Abstract
The addition of novel amino acids to the genetic code of Escherichia coli involves the generation of an aminoacyl-tRNA synthetase and tRNA pair that is ‘orthogonal’, meaning that it functions independently of the synthetases and tRNAs endogenous to E.coli. The amino acid specificity of the orthogonal synthetase is then modified to charge the corresponding orthogonal tRNA with an unnatural amino acid that is subsequently incorporated into a polypeptide in response to a nonsense or missense codon. Here we report the development of an orthogonal glutamic acid synthetase and tRNA pair. The tRNA is derived from the consensus sequence obtained from a multiple sequence alignment of archaeal tRNAGlu sequences. The glutamyl-tRNA synthetase is from the achaebacterium Pyrococcus horikoshii. The new orthogonal pair suppresses amber nonsense codons with an efficiency roughly comparable to that of the orthogonal tyrosine pair derived from Methanococcus jannaschii, which has been used to selectively incorporate a variety of unnatural amino acids into proteins in E.coli. Development of the glutamic acid orthogonal pair increases the potential diversity of unnatural amino acid structures that may be incorporated into proteins in E.coli.
INTRODUCTION
The ability to genetically incorporate unnatural amino acids into proteins offers new opportunities to investigate protein structure and function in vitro and in vivo, as well as produce large quantities of proteins with novel properties. This approach involves the directed evolution of the amino acid specificity of an aminoacyl-tRNA synthetase and tRNA pair that is orthogonal in Escherichia coli (1–3). Such orthogonal pairs satisfy several criteria: the tRNA is not a substrate for any of the endogenous E.coli synthetases but functions efficiently in protein translation; the orthogonal synthetase efficiently aminoacylates the orthogonal tRNA, the anticodon of which has been modified to recognize an amber (UAG) or opal (UGA) stop codon or a four-base codon; the synthetase does not aminoacylate any of the endogenous E.coli tRNAs. To date, several orthogonal pairs have been developed for use in E.coli, including glutamine (1), aspartic acid (4) and tyrosine (5,6) pairs from Saccharomyces cerevisiae, a tyrosine pair from Methanococcus jannaschii (7) and a leucine pair from Methanobacterium thermoautotrophicum (8). In addition, glutamine (6) and tyrosine (9) synthetase-tRNA pairs from E.coli have been developed for use as orthogonal pairs in S.cerevisiae.
To date, only the orthogonal tyrosine pair from M.jannaschii has been successfully used to incorporate unnatural amino acids into proteins in E.coli. The explanation for this is not entirely clear, but may be related, at least in part, to the low amber suppression efficiencies exhibited by some of the other orthogonal synthetase–tRNA pairs, particularly those for glutamine (1) and aspartic acid (4). Moreover, all of the unnatural amino acids that have been incorporated thus far using variants of the tyrosyl-tRNA synthetase are aryl derivatives, including: p-azido- (10), p-benzoyl- (11), p-amino- (3), p-isopropyl- (3), m-acetyl- (12) and p-acetyl-phenylalanine (12); O-methyl- (2) and O-allyl-tyrosine (3,13); 3-(2-naphthyl)alanine (14). In order to increase the structural and chemical diversity of unnatural amino acids that can be incorporated into proteins expressed by E.coli, there is a need to develop additional active orthogonal pairs.
Archaea appear to be an especially good source of orthogonal pairs for E.coli. Previous studies revealed that several archaeal tRNAs are not recognized by E.coli aminoacyl-tRNA synthetases (15). Two such synthetase–tRNA pairs derived from archaea, for tyrosine (7) and leucine (8), were developed for use in E.coli. Both exhibited high levels of orthogonality and amber suppression efficiency. Moreover, these archaeal synthetases and tRNAs are efficiently produced and processed in E.coli. Finally, the recent proliferation of genomic sequence data for archaeal species and the commercial availability of their genomic DNA have facilitated the cloning of synthetases from these organisms as well as the design of compatible tRNAs.
Archaeal glutamyl-tRNA synthetases and tRNAs are attractive starting points for the generation of additional orthogonal pairs for several reasons. Archaeal glutamyl-tRNA synthetases are expected to tolerate changes in the anticodon loop of their substrate tRNAs, since the enzyme naturally accommodates four different tRNA anticodons: UUC, CUC, UUG and CUG. This is due to the fact that in archaeal species, glutamyl-tRNA synthetases must recognize and acylate both tRNAGlu and tRNAGln (16). It follows that an archaeal glutamyl-tRNA synthetase should tolerate variance within at least two positions of the anticodon of its tRNA substrate, which should permit efficient aminoacylation of a tRNA bearing an amber-suppressing CUA anticodon. In addition, in vitro aminoacylation data (15) predict that archaeal glutamic acid and glutamine tRNAs should be orthogonal in E.coli. Finally, the three-dimensional structure of the glutamyl-tRNA synthetase from Thermus thermophilus is known (17,18), which should facilitate the design of libraries of synthetase variants for use in directed evolution experiments.
