ceh-30 function, fail to inappropriately die, we analyzed egl-1 transcription in the CEMs in masculinized hermaphrodites lacking unc-86, lrs-1 or unc-132. To our surprise, we found that the CEMs in these animals transcribe egl-1 (Figure 3A, B). This observation indicates that, unlike in masculinized hermaphrodites lacking ceh-30 function, in masculinized hermaphrodites lacking unc-86, lrs-1 or unc-132 function, the transcriptional upregulation of egl-1 in the CEMs is not sufficient for their apoptotic death.
ced-3 is transcribed in the CEMs in hermaphrodites and its transcription in the CEMs in masculinized hermaphrodites is repressed by ceh-30
Using a reporter for the caspase gene ced-3, it has been shown that ced-3 is not transcribed in most of the 131 cells that are programmed to die (10). However, ced-3 is actively transcribed in at least one cell that is programmed to die, the tail spike cell (10). Like the CEMs, the tail spike cell survives much longer than most cells programmed to die (~300 min) before it dies. For this reason, we explored whether the ced-3 gene is transcribed in the CEMs prior to their deaths in hermaphrodites.
Using a transcriptional reporter (Pced-3gfp), we found that ced-3 is transcribed in most CEMs in wild-type hermaphrodites, in which the CEMs die (Figure 4 A, B, +/+). Therefore, the ced-3 gene is transcriptionally active in the CEMs. Furthermore, we found that in masculinized hermaphrodites, in which the CEMs survive, ced-3 is not transcribed (Figure 4 A, B, sel-10(n1077)). These observations are consistent with the notion that, like egl-1 transcription, the transcription of ced-3 in the CEMs correlates with CEM death. Next we determined the role of ceh-30 in ced-3 transcription in the CEMs. We found that in masculinized hermaphrodites lacking ceh-30 function, in which the CEMs inappropriately die, ced-3 was expressed in the CEMs (Figure 4 A, B). Therefore, we conclude that ceh-30 functions to repress ced-3 transcription in the CEMs in masculinized hermaphrodites (Figure 5).
Fig. 4. Analysis of ced-3 transcription in the CEMs.
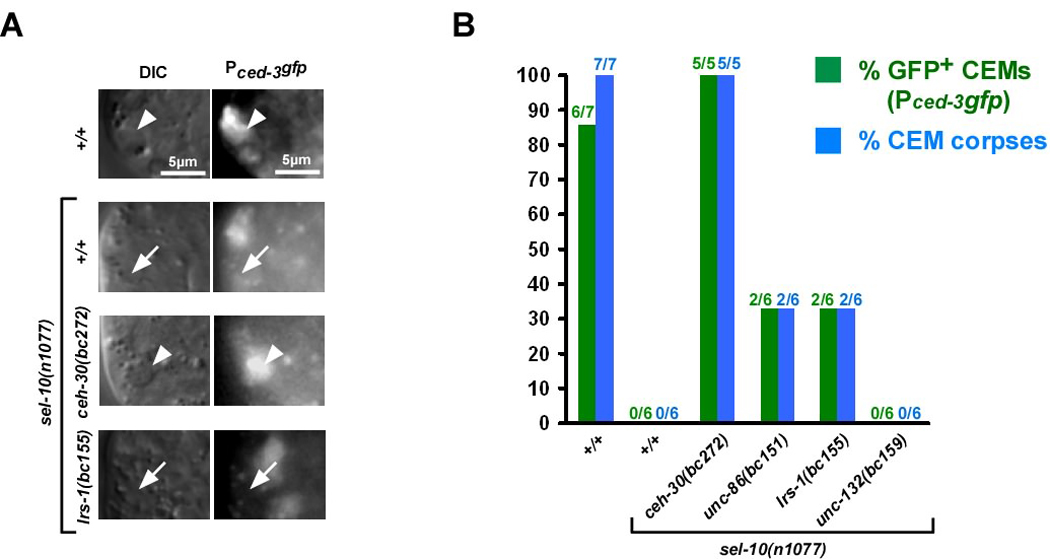
(A) DIC and fluorescence images (Pced-3gfp) of CEM corpses (white arrow heads) or CEMs (white arrows) in wild-type (+/+), sel-10(n1077), sel-10(n1077); ceh-30(bc272) or lrs-1(bc155); sel-10(n1077) hermaphrodites. (B) Summary of data obtained on Pced-3gfp expression. Green bars indicate the percentage of CEMs that were GFP-positive and blue bars indicate the percentage of CEMs that acquired a corpse-like morphology. All strains analyzed were homozygous for the integrated Ppkd-2gfp array bcIs9 and carried extrachromosomal arrays of Pced-3gfp (+/+ [bcEx834, bcEX836], sel-10(n1077) [bcEx839, bcEx840], ceh-30(bc272); sel-10(n1077) [bcEx876], unc-86(bc151); sel-10(n1077) [bcEx870, bcEx871], lrs-1(bc155); sel-10(n1077) [bcEx872, bcEx873], unc-132(bc159); sel-10(n1077) [bcEx874, bcEx875]).