MATERIALS AND METHODS
Bacterial strains, plasmids and reagents
Escherichia coli strains DH5α and DH10B were purchased from Stratagene (La Jolla, CA) and Invitrogen Life Sciences (Carlsbad, CA), respectively. Archaeoglobus fulgidus (Af), Aeropyrum pernix (Ap), M.jannaschii (Mj), Methanosarcina mazei Goe1 (Mm), M.thermoautotrophicum (Mt), Pyrococcus horikoshii (Ph) and Sulfolobus solfataricus (Ss) genomic DNA was obtained from ATCC (Manassas, VA). Escherichia coli genomic DNA was prepared from strain DH5α. Plasmid pKQ, which contains a ColE1 origin of replication, a kanamycin resistance marker and cloning sites for an aminoacyl-tRNA synthetase gene, has been described previously (8). Plasmid pACKO-A184TAG, which contains a p15A origin of replication, a chloramphenicol resistance marker, a β-lactamase ampicillin resistance gene containing an amber stop codon at position Ala184 and tRNA cloning sites has been described previously (8). Plasmid pArgU218, which contains a p15A origin of replication and encodes a kanamycin resistance marker and the isoacceptor of tRNAArg, corresponding to the codons AGA and AGG, has been described previously (19). Plasmid pBAD-Myc/HisA was purchased from Invitrogen. PCR reactions were carried out using the Expand Kit (Roche, Indianapolis, IN). Whole E.coli tRNA was purchased from Roche. Whole halobacterial tRNA was isolated from cultures of Halobacterium sp. NRC-1 using an RNA/DNA Extraction Kit (Qiagen). Overlap extensions were carried out using Taq polymerase (Stratagene). Restriction enzymes were purchased from New England Biolabs (Beverly, MA). DNA was purified using miniprep, gel purification or PCR purification kits (Qiagen). Oligodeoxyribonucleotides were prepared by Qiagen Operon (Alameda, CA) and their sequences can be found in Supplementary Material.
Cloning of aminoacyl-tRNA synthetase genes
The open reading frames of glutamyl-tRNA synthetase from A.fulgidus (AfERS), A.pernix (ApERS), M.jannaschii (MjERS), M.mazei (MmERS), M.thermoautotrophicum (MtERS), P.horikoshii (PhERS) and S.solfataricus (SsERS) were amplified by PCR using genomic DNA as template and the appropriate oligodeoxyribonucleotide primers (see Supplementary Material) to yield products of ∼1.7 kb. Amplification of the MjERS gene was unsuccessful. The AfERS PCR product was digested with BbsI, the ApERS and SsERS products with EcoRI and NcoI, the MmERS and MtERS products with BsmBI and the PhERS product with BsaI, to generate EcoRI/NcoI compatible cohesive ends for all products. Construction of plasmids pKQ-AfERS, pKQ-ApERS, pKQ-MjERS, pKQ-MmERS, pKQ-MtERS, pKQ-PhERS and pKQ-SsERS was accomplished by insertion of restriction-digested, gel-purified glutamyl-tRNA synthetase gene PCR fragments into plasmid pKQ. Plasmid constructs were confirmed by restriction mapping and sequencing of individual clones.
Design and cloning of tRNAs
Nomenclature of tRNAs: AQ(GU), A archaeal, Q glutaminyl, GU base pair at postion 10–26; AQ(GC), A archaeal, Q glutaminyl, GC base pair at position 10–26; AE(GU), A archaeal, E glutamyl, GU base pair at position 10–28; AE(GC), A archaeal, E glutamyl, GC base pair at position 10–28; AE(NN), A archaeal, E glutamyl, NN base pair at position 10–28.
Consensus archaeal tRNA genes were identified by alignment using the program pileup from the Genetics Computer Group Inc. Genes for tRNAs were constructed by overlap extension of the appropriate forward and reverse primers (see above). EcoRI + PstI-digested extension fragments were inserted into a 4.2 kb fragment of plasmid pACKO-A184TAG to construct plasmids pAC-AE(GU), pAC-AQ(GU) and pAC-AQ(GC). The AE(NN) tRNA library was constructed by PCR amplification using primers AE(NN)-RNA.F and pAC-tRNA.R with plasmid pAC-AE(GU) as template. The pAC-AE(NN) plasmid library was constructed by insertion of the EcoRI + PstI-digested AE(NN) tRNA library PCR fragment into the tRNA cloning site of pACKO-A184TAG. Plasmid constructs were confirmed by sequence analysis.
Ampicillin IC50 value measurements
To examine the orthogonality of tRNAs, 10 µl of E.coli DH10B cells (1 c.f.u./µl) carrying a tRNA-expressing plasmid were plated on a series of LB agar plates containing chloramphenicol and varying concentrations of ampicillin. To measure the IC50 values for cells carrying both tRNA- and synthetase-expressing plasmids, cells were plated on LB agar plates containing chloramphenicol, kanamycin and varying concentrations of ampicillin. In the initial series, cells were plated at five ampicillin concentrations varying from 0 to 1000 µg/ml and incubated overnight at 37°C. In subsequent series, cells were plated at a narrower ampicillin concentration range surrounding the initial ampicillin IC50 value estimate. Typically, 10 ampicillin concentrations spanning a range of ±45% of the initial IC50 estimate were tested in 10% increments. Measurements were made in duplicate or triplicate for each cell strain.
Optimization of the AEGU tRNA
Escherichia coli DH10B cells carrying plasmid pKQ-MmERS were transformed with the pAC-AE(NN) plasmid library to yield ∼106 independent transformants. Cells were plated (∼1000 c.f.u./plate) on a series of 10 LB agar plates containing chloramphenicol, kanamycin and 0–1000 µg/ml ampicillin. Ten individual colonies from the plate containing 500 µg/ml ampicillin were picked and their pAC-AE(NN) plasmids amplified, miniprepped and sequenced.
Synthetase overexpression
Aminoacyl-tRNA synthetase genes were PCR amplified using the appropriate N- and C-terminal pBAD construct primers and the appropriate synthetase pKQ construct templates. The AfERS PCR product was digested with BbsI, the EcERS product with EcoRI and NcoI, the MmERS and MtERS products with BsmBI and the PhERS product with BsaI to generate EcoRI and NcoI compatible cohesive ends for all products. Construction of the synthetase pBAD plasmids was accomplished by insertion of restriction enzyme digested, gel purified PCR fragments into the EcoRI and NcoI cloning sites of plasmid pBAD. Plasmid constructs were confirmed by restriction mapping and sequencing of individual clones. The synthetase pBAD plasmids were transferred to DH10B cells carrying plasmid pArgU218. Individual colonies were used to inoculate 500 ml cultures of 2YT containing ampicillin and kanamycin. Protein was isolated according to the QIAexpressionist (Qiagen) protocol under native conditions and dialyzed overnight against 20 mM Tris–HCl (pH 8), 200 mM NaCl and 1 mM dithiothreitol. Glycerol was added to protein samples to a final concentration of 50%. Glycerol-containing samples were stored at –20°C. Protein concentrations were determined using SDS–PAGE by comparison to a BSA standard.
In vitro aminoacylation assays
Reactions were carried out as described previously (20). Each 20 µl reaction contained 50 mM Tris–HCl (pH 7.5), 30 mM KCl, 20 mM MgCl2, 0.1 mg/ml BSA, 5 mM glutathione, 2.5 mM ATP, 100 nM l-[G-3H]glutamic acid (Amersham), 750 nM aminoacyl-tRNA synthetase, and 40 µM whole tRNA. Reactions were allowed to proceed for 15 min or 1 h at 37°C.
RESULTS
Development of an orthogonal tRNA
Previous studies have demonstrated that an orthogonal tRNA derived from the consensus sequence of archaeal leucine tRNAs can serve as an efficient substrate for archaeal leucyl-tRNA synthetases (8). The design of an orthogonal tRNA using this strategy involves: alignment of a group of related tRNA sequences to determine the consensus sequence; analysis of the secondary structure of the consensus-derived tRNA to identify and correct potentially deleterious non-canonical base pairs; conversion of the anticodon of the consensus tRNA to CUA to allow recognition and suppression of an amber stop codon within the target mRNA. Given the previous success of this approach in the generation of an orthogonal tRNALeu (8), we applied the same strategy to the construction of a cognate orthogonal tRNA for archaeal glutamyl-tRNA synthetases.
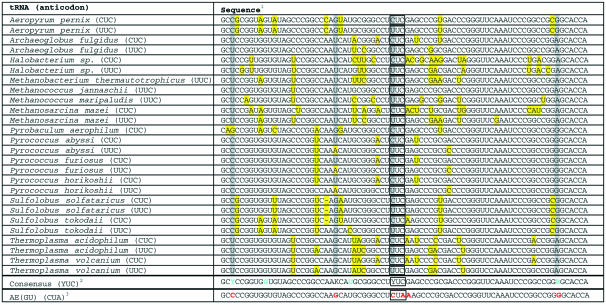
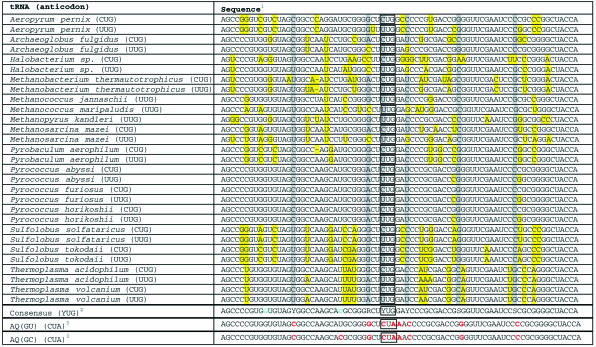
In archaebacteria, a single enzyme, glutamyl-tRNA synthetase, aminoacylates both tRNAGlu and tRNAGln. The product of the latter reaction, glu-tRNAGln, is subsequently converted to gln-tRNAGln by transamidation (16). Previous reports suggest that both archaeal tRNAGlu and archaeal tRNAGln are not substrates for endogenous E.coli synthetases (15). Thus, both tRNAs were viewed as attractive starting points for the development of an orthogonal tRNA using the consensus sequence design strategy. To test this hypothesis, the available archaeal tRNAGlu and tRNAGln sequences were separately aligned, revealing a high degree of homology within each family (Tables 1 and 2). The consensus sequence of each family was derived by identification of the consensus base at each position within the primary sequence. The consensus tRNAGlu sequence contains three positions outside the anticodon (positions 3, 25 and 72) at which two bases predominate with approximately equal frequency (Table 1). The consensus tRNAGln sequence contains five positions outside the anticodon (positions 16, 32, 40, 51 and 63) at which two bases predominate with approximately equal frequency and two positions (positions 6 and 67) at which one base predominates but does not represent a majority at its position (Table 2).
Table 1. Archaeal glutamic acid and consensus tRNAs.
1Bases highlighted in yellow deviate from the identity of their consensus bases. Bases highlighted in gray correspond to positions at which two different bases predominate with approximately equal frequency. Anticodon positions are boxed.
2Bases in blue correspond to positions involved in non-canonical base pairs (Fig. 1A). Y corresponds to bases C or U; R corresponds to bases A or G; S corresponds to bases C or G.
3Bases in red correspond to positions that were varied from the consensus sequence in the design of a prospective orthogonal tRNA (Fig. 1B).
Table 2. Archaeal glutamine and consensus tRNAs.
1Bases highlighted in yellow deviate from the identity of their consensus bases. Bases highlighted in gray correspond to positions at which two different bases predominate with approximately equal frequency. Anticodon positions are boxed.
2Bases in blue correspond to positions involved in non-canonical base pairs (Fig. 1A). Y corresponds to bases C or U; R corresponds to bases A or G; S corresponds to bases C or G.
3Bases in red correspond to positions that were varied from the consensus sequence in the design of a prospective orthogonal tRNA (Fig. 1B).