The loss of unc-86, lrs-1 or unc-132 function does not cause de-repression of ced-3 transcription in the CEMs in masculinized hermaphrodites
To determine the role of unc-86, lrs-1 and unc-132 in ced-3 transcriptional control in the CEMs, we analyzed the expression of the Pced-3gfp reporter in masculinized hermaphrodites lacking unc-86, lrs-1 or unc-132 function. We found that, like in masculinized hermaphrodites, the Pced-3gfp reporter was not expressed in most CEMs (Figure 4 A, B). Therefore, while the loss of unc-86, lrs-1 or unc-132 function results in the de-repression of egl-1 transcription in the CEMs in masculinized hermaphrodites, it does not result in the de-repression of ced-3 transcription. Based on this observation we conclude that unc-86, lrs-1 and unc-132 not only repress ced-3 transcription in a ceh-30-dependent manner but also promote ced-3 transcription in a ceh-30-independent manner (Figure 5). Furthermore, we propose that the CEMs in masculinized hermaphrodites lacking unc-86, lrs-1 or unc-132 function fail to die because ced-3 transcription in the CEMs is not de-repressed.
ceh-30 expression inversely correlates with egl-1 and ced-3 transcription
Our results indicate that the death of the CEMs in wild-type hermaphrodites is the result of TRA-1-dependent repression of ceh-30 transcription, which results in the de-repression of egl-1 and ced-3 transcription in the CEMs. To test this hypothesis, we monitored ceh-30 expression as well as egl-1 and ced-3 transcription throughout the life span of the CEMs in hermaphrodites using 4D lineaging analysis and appropriate reporters (Pceh-30ceh-30∷gfp, Pegl-1his-24∷gfp, Pced-3gfp). We found that ceh-30 expression in the CEMs in wild-type hermaphrodites can only be detected right after the cells are generated (~340 min) (Figure 6, Figure S1). ceh-30 expression cannot be detected in the mother cell or in the CEMs at ~360 min or later time points. In contrast, egl-1 and ced-3 transcription can only be detected starting at ~390 and ~460 min, respectively (Figure 6, Figure S1). These observations demonstrate that ceh-30 expression in the CEMs inversely correlates with egl-1 and ced-3 transcription, which supports the model that TRA-1-dependent repression of ceh-30 transcription causes the de-repression of egl-1 and ced-3 transcription in the CEMs in wild-type hermaphrodites and, hence, CEM death.
Fig. 6. ceh-30, egl-1 and ced-3 expression in embryonic CEMs in wild-type hermaphrodites.
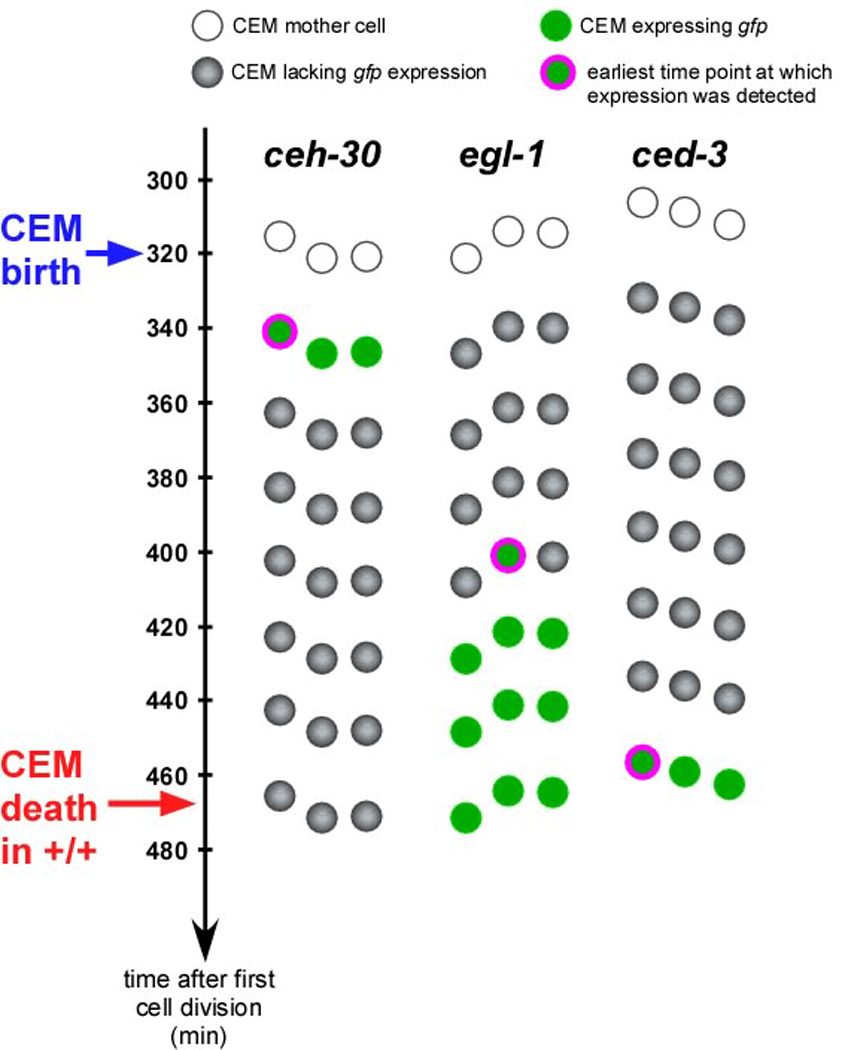
Schematic representation of time course analyses of the expression of the reporters Pceh-30ceh-30∷gfp, Pegl-1his-24∷gfp, and Pced-3gfp in the ventral left embryonic CEM (CEMVL) in wild-type hermaphrodites. Time (min) is indicated on the left. Three embryos were analyzed. White dots represent the CEM mother cells, green dots indicate CEMs expressing a particular gfp reporter. Green dots surrounded by purple circles indicate CEMs at the time when expression of the gfp reporter was first detected. (See Figure S1 for corresponding DIC and fluorescent images.) Gray dots indicate CEMs lacking reporter expression. Embryos were prepared for 4-D microscopy and lineaged starting at the 2- or 4-cell stage as described in Materials and Methods. Fluorescent stacks were taken every ~20 min after the CEM mother cell was born. No expression was detected for any of the reporters in the CEM mother cell.