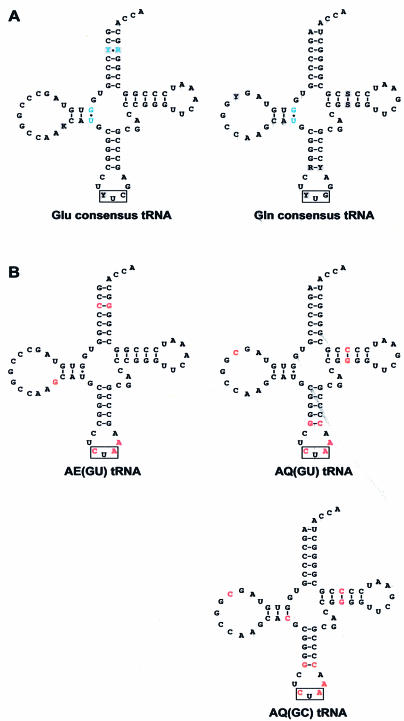
The secondary structures of the consensus archaeal tRNAGlu and tRNAGln sequences (Fig. 1A) were used to design three candidate orthogonal tRNAs (Fig. 1B). The anticodon loops contained the sequence CUA for suppression of an amber stop codon (UAG). The nucleotide to the 3′ side of each anticodon was converted to A to improve suppression efficiency in E.coli (8). Consensus tRNAGlu and tRNAGln structures contain a conserved non-canonical G-U base pair at positions 10–28 and 10–26, respectively (Fig. 1A). Although it has been demonstrated that alteration of non-canonical base pairs within orthogonal tRNAs can improve amber stop codon suppression efficiency in E.coli (8), the conserved nature of this pair suggested that it might constitute an important identity element for recognition by archaeal glutamyl-tRNA synthetases. Therefore, in designing the tRNAGlu orthogonal tRNA, AE(GU) (see Materials and Methods for tRNA nomenclature), the 10–28 G-U base pair was preserved (Fig. 1B). In designing the tRNAGln orthogonal tRNAs, AQ(GU) and AQ(GC), the 10–26 pair was either preserved or changed to a G-C pair, respectively.
Figure 1.
Design of orthogonal tRNAs. (A) Consensus archaeal tRNAGlu and tRNAGln sequences. Non-canonical base pairs are in blue; tRNA anticodons are boxed. Bases highlighted in gray correspond to positions at which two different bases predominate approximately equally. K corresponds to bases G or U; Y corresponds to bases C or U; R corresponds to bases A or G; S corresponds to bases C or G. (B) Initial candidate orthogonal tRNAs. Bases in red correspond to positions that vary from the consensus sequence.
Orthogonality of tRNAs
To test the activity and cross-reactivity of the AE(GU), AQ(GU) and AQ(GC) tRNAs, their genes were inserted into plasmid pACKO-A184TAG. Escherichia coli cells were transformed with the resulting plasmids, pAC-AE(GU), pAC-AQ(GU) and pAC-AQ(GC) and the ampicillin IC50 values for the cells were assayed. The tRNA-expressing pAC-based plasmids carry a β-lactamase reporter gene containing an amber mutation at the permissive site Ala184, which serves as a useful reporter for amber codon suppression. Acylation of an amber suppressor tRNA causes the incorporation of the activated amino acid at position 184 of β-lactamase, resulting in suppression of the amber stop codon, production of active full-length β-lactamase and resistance to ampicillin. Because an orthogonal tRNA should not cross-react with endogenous aminoacyl-tRNA synthetases, strains transformed with an orthogonal suppressor tRNA alone should survive only on low concentrations of ampicillin. To assess the orthogonality of designed tRNAs, E.coli transformants were plated on LB agar plates containing varying concentrations of ampicillin (Fig. 2). Cells expressing the AQ(GU) and AQ(GC) tRNAs exhibited ampicillin IC50 values of 60 ± 12 and 20 ± 4 µg/ml, respectively. Cells expressing the AE(GU) tRNA exhibited a relatively lower ampicillin IC50 value of 5 ± 1 µg/ml, suggesting that this tRNA is the least cross-reactive of the three.
Figure 2.
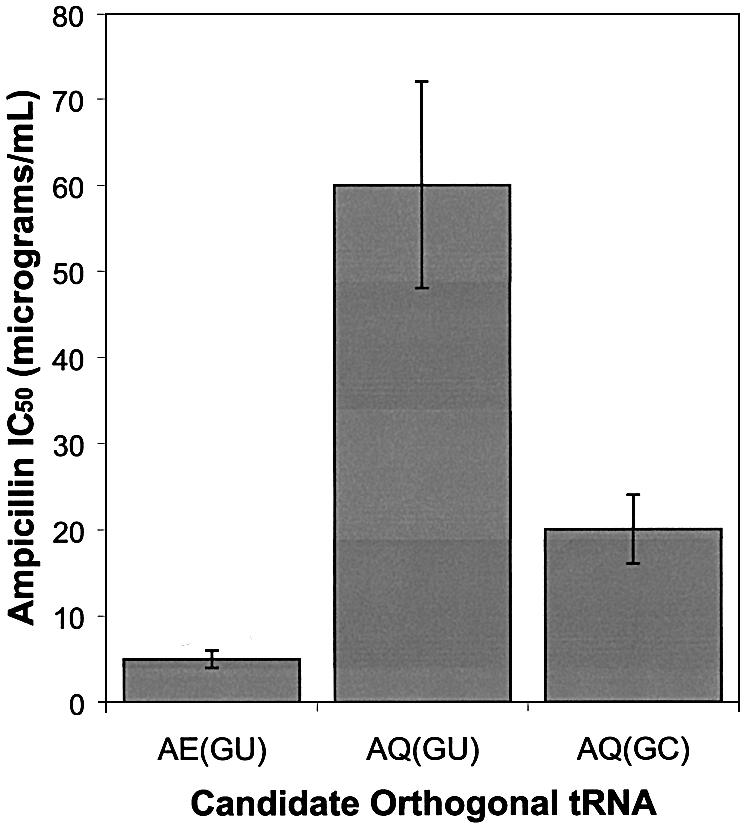
Orthogonality of candidate tRNAs in E.coli. The β-lactamase amber suppression assay was used to measure the ampicillin IC50 values for E.coli expressing the indicated tRNA.
Cloning and assaying archaeal glutamyl-tRNA synthetases in E.coli
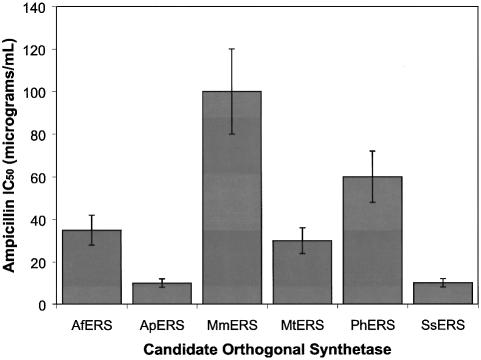
Based on previous experiments (8), it was anticipated that the AE(GU), AQ(GU) and AQ(GC) tRNAs might serve as substrates for archaeal glutamyl-tRNA synthetases. To investigate this, suppression efficiencies corresponding to various synthetases-tRNA pair combinations were assessed using the ampicillin IC50 assay. If the designed tRNAs are recognized by the archaeal glutamyl-tRNA synthetases, then E.coli cells co-expressing the synthetase-tRNA pairs should exhibit ampicillin IC50 values greater than those displayed by cells expressing tRNAs alone. The AfERS, ApERS, MmERS, MtERS, PhERS and SsERS genes were each inserted into the vector pKQ, which can be co-maintained in E.coli in combination with the tRNA-expressing plasmids. Following co-transformation of E.coli with all pair-wise combinations of synthetase- and tRNA-expressing plasmids, cells were grown on LB agar plates containing varying concentrations of ampicillin. Cells expressing synthetase–AQ(GU) or synthetase–AQ(GC) combinations exhibited IC50 values equivalent to those of cells expressing AQ(GU) alone or AQ(GC) alone, respectively, suggesting that neither of these tRNAs is recognized by any of the archaeal glutamyl-tRNA synthetases when co-expressed in E.coli. In contrast, cells expressing combinations of synthetase and AE(GU) tRNA exhibited ampicillin IC50 values 2- to 20-fold higher than those displayed by cells expressing AE(GU) alone (Fig. 3). The greatest IC50 value differences were exhibited by cells co-expressing the AE(GU) tRNA together with MtERS (6-fold), AfERS (7-fold), PhERS (12-fold) and MmERS (20-fold). Cells co-expressing the best pair, MmERS and AE(GU), exhibited an IC50 value of 100 µg/ml, a value still 4.5-fold lower than that displayed by the M.jannaschii-derived tyrosine pair (2) and 10-fold lower than that displayed by the M.thermoautotrophicum-derived leucine pair (8). These results indicate that AE(GU) tRNA combined with MtERS, AfERS, PhERS or MmERS constitute active but inefficient orthogonal pairs in E.coli.
Figure 3.
Aminoacylation of the AE(GU) tRNA by orthogonal synthetase candidates in E.coli. The β-lactamase amber suppression assay was used to measure the ampicillin IC50 values of E.coli expressing the indicated glutamyl-tRNA synthetase together with the AE(GU) tRNA. Ampicillin IC50 values reflect amber suppression efficiency in E.coli.
Selection and characterization of an optimized consensus tRNAGlu
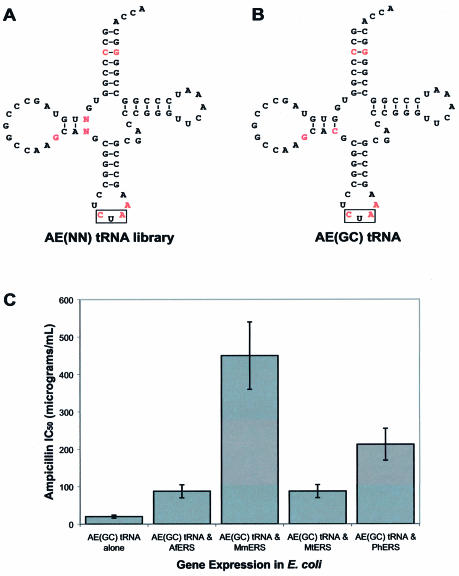
In order to increase the efficiency of the archaeal glutamic acid orthogonal pairs, improved variants of the AE(GU) tRNA were sought. The presence of the non-canonical G10-U28 base pair was regarded as a possible explanation for the low amber suppression efficiency exhibited by synthetase–AE(GU) pairs. An attempt to investigate the importance of this G-U base pair had already been made with the construction of the two tRNAGln-based consensus tRNAs, AQ(GU) and AQ(GC), which differed only with respect to this base pair. However, because both AQ(GU) and AQ(GC) lacked the ability to be aminoacylated by archaeal glutamyl-tRNA synthetases in E.coli, the importance of the G-U base pair was still unknown. To further address this issue, a small library of variants of the consensus tRNAGlu, AE(GU), was constructed in which nucleotides in the 10 and 28 positions were randomized (Fig. 4A). This ‘AE(NN)’ tRNA library was constructed by PCR using a degenerate oligoribonucleotide primer and inserted into the tRNA cloning site of plasmid pACKO-A184TAG, to afford the pAC-AE(NN) plasmid library. Escherichia coli cells co-transformed with plasmid pKQ-MmERS and plasmid library pAC-AE(NN) were then plated on a series of LB agar plates containing varying concentrations of ampicillin. Bacterial colonies were observed on plates containing ampicillin concentrations as high as 500 µg/ml, indicating that at least one variant of the AE(GU) tRNA exhibited enhanced amber suppression efficiency compared to the parent when co-expressed with MmERS in E.coli. Ten bacterial colonies were picked from the 500 µg/ml ampicillin plate and the pAC-AE(NN) plasmids were sequenced. All 10 of the clones contained a G10-C28 base pair (Fig. 4B), indicating that this ‘AE(GC)’ tRNA is the most active variant within the AE(NN) library.
Figure 4.