Discussion
Transcriptional upregulation of both egl-1 and ced-3 is required for the apoptotic death of the CEMs in hermaphrodites
The transcriptional upregulation of egl-1 is thought to be necessary and sufficient for many of the cell death events that take place during development (14–16). We found that egl-1 transcriptional upregulation also correlates with cell death in the CEMs. Specifically, egl-1 is transcriptionally active in the CEMs in hermaphrodites in which the CEMs die but not in masculinized hermaphrodites (and most probably males) in which the CEMs survive. However, our results demonstrate that the transcriptional upregulation of egl-1 in the CEMs in hermaphrodites is necessary, but not sufficient for their death.
The ced-3 gene does not appear to be transcriptionally active in many of the cells that are programmed to die during development (10). However, we found that ced-3 is transcriptionally active in the CEMs in hermaphrodites in which the CEMs die but not in masculinized hermaphrodites in which the CEMs survive. Furthermore, we present data that indicate that transcriptional activation of ced-3 in the CEMs is necessary for their death in hermaphrodites. Based on these findings we propose that in contrast to most cells that are programmed to die during development, in the CEMs, the transcriptional upregulation of egl-1 and ced-3 is necessary for their death in hermaphrodites. At least to our knowledge, this is the first demonstration that the transcriptional upregulation of both a BH3-only gene as well as a caspase gene is required for apoptosis induction.
Why would the transcriptional upregulation of not only egl-1 but ced-3 be necessary for apoptosis induction in the CEMs? What distinguishes the CEMs from the majority of cells that die during development is that rather than surviving for only ~30 min after being generated, they survive for ~150 min (12). In the majority of cells that are programmed to die proCED-3 protein is inherited from progenitors (10). Based on this observation, we propose that at the time apoptosis induction takes place in the CEMs, as a result of proCED-3 turn over, the level of progenitor-derived proCED-3 has reached a level in the CEMs that is no longer sufficient for apoptosis induction (Figure 6A). For this reason, the transcriptional upregulation of not only the egl-1 gene but also the ced-3 gene is necessary for apoptosis induction in the CEMs.
Our model is supported by findings on the death of the tail spike cell. The tail spike cell, which may play a role in the morphogenesis of the tail, does not die until ~300 min after being generated (12). As is the case for the majority of cell death events as well as the death of the CEMs, the death of the tail spike cell is absolutely dependent on ced-3 (10). However, in contrast to the majority of cell death events as well as the death of the CEMs, it is only partially dependent on egl-1 (10). In addition, as in the CEMs, the ced-3 gene is transcriptionally active in the tail spike cell prior to its death. Furthermore, mutations in the gene pal-1, which encodes a caudal-like transcription factor, can prevent ced-3 transcriptional activation in the tail spike cell and tail spike cell death (10). What is unclear at the moment is whether egl-1 is also transcriptionally activated in the tail spike cell prior to its death. The over-expression of the ced-3 gene can be sufficient for apoptosis induction in the absence of apoptotic stimuli and egl-1 transcriptional upregulation (i.e. in cells programmed to live) (11). Therefore, we speculate that in the tail spike cell, ced-3 transcription might be upregulated to a level that results in amounts of proCED-3 that can be sufficient for EGL-1-independent apoptosis induction.
unc-86, lrs-1 and unc-132 promote ceh-30 transcription in the CEMs in masculinized hermaphrodites thereby causing repression of egl-1 and ced-3 transcription
Loss-of-function mutations of ceh-30 cause the CEMs to inappropriately undergo apoptosis in males and masculinized hermaphrodites through a yet unknown mechanism (21, 22) (Grote, P. and Conradt, B., unpublished data). The inappropriate death of CEMs in males lacking ceh-30 function is suppressed by the loss of egl-1 function (Schwartz and Horvitz, 2007). This suggests that the egl-1 gene acts downstream of the ceh-30 gene and is a potential target of the CEH-30 transcription factor. However, analyses performed with a gain-of-function mutation of ceh-30, which results in the mis-expression of the CEH-30 protein in the CEMs in hermaphrodites and, consequently, inappropriate survival, suggest that the ceh-30 gene blocks the death of the CEMs by acting in parallel to or downstream of egl-1 (22). We present data that indicate that the inappropriate death of CEMs in masculinized hermaphrodites lacking ceh-30 function is the result of the transcriptional de-repression of the pro-apoptotic genes egl-1 and ced-3. In other words, we show that the ceh-30 gene acts both upstream of as well as in parallel to egl-1 to prevent the activation of the central cell death machinery. We propose that CEH-30 is required to repress egl-1 and ced-3 transcription in the CEMs of masculinized hermaphrodites, thereby causing their survival (Figure 6B). Whether CEH-30 represses egl-1 and ced-3 transcription directly or indirectly remains to be determined.
The ceh-30 gene is transcriptionally active in the CEMs in males and masculinized hermaphrodites, but not in hermaphrodites. In hermaphrodites, ceh-30 transcription in the CEMs is directly repressed by the GLI-like transcription factor TRA-1, which is the terminal, global regulator of somatic sexual fate (20–22). Our results indicate that the genes unc-86, lrs-1, and unc-132, which are required for correct CEM specification, are necessary for ceh-30 transcriptional activation in the CEMs in masculinized hermaphrodites. The sex-specific and cell-specific signals that determine the life-versus-death decision in the CEMs are therefore integrated at the level of ceh-30 transcriptional control (Figure 5).
unc-86 encodes a POU homeodomain transcription factor that has previously been implicated in CEM specification and differentiation (27, 28). Our results demonstrate that the UNC-86 protein is required for ceh-30 transcription in the CEMs, which is consistent with previous studies that suggest that UNC-86 acts as a direct activator of ceh-30 transcription (21). Surprisingly, lrs-1 encodes a leucyl-tRNA synthetase. Based on our analyses, compromising translation by decreasing the level of at least certain aminoacyl-tRNA synthetases abrogates ceh-30 transcription in the CEMs. Therefore, we propose that ceh-30 transcription is particularly sensitive to perturbations of the translational machinery. For example, the translation of factors required for ceh-30 transcription (such as UNC-86 or UNC-132 or regulators thereof) may be particularly sensitive to such perturbations. Finally, unc-132 encodes a novel protein of unknown function with a P-loop domain, which suggests that it is capable of nucleotide binding. The D. melanogaster orthologue of unc-132, CG30118, was identified in a screen for genes involved in Drosophila hematopoiesis and is required for the correct localization of crystal cells and plasmatocytes (two classes of terminally differentiated blood cells) in Drosophila embryos (30). However, the mechanism through which CG30118 affects the localization of these cells has so far not been determined. Furthermore, we found that the N-terminal region of the UNC-132 protein shares sequence similarity with the N-terminal region of the human proto-oncoprotein PIM-1 (31). PIM-1 is a serine/threonine kinase, which is involved in various cellular processes such as cell metabolism, cell proliferation and differentiation and cell survival (35, 36). However, the roles of PIM-1 in these cellular processes are thought to be dependent on its kinase activity. Since UNC-132 does not contain a classical kinase domain, it is therefore unclear how extensive the functional similarities between PIM-1 and UNC-132 are. In summary, while UNC-86 may be a direct activator of ceh-30 transcription, the mechanisms through which LRS-1 and UNC-132 promote ceh-30 transcription in the CEMs remain to be elucidated.
unc-86, lrs-1 and unc-132 promote ced-3 transcription in the CEMs of hermaphrodites in a ceh-30-independent manner thereby promoting their death
unc-86, lrs-1, and unc-132 are required for ceh-30 transcriptional activation in the CEMs in masculinized hermaphrodites. However, while we found that the loss of ceh-30 function results in the de-repression of both egl-1 and ced-3 transcription, the loss of unc-86, lrs-1, or unc-132 function only results in the de-repression of egl-1 transcription. For this reason, we propose that apart from blocking egl-1 and ced-3 transcription in a ceh-30-dependent manner, unc-86, lrs-1, and unc-132 also promote ced-3 transcription in a ceh-30-independent manner (Figure 5). Furthermore, we hypothesize that the ability of unc-86, lrs-1, and unc-132 to promote ced-3 transcription in a ceh-30-independent manner is relevant in hermaphrodites in which the ced-3 gene is relieved of ceh-30-dependent transcriptional repression. Conversely, their ability to block ced-3 transcription in a ceh-30-dependent manner may be relevant in males and masculinized hermaphrodites in which the ceh-30 gene is relieved of TRA-1-dependent transcriptional repression (Figure 5). How UNC-86, LRS-1, and UNC-132 promote ced-3 transcription in the CEMs in hermaphrodites remains to be determined.
Why do the CEMs survive for 150 min?
One obvious question that arises from our studies is why CEMs survive for ~150 rather than ~30 min before they undergo apoptosis. The CEMs are born ~320 min after the first cell division and die in hermaphrodites at ~470 min (12). The TRA-1 protein plays a pivotal role in the life-versus-death decision of the CEMs. In the CEMs, TRA-1 represses ceh-30 transcription thereby causing egl-1 and ced-3 de-repression and apoptosis induction. The activity of TRA-1 is regulated at the post-translational level by sex-specific cleavage and proteolysis (37, 38). Specifically, in males, TRA-1 is subject to proteasomal degradation. In hermaphrodites, however, TRA-1 is processed and phosphorylated, which generates a stable and active TRA-1 fragment capable of controlling transcription. Currently it is unclear when during embryonic development active TRA-1 protein is first generated in hermaphrodites. However, experiments using a temperature-sensitive loss-of-function mutation of the gene tra-2, which acts upstream of tra-1 in somatic sex determination, suggest that active TRA-1 might be generated in hermaphrodites starting at ~320 min after the first cell division (39). This notion is supported by the finding that the ceh-30 gene, whose expression is directly repressed by TRA-1, is no longer expressed at ~360 min (Figure 5, Figure 7). Based on these observations, we propose that the CEMs die only at ~470 min, because a level of active TRA-1 first has to be generated in the CEMs in hermaphrodites that is sufficient for the transcriptional repression of ceh-30, which is a prerequisite for the transcriptional de-repression of egl-1 and ced-3 transcription at ~390 and 460 min, respectively, and, hence, apoptosis induction (Figure 7). Hence, we propose that the timing of the CEM death is controlled by the sex determination pathway and, in particular, its terminal, global regulator, the Gli-like transcription factor TRA-1.
Fig. 7. Model of the life-versus-death decision in the CEMs.