Optimization of the AE(GU) tRNA by selection. (A) Sequence and secondary structure of the AE(NN) tRNA library, which differs from the AE(GU) tRNA only at positions 10 and 28, which have been randomized. Bases in red correspond to positions that vary from the consensus sequence; tRNA anticodons are boxed. (B) Sequence and secondary structure of the AE(GC) tRNA, which was identified as the most active tRNA within the AE(NN) library. (C) Aminoacylation of the AE(GC) tRNA in E.coli. The β-lactamase assay was used to measure the ampicillin IC50 values for E.coli expressing the indicated glutamyl-tRNA synthetases together with the AE(GC) tRNA or the AE(GC) tRNA with no synthetase.
To measure the amber suppression efficiency of the selected AE(GC) tRNA, E.coli cells carrying either pAC-AE(GC) alone or in combination with plasmid pKQ-AfERS, pKQ-MmERS, pKQ-MtERS or pKQ-PhERS were plated on a series of LB agar plates containing varying concentrations of ampicillin. In all cases, E.coli cells co-expressing the AE(GC) tRNA were found to exhibit ampicillin IC50 values 4- to 5-fold greater than those of cells co-expressing the AE(GU) tRNA (Fig. 4C). Cells co-expressing the MmERS and AE(GC) pair exhibited an ampicillin IC50 value of 450 µg/ml, 4.5-fold greater than that displayed by cells co-expressing the MmERS and AE(GU) combination. Similarly, cells co-expressing the PhERS and AE(GC) pair exhibited an IC50 value of 212 µg/ml, ∼3.5-fold greater than those co-expressing PhERS and AE(GU). The ampicillin IC50 value for cells expressing the AE(GC) tRNA alone also increased 4-fold relative to cells expressing the AE(GU) tRNA alone (20 versus 5 µg/ml ampicillin), suggesting that the U28→G replacement enhanced the overall amber suppression efficiency of the tRNA. Such an enhancement may have resulted from improved stability or E.coli processing of the AE(GC) tRNA compared to AE(GU) (8). Alternatively, the increased IC50 value could be due to an increased rate of background aminoacylation of the AE(GC) tRNA compared to AE(GU).
Orthogonality of archaeal glutamyl-tRNA synthetases in E.coli
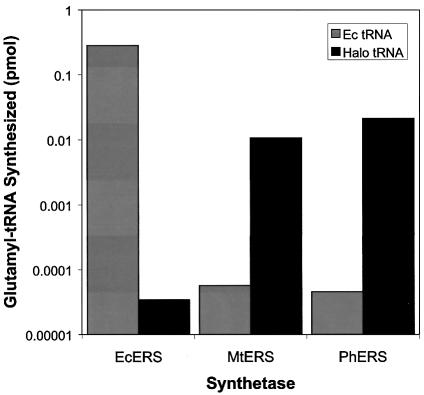
The ampicillin IC50 measurements described above provide insights into the orthogonality of consensus-designed tRNAs, but provide no information about the orthogonality of archaeal glutamyl-tRNA synthetases in E.coli. To investigate synthetase orthogonality, the AfERS, MmERS, MtERS and PhERS genes were inserted into a protein overexpression vector. Escherichia coli glutamyl-tRNA synthetase (EcERS) was inserted into the same vector for use as a control. Synthetases were overexpressed in E.coli, purified to homogeneity and used in an in vitro aminoacylation assay with whole tRNA prepared from either E.coli or the halophilic archaebacterium Halobacterium sp. NRC-1. As expected, EcERS robustly aminoacylated whole E.coli tRNA (0.28 pmol) and exhibited virtually no activity toward whole archaeal tRNA (3.4 × 10–5 pmol) (Fig. 5). In contrast, none of the four archaeal glutamyl-tRNA synthetases exhibited measurable aminoacylation activity with whole E.coli tRNA (≤5.7 × 10–5 pmol), while two of the four synthetases actively aminoacylated whole archaeal tRNA (0.011 and 0.021 pmol for MtERS and PhERS, respectively). It is unclear why AfERS and MmERS could not aminoacylate whole halobacterial tRNA in vitro (≤10–4 pmol) (not shown). One possible explanation is that the enzyme purification and/or assay conditions employed were deleterious to the aminoacylation activity of AfERS and MmERS. A second possibility is that subtle differences in tRNA identity elements might preclude recognition of a given archaeal tRNAGlu by all archaeal glutamyl-tRNA synthetases. To test the latter hypothesis, the activity of MmERS was investigated using the in vitro aminoacylation assay with whole tRNA derived from E.coli expressing the AE(GC) tRNA. Again, no activity was observed (data not shown), suggesting that the C-terminally His-tagged AfERS and MmERS enzymes are not active following purification. The above data demonstrate the orthogonality of MtERS and PhERS. The orthogonality of AfERS and MmERS has yet to be determined.
Figure 5.
Orthogonality of archaeal glutamyl-tRNA synthetases in vitro. Aminoacylation of whole tRNA from either E.coli (Ec) or Halobacterium sp. NRC-1 (halo) with [3H]glutamic acid was assayed in vitro.
DISCUSSION
The consensus sequence strategy for the design of orthogonal tRNAs was successfully applied to the development of an orthogonal tRNA based on the family of tRNAGlu sequences from archaea. The initial candidate tRNAGlu-based tRNA, AE(GU), was found to be highly orthogonal in E.coli, with cells expressing the tRNA alone displaying an ampicillin IC50 value of just 5 µg/ml. However, the AE(GU) tRNA inefficiently suppressed amber codons when expressed in combination with archaeal glutamyl-tRNA synthetases in E.coli. Cells containing this tRNA as part of a synthetase–tRNA pair displayed ampicillin IC50 values of only 60 and 100 µg/ml for the two best synthetases, PhERS and MmERS, respectively. In order to improve the amber suppression efficiency of the AE(GU) tRNA, a small library of variants, randomized at the 10 and 28 positions, was constructed. Library members were selected for the ability to efficiently suppress an amber stop codon in E.coli when expressed in combination with MmERS. The resulting AE(GC) tRNA, when paired with PhERS and MmERS, suppresses amber codons with high efficiency. Cells co-expressing this tRNA with PhERS and MmERS display ampicillin IC50 values of 212 and 450 µg/ml, respectively.
The consensus sequence strategy was not successful in developing an orthogonal tRNA based on the tRNAGln family of sequences from the same organisms. One possible explanation for this result is that, in general, members of the family of archaeal tRNAGln sequences deviate more from their consensus tRNA than do tRNAGlu sequences from theirs (Tables 1 and 2). This deviation suggests that individuals within the family of tRNAGln may cluster less tightly in their evolutionary relatedness compared to tRNAs within the archaeal tRNAGlu family. Moreover, the degree of deviation within the tRNAGln family may prevent individual archaeal glutamyl-tRNA synthetases from productively recognizing the consensus tRNA. A less likely explanation for the failure of the tRNAGln-based consensus design is that the AQ(GU) and AQ(GC) tRNAs may lack unknown elements that permit efficient processing in E.coli. Escherichia coli cells expressing the AQ(GU) and AQ(GC) tRNAs without co-expressed archaeal glutamyl-tRNA synthetases exhibit relatively high ampicillin IC50 values compared to cells expressing the AE(GU) tRNA or no exogenous tRNA (Fig. 2). These data suggest that the AQ(GU) and AQ(GC) tRNAs are processed to at least some extent in E.coli and that their inability to be charged by archaeal glutamyl-tRNA synthetases in E.coli is due to the fact that they are not recognized as substrates.
The glutamyl-tRNA synthetase from P.horikoshii functions as part of an orthogonal tRNA synthetase and amber suppressor tRNA pair in E.coli. The development of this pair, however, is only the first step in the process of incorporating unnatural amino acids into proteins in vivo. Synthetase variants must be evolved to selectively recognize unnatural amino acids and catalyze their condensation with the orthogonal AE(GC) tRNA. The evolved synthetases may then be used in unnatural amino acid mutagenesis of target proteins in E.coli.
In addition to synthetase and tRNA orthogonality, it is likely that the amber suppression efficiency of the orthogonal synthetase and amber suppressor tRNA pair in E.coli is an important determinant of synthetase evolvability. Amber suppression efficiency, in turn, is determined by several kinetic parameters, including the rate of synthetase and tRNA production and processing in E.coli and the catalytic efficiency of tRNA aminoacylation. Altering the amino acid substrate specificity of a synthetase through the variation of active site residues likely reduces amber suppression efficiency by affecting either the kcat or Km of the variant enzyme (2,21). Thus, the suppression efficiency of the parent synthetase-tRNA pair must be high enough to withstand the reduction in catalytic efficiency that accompanies protein evolution and allow the selection criteria to be met.
Amber suppression efficiencies can be determined by measurement of ampicillin IC50 values for E.coli bearing a synthetase–tRNA pair. The only orthogonal synthetase that has been evolved to accept unnatural amino acids in E.coli, the tyrosyl-tRNA synthetase from M.jannaschii, exhibits an ampicillin IC50 value of 440 µg/ml (2). In contrast, the glutamine pair derived from S.cerevisiae, for which variants capable of accepting unnatural amino acids have not yet been found, displays an ampicillin IC50 value of 140 µg/ml (4). The difference in suppression efficiency between these two pairs may explain the apparent difference in the evolvability of their synthetases. On the basis of these data, PhERS, which, together with the AE(GC) tRNA, exhibits an amber suppression efficiency in E.coli ∼2-fold less than that of the tyrosyl-tRNA synthetase, may prove amenable to selection experiments to alter amino acid specificity. MmERS, which has an ampicillin IC50 value roughly comparable to that of the tyrosyl-tRNA synthetase, may also prove amenable to selection, although its orthogonality in E.coli awaits demonstration. Besides catalytic efficiency, however, other factors are likely to play a role in synthetase evolvability. The extensive hydrogen bonding network within the active site of the glutamine synthetase, for example, may affect evolvability of the enzyme. Our understanding of the relative importance of activity versus other, perhaps more subtle, features will improve our ability to generate and evolve additional orthogonal pairs.
The M.jannaschii-derived tyrosine orthogonal pair has been evolved to recognize a variety of unnatural amino acids containing aryl side chains. Efforts to develop variants of this synthetase that accept more diverse chemical structures are in progress, however, the degree of difficulty with which such variants will be developed is unclear. The availability of additional orthogonal pairs opens the possibility of expanding the diversity of unnatural amino acids that can be incorporated in E.coli. The glutamic acid pairs described here, for example, may facilitate the development of synthetase variants that recognize negatively charged amino acids. This involves the generation of a large library of synthetase variants containing random amino acids substitutions at specific residues involved in binding the natural amino acid. Therefore, an important requirement for the directed evolution of a synthetase is the availability of high resolution structural information about the amino acid substrate-binding site. Although the X-ray crystal structure of the glutamyl-tRNA synthetase from P.horikoshii has not yet been solved, the structure of the homologous enzyme from Thermus thermophilus is available (17,18). This information is supplemented by co-crystal structural data on the glutaminyl-tRNA synthetase from E.coli with a glutaminyl-adenylate analog (22). Although they recognize somewhat different amino acid structures, the amino acid-binding pockets of these enzymes are superimposable.
For many applications, the ability to incorporate multiple distinct unnatural amino acids into a protein in vivo is highly desirable. For example, the incorporation of two different fluorescent amino acids into a protein molecule would enable the use of fluorescence resonance energy transfer to study the conformational dynamics of proteins in vivo. The incorporation of two distinct unnatural amino acids into a protein requires the use of two mutually orthogonal synthetases that have been evolved to accept different unnatural amino acids. Thus, the incorporation of more than one unnatural amino acid is presently limited by the lack of evolvable orthogonal synthetase and tRNA pairs. The development of an orthogonal glutamyl-tRNA synthetase and tRNA pair further advances the potential for multiple distinct unnatural amino acids to be incorporated into a single protein chain in living cells.