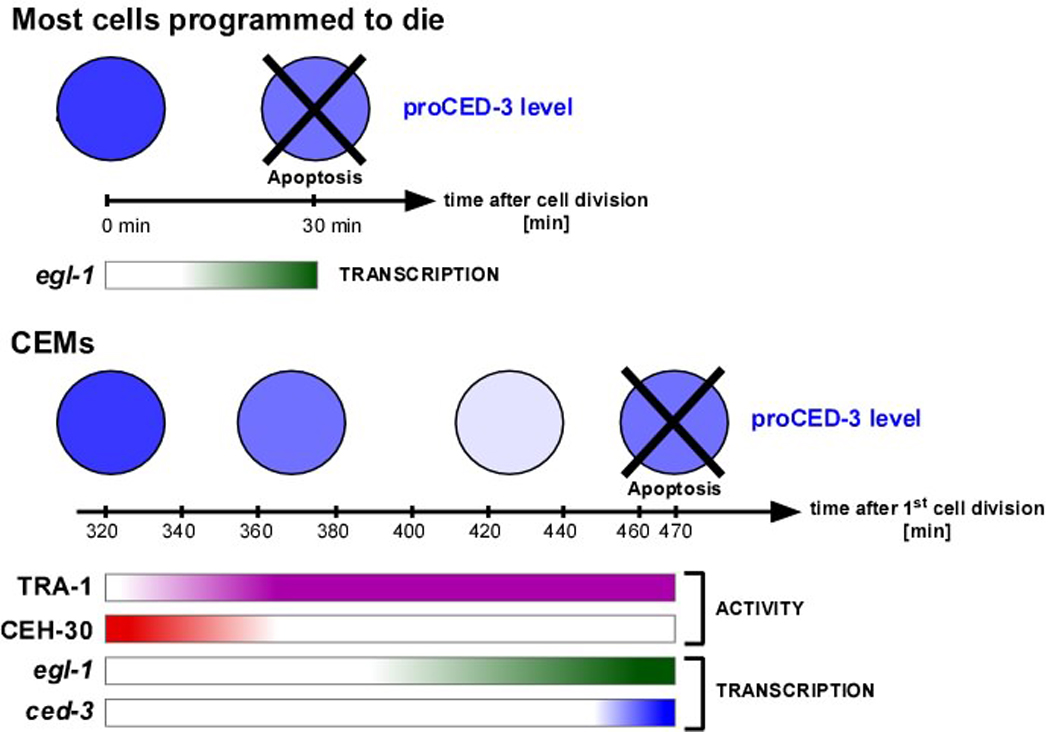
Top panel. Most cells programmed to die do so ~30 min after being generated. In these cells, proCED-3 protein inherited from the progenitor is sufficient for apoptosis induction in response to egl-1 transcriptional activation. Bottom panel. The CEMs in hermaphrodites die ~150 min after being generated (~470 min after the first cell division). Increasing TRA-1 activity results in decreasing CEH-30 activity, which is required for the de-repression of egl-1 and ced-3 transcription. The activation at the transcriptional level of the ced-3 gene compensates for decreased levels of proCED-3. See text for details.
Potential role for the co-expression of BH3-only and caspase genes in the developmental control of apoptosis
At least during animal development, pro-caspases are thought to be present in most if not all cells (2–5). Furthermore, pro-caspases are thought to be present at levels that, once they become matured and activated, are sufficient for apoptosis induction. Since the maturation and activation of pro-caspases generally represents the commitment of a cell to the apoptotic fate, it is a highly regulated process (40, 41). In contrast, very little is known thus far about the contribution of transcriptional control in the regulation of pro-caspases. The most comprehensive studies available to date were performed in D. melanogaster. These studies suggest that transcriptional regulation of caspase genes plays an important role in apoptosis induction at least during D. melanogaster development (42). Specifically, it was found that ecdysone-induced apoptosis during Drosophila metamorphosis is dependent on the transcriptional upregulation of the caspase genes DRONC and DRICE (43–45). The data presented here demonstrate that the transcriptional up-regulation of the C. elegans caspase gene ced-3 is required for the sexually-dimorphic, apoptotic death of the CEM neurons. Based on these findings, we propose that the transcriptional up-regulation of caspase genes is a conserved aspect of apoptosis induction during animal development. Furthermore, our data demonstrate that in C. elegans, genes encoding BH3-only proteins and pro-caspases can be under the same transcriptional control. We hypothesize that such co-regulatory mechanisms have evolved to ensure efficient apoptosis induction during development.
Materials and Methods
Strains and general methods
C. elegans strains were cultured as described (46). Bristol N2 was used as the wild-type strain. N2 and the CB4856 (Hawaii) strain were used for SNP mapping. Mutations and integrated transgenes used in this study are listed below and are described (47) except where noted otherwise: LG II: rrf-3(pk1426) (48). LG III: ced-4(n1162), ced-3(n717), unc-86(bc151) (this study), lrs-1(bc155) (this study), dpy-17(e164), unc-93(e1500sd). LG V: bcIs37 (Pegl-1his-24∷gfp) (16), egl-1(n1084 n3082) (13), sel-10(n1077), unc-132(bc159) (this study), unc-76(e911), him-5(e1490), him-5(e1467ts), dpy-11(e224). LG X: ceh-30(bc272) (P. Grote and B. Conradt; unpublished data), lin-15(n765ts), bcIs9 (Ppkd-2gfp) (49). The following extrachromosomal arrays were used: nEx1171 (Pceh-30ceh-30∷gfp) (22), bgEx21 (Punc-53gfp) (34), syEx301 (Plov-1gfp) (26), nsEx37 (Pcfi-1gfp) (33), akEx32 (Pglr-4glr-4∷gfp) (32).
RNAi by feeding was performed as described using 6 mM IPTG (50). The plasmids pBC966 (W08A12.1b(RNAi)) and pBC967 (W08A12.1c(RNAi)) contained full-length cDNA clones cloned as NcoI-NheI fragments into vector L4440. sel-10(n1077); bcIs9 animals were mutagenized with EMS (ethyl-methanesulfonate) as described (46). Germline transformations were performed as described (51). Cosmids and fosmids were injected at a concentration of 10 ng/µl using pRF4 (rol-6(su1006)) at 50 ng/µl as coinjection marker. The plasmid Pced-3gfp was injected at 40 ng/ul using pRF4 (rol-6(su1006)) (50 ng/µl) as coinjection marker. In the case of bcIs9 (Ppkd-2gfp), syEx301 (Plov-1gfp), bgEx21 (Punc-53gfp), nsEx37 (Pcfi-1gfp), akEx32 (Pglr-4glr-4∷gfp), bcIs37 (Pegl-1his-24∷gfp) and nEx1171 (Pceh-30ceh-30∷gfp), transgenic animals were crossed with unc-86(bc151), lrs-1(bc155) or unc-132(bc159) mutants to generate unc-86(bc151), lrs-1(bc155) and unc-132(bc159) strains carrying these arrays. pBC957 (Pced-3gfp) was constructed by removing an internal, 4.6 kb EcoRV fragment containing most of the ced-3 coding sequence from a ced-3 reporter (10).
Cloning of unc-86(bc151), lrs-1(bc155) and unc-132(bc159)
Standard genetic techniques were used to map bc151 to the right of dpy-17 on LG III, bc155 between unc-93 and dpy-17 on LG III, and bc159 to the left of dpy-11 on LG V.
bc151: bc151 failed to complement unc-86(n846), a lf allele of the gene unc-86, indicating that bc151 is a loss-of-function mutation in the unc-86 gene.
bc155: SNP mapping was used to locate bc155 between the SNPs C30D11:9408 and R10E4:10469. The fosmid WRM0638-J8 rescued the phenotype observed in bc155; sel-10(n1077); bcIs9 animals.
bc159: SNP mapping was used to map bc159 to the left of SNP H10D18:26865. The cosmid W08A12 rescued the phenotype observed in bc159; sel-10(n1077); bcIs9 animals. The bc159 phenotype was also rescued by a 31.3 kb XcmI subclone of W08A12 (pBC430, containing the genes W08A12.1, W08A12.2 and egr-1) and was partially rescued by a 23.9 kb ApaI subclone of W08A12 (pBC431, spanning the genes W08A12.4, egr-1, and W08A12.1).
Phenotypic analysis
The presence of CEMs in adults was analyzed using bcIs9 (Ppkd-2gfp) and a Zeiss Axioskop2 equipped with epifluorescence as described (49). The ɑ GFP-positive CEMs was calculated by dividing the number of CEMs observed by the maximum number of possible CEMs (four CEMs per animal). The presence of CEMs in L4 larvae was analyzed using DIC as described (17). The presence of CEMs in embryos was analyzed as follows. CEMs were identified by DIC in 1½ -fold stage embryos (~450 min) based on their positions and observed for at least ~30 min. (Since the positions of the two dorsal CEMs are less characteristic, these cells could not always be examined.) CEM corpses appear in 2-fold embryos (~465 min). Microscopy of living embryos was performed by mounting embryos on 2% agar pads in M9 buffer, using a Zeiss Axioskop2 equipped with epifluorescence, a Micromax CCD camera (Princeton Instruments), and Metamorph software. DIC and epifluorescence images were taken every 5 min between the 1½- and 2-fold stage to determine the fate of the CEMs and the expression of the GFP reporters in embryonic CEMs.
The presence of the AMso (amphid socket) cells was scored in adults using Punc-53gfp as described (32). The presence of the URA cells was determined in adults using two different reporters, Pglr-4grl-4∷gfp and Pcfi-1gfp, as described (33).
4-D microscopy and lineage analysis
4-D microscopy and lineaging analysis was performed on C. elegans embryos as previously described (52, 53). Embryos were recorded at 20°C, and GFP expression of the reporters Pceh-30ceh-30∷gfp, Pegl-1his-24∷gfp and Pced-3gfp was recorded after every 30 DIC stacks. The 4-D recordings were analyzed using the SIMI BioCell software as previously described (52, 54). The ventral left CEM (CEMVL, ABplpaapapp) was lineaged in all embryos. Starting at the 2 to 4-cell stage, cells were tracked and their 3-D coordinates were saved approximately every 2 min, until at least ~465 min after the first cell division. Additionally, the 3-D coordinates of the CEMVL progenitor (ABplpaapap) or CEMVL were saved at the time points when GFP expression of the reporters Pceh-30ceh-30∷gfp, Pegl-1his-24∷gfp or Pced-3gfp was recorded. Using this approach, the status of expression of these reporters in the CEM could be determined. The cell death fate of the CEMVL was determined at ~465 min by DIC.
Supplementary Material
Acknowledgements
We thank E. Lambie and S. Rolland for comments on the manuscript; H. Schwartz, M. Saito, J. Dunlap and members of the Conradt lab for discussions; D. Mayka and C. Huber for excellent technical support; E. Lambie for use of the micro-injection set-up; H. Schwartz and B. Horvitz for array nEx1171 (Pceh-30ceh-30∷gfp); S. Shaham for array nsEx37 (Pcfi-1gfp) and plasmid Pced-3gfp; N. Pujol for array bgEx21 (Punc-53gfp); V. Maricq for array akEx32 (Pglr-4glr-4∷gfp); M. Barr for array syEx301 (Plov-1gfp) and plasmid Ppkd-2gfp; the Sanger Centre (Hinxton, UK) for cosmids; and the C. elegans Genetics Center (CGC, supported by the NIH National Center for Research Resources) for strains. This work was supported by funding from the Max Planck Society, Howard Hughes Medical Institute Award 76200–560801 to Dartmouth Medical School under the Biomedical Research Support Program for Medical Schools, and National Institute of Health grant R01-GM069950.
References
- 1.Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96(2):245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 2.Adams JM. Ways of dying: multiple pathways to apoptosis. Genes Dev. 2003 Oct 15;17(20):2481–2495. doi: 10.1101/gad.1126903. [DOI] [PubMed] [Google Scholar]
- 3.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004 Jan 23;116(2):205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 4.Horvitz HR. Nobel lecture. Worms, life and death. Biosci Rep. 2003 Oct-Dec;23(5–6):239–303. doi: 10.1023/b:bire.0000019187.19019.e6. [DOI] [PubMed] [Google Scholar]
- 5.Lettre G, Hengartner MO. Developmental apoptosis in C. elegans: a complex CEDnario. Nature Reviews Molecular Cell Biology. 2006;7:97–108. doi: 10.1038/nrm1836. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher JI, Huang DC. BH3-only proteins: orchestrating cell death. Cell Death Differ. 2006 Aug;13(8):1268–1271. doi: 10.1038/sj.cdd.4401995. [DOI] [PubMed] [Google Scholar]
- 7.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008 Jan;9(1):47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 8.Conradt B, Xue D. Programmed cell death. WormBook. 2005:1–13. doi: 10.1895/wormbook.1.32.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, Gruenbaum Y, et al. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000;287(5457):1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- 10.Maurer CW, Chiorazzi M, Shaham S. Timing of the onset of a developmental cell death is controlled by transcriptional induction of the C. elegans ced-3 caspase-encoding gene. Development. 2007 Apr;134(7):1357–1368. doi: 10.1242/dev.02818. [DOI] [PubMed] [Google Scholar]
- 11.Shaham S, Horvitz HR. Developing Caenorhabditis elegans neurons may contain both cell-death protective and killer activities. Genes Dev. 1996;10(5):578–591. doi: 10.1101/gad.10.5.578. [DOI] [PubMed] [Google Scholar]
- 12.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100(1):64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 13.Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998 May 15;93(4):519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 14.Conradt B, Horvitz HR. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 1999 Aug 6;98(3):317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, Strauss TJ, Potts MB, Cameron S. Direct regulation of egl-1 and of programmed cell death by the Hox protein MAB-5 and by CEH-20, a C. elegans homolog of Pbx1. Development. 2006 Feb;133(4):641–650. doi: 10.1242/dev.02234. [DOI] [PubMed] [Google Scholar]
- 16.Thellmann M, Hatzold J, Conradt B. The Snail-like CES-1 protein of C. elegans can block the expression of the BH3-only cell-death activator gene egl-1 by antagonizing the function of bHLH proteins. Development. 2003 Sep;130(17):4057–4071. doi: 10.1242/dev.00597. [DOI] [PubMed] [Google Scholar]
- 17.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56(1):110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 18.White JQ, Nicholas TJ, Gritton J, Truong L, Davidson ER, Jorgensen EM. The Sensory Circuitry for Sexual Attraction in C. elegans Males. Curr Biol. 2007 Nov 6;17(21):1847–1857. doi: 10.1016/j.cub.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44(6):817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 20.Wolff JR, Zarkower D. Chapter 1 Somatic Sexual Differentiation in Caenorhabditis elegans. Curr Top Dev Biol. 2008;83:1–39. doi: 10.1016/S0070-2153(08)00401-8. [DOI] [PubMed] [Google Scholar]
- 21.Peden E, Kimberly E, Gengyo-Ando K, Mitani S, Xue D. Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho. Genes Dev. 2007 Dec 1;21(23):3195–3207. doi: 10.1101/gad.1607807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz HT, Horvitz HR. The C. elegans protein CEH-30 protects male-specific neurons from apoptosis independently of the Bcl-2 homolog CED-9. Genes Dev. 2007 Dec 1;21(23):3181–3194. doi: 10.1101/gad.1607007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai C, Horvitz HR. Caenorhabditis elegans mutants defective in the functioning of the motor neurons responsible for egg laying. Genetics. 1989;121(4):703–721. doi: 10.1093/genetics/121.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doniach T. Activity of the sex-determining gene tra-2 is modulated to allow spermatogenesis in the C. elegans hermaphrodite. Genetics. 1986 Sep;114(1):53–76. doi: 10.1093/genetics/114.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jager S, Schwartz HT, Horvitz HR, Conradt B. The Caenorhabditis elegans F-box protein SEL-10 promotes female development and may target FEM-1 and FEM-3 for degradation by the proteasome. Proc Natl Acad Sci U S A. 2004 Aug 24;101(34):12549–12554. doi: 10.1073/pnas.0405087101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barr MM, Sternberg PW. A polycystic kidney-disease gene homologue required for male mating behaviour in C. elegans. Nature. 1999;401(6751):386–389. doi: 10.1038/43913. [DOI] [PubMed] [Google Scholar]
- 27.Finney M, Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63(5):895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 28.Finney M, Ruvkun G, Horvitz HR. The C. elegans cell lineage and differentiation gene unc-86 encodes a protein with a homeodomain and extended similarity to transcription factors. Cell. 1988;55(5):757–769. doi: 10.1016/0092-8674(88)90132-8. [DOI] [PubMed] [Google Scholar]
- 29.Schimmel P. Development of tRNA synthetases and connection to genetic code and disease. Protein Sci. 2008 Oct;17(10):1643–1652. doi: 10.1110/ps.037242.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milchanowski AB, Henkenius AL, Narayanan M, Hartenstein V, Banerjee U. Identification and characterization of genes involved in embryonic crystal cell formation during Drosophila hematopoiesis. Genetics. 2004 Sep;168(1):325–339. doi: 10.1534/genetics.104.028639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagarajan L, Louie E, Tsujimoto Y, ar-Rushdi A, Huebner K, Croce CM. Localization of the human pim oncogene (PIM) to a region of chromosome 6 involved in translocations in acute leukemias. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2556–2560. doi: 10.1073/pnas.83.8.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J Neurosci. 2001 Mar 1;21(5):1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaham S, Bargmann CI. Control of neuronal subtype identity by the C. elegans ARID protein CFI-1. Genes Dev. 2002 Apr 15;16(8):972–983. doi: 10.1101/gad.976002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stringham E, Pujol N, Vandekerckhove J, Bogaert T. unc-53 controls longitudinal migration in C. elegans. Development. 2002 Jul;129(14):3367–3379. doi: 10.1242/dev.129.14.3367. [DOI] [PubMed] [Google Scholar]
- 35.Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. J Clin Invest. 2005 Oct;115(10):2618–2624. doi: 10.1172/JCI26273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bachmann M, Moroy T. The serine/threonine kinase Pim-1. Int J Biochem Cell Biol. 2005 Apr;37(4):726–730. doi: 10.1016/j.biocel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Schvarzstein M, Spence AM. The C. elegans sex-determining GLI protein TRA-1A is regulated by sex-specific proteolysis. Dev Cell. 2006 Nov;11(5):733–740. doi: 10.1016/j.devcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev Cell. 2007 Jul;13(1):127–139. doi: 10.1016/j.devcel.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klass M, Wolf N, Hirsh D. Development of the male reproductive system and sexual transformation in the nematode Caenorhabditis elegans. Dev Biol. 1976 Aug;52(1):1–18. doi: 10.1016/0012-1606(76)90002-6. [DOI] [PubMed] [Google Scholar]
- 40.Boyce M, Degterev A, Yuan J. Caspases: an ancient cellular sword of Damocles. Cell Death Differ. 2004 Jan;11(1):29–37. doi: 10.1038/sj.cdd.4401339. [DOI] [PubMed] [Google Scholar]
- 41.Schafer ZT, Kornbluth S. The apoptosome: physiological, developmental, and pathological modes of regulation. Dev Cell. 2006 May;10(5):549–561. doi: 10.1016/j.devcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 42.Kumar S, Cakouros D. Transcriptional control of the core cell-death machinery. Trends Biochem Sci. 2004 Apr;29(4):193–199. doi: 10.1016/j.tibs.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Cakouros D, Daish T, Martin D, Baehrecke EH, Kumar S. Ecdysone-induced expression of the caspase DRONC during hormone-dependent programmed cell death in Drosophila is regulated by Broad-Complex. J Cell Biol. 2002 Jun 10;157(6):985–995. doi: 10.1083/jcb.200201034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cakouros D, Daish TJ, Kumar S. Ecdysone receptor directly binds the promoter of the Drosophila caspase dronc, regulating its expression in specific tissues. J Cell Biol. 2004 Jun 7;165(5):631–640. doi: 10.1083/jcb.200311057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kilpatrick ZE, Cakouros D, Kumar S. Ecdysone-mediated up-regulation of the effector caspase DRICE is required for hormone-dependent apoptosis in Drosophila cells. J Biol Chem. 2005 Mar 25;280(12):11981–11986. doi: 10.1074/jbc.M413971200. [DOI] [PubMed] [Google Scholar]
- 46.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riddle DL. C. elegans II. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 48.Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002 Aug 6;12(15):1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 49.Grote P, Conradt B. The PLZF-like protein TRA-4 cooperates with the Gli-like transcription factor TRA-1 to promote female development in C. elegans. Dev Cell. 2006 Oct;11(4):561–573. doi: 10.1016/j.devcel.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 50.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263(1–2):103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 51.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 52.Bischoff M, Schnabel R. A posterior centre establishes and maintains polarity of the Caenorhabditis elegans embryo by a Wnt-dependent relay mechanism. PLoS Biol. 2006 Nov;4(12):e396. doi: 10.1371/journal.pbio.0040396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schnabel R, Hutter H, Moerman D, Schnabel H. Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Dev Biol. 1997;184(2):234–265. doi: 10.1006/dbio.1997.8509. [DOI] [PubMed] [Google Scholar]
- 54.Schnabel R, Bischoff M, Hintze A, Schulz AK, Hejnol A, Meinhardt H, et al. Global cell sorting in the C. elegans embryo defines a new mechanism for pattern formation. Dev Biol. 2006 Jun 15;294(2):418–431. doi: 10.1016/j.ydbio.2006.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