In conclusion, the development of orthogonal glutamic acid glutamyl-tRNA synthetase and tRNA pairs underscores that archaeal species represent a rich source of pairs for use in unnatural amino acid mutagenesis in E.coli. Moreover, these results provide insight into the rational design of efficiently charged orthogonal tRNAs.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
S.W.S. is supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. J.C.A. is an NSF pre-doctoral fellow. This work was supported by NIH grant GM62159. This is manuscript 15788-CH of the Scripps Research Institute.
REFERENCES
- 1.Liu D.R. and Schultz,P.G. (1999) Progress toward the evolution of an organism with an expanded genetic code. Proc. Natl Acad. Sci. USA, 96, 4780–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L., Brock,A., Herberich,B. and Schultz,P.G. (2001) Expanding the genetic code of Escherichia coli. Science, 292, 498–500. [DOI] [PubMed] [Google Scholar]
- 3.Santoro S.W., Wang,L., Herberich,B., King,D.S. and Schultz,P.G. (2002) An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat. Biotechnol., 20, 1044–1048. [DOI] [PubMed] [Google Scholar]
- 4.Pastrnak M., Magliery,T.J. and Schultz,P.G. (2000) A new orthogonal suppressor tRNA/aminoacyl-tRNA synthetase pair for evolving an organism with an expanded genetic code. Helv. Chim. Acta, 83, 2277–2286. [Google Scholar]
- 5.Ohno S., Yokogawa,T., Fujii,I., Asahara,H., Inokuchi,H. and Nishikawa,K. (1998) Co-expression of yeast amber suppressor tRNATyr and tyrosyl-tRNA synthetase in Escherichia coli: possibility to expand the genetic code. J. Biochem. (Tokyo), 124, 1065–1068. [DOI] [PubMed] [Google Scholar]
- 6.Kowal A.K., Kohrer,C. and RajBhandary,U.L. (2001) Twenty-first aminoacyl-tRNA synthetase-suppressor tRNA pairs for possible use in site-specific incorporation of amino acid analogues into proteins in eukaryotes and in eubacteria. Proc. Natl Acad. Sci. USA, 98, 2268–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L., Magliery,T.J., Liu,D.R. and Schultz,P.G. (2000) A new functional suppressor tRNA/aminoacyl-tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins. J. Am. Chem. Soc., 122, 5010–5011. [Google Scholar]
- 8.Anderson J.C. and Schultz,P.G. (2003) Adaptation of an orthogonal archaeal leucyl-tRNA and synthetase pair for four-base, amber and opal suppression. Biochemistry, 42, 9598–9608. [DOI] [PubMed] [Google Scholar]
- 9.Edwards H. and Schimmel,P. (1990) A bacterial amber suppressor in Saccharomyces cerevisiae is selectively recognized by a bacterial aminoacyl-tRNA synthetase. Mol. Cell. Biol., 10, 1633–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin J.W., Santoro,S.W., Martin,A.B., King,D.S., Wang,L. and Schultz,P.G. (2002) Addition of p-azido-l-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc., 124, 9026–9027. [DOI] [PubMed] [Google Scholar]
- 11.Chin J.W., Martin,A.B., King,D.S., Wang,L. and Schultz,P.G. (2002) Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl Acad. Sci. USA, 99, 11020–11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L., Zhang,Z., Brock,A. and Schultz,P.G. (2003) Addition of the keto functional group to the genetic code of Escherichia coli. Proc. Natl Acad. Sci. USA, 100, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z., Wang,L., Brock,A. and Schultz,P.G. (2002) The selective incorporation of alkenes into proteins in Escherichia coli. Angew. Chem. Int. Ed. Engl., 41, 2840–2842. [DOI] [PubMed] [Google Scholar]
- 14.Wang L., Brock,A. and Schultz,P.G. (2002) Adding l-3-(2-naphthyl) alanine to the genetic code of E. coli. J. Am. Chem. Soc., 124, 1836–1837. [DOI] [PubMed] [Google Scholar]
- 15.Kwok Y. and Wong,J.T. (1980) Evolutionary relationship between Halobacterium cutirubrum and eukaryotes determined by use of aminoacyl-tRNA synthetases as phylogenetic probes. Can. J. Biochem., 58, 213–218. [DOI] [PubMed] [Google Scholar]
- 16.Tumbula D.L., Becker,H.D., Chang,W.Z. and Soll,D. (2000) Domain-specific recruitment of amide amino acids for protein synthesis. Nature, 407, 106–110. [DOI] [PubMed] [Google Scholar]
- 17.Nureki O., Vassylyev,D.G., Katayanagi,K., Shimizu,T., Sekine,S., Kigawa,T., Miyazawa,T., Yokoyama,S. and Morikawa,K. (1995) Architectures of class-defining and specific domains of glutamyl-tRNA synthetase. Science, 31, 1958–1965. [DOI] [PubMed] [Google Scholar]
- 18.Sekine S., Nureki,O., Shimada,A., Vassylyev,D.G. and Yokoyama,S. (2001) Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nature Struct. Biol., 8, 203–206. [DOI] [PubMed] [Google Scholar]
- 19.Bullard J.M., Cai,Y.C. and Spremulli,L.L. (2000) Expression and characterization of the human mitochondrial leucyl-tRNA synthetase. Biochim. Biophys. Acta, 1490, 245–258. [DOI] [PubMed] [Google Scholar]
- 20.Hoben P. and Soll,D. (1985) Glutaminyl-tRNA synthetase of Escherichia coli. Methods Enzymol., 113, 55–59. [DOI] [PubMed] [Google Scholar]
- 21.Agou F., Quevillon,S., Kerjan,P. and Mirande,M. (1998) Switching the amino acid specificity of an aminoacyl-tRNA synthetase. Biochemistry, 11, 11309–11314. [DOI] [PubMed] [Google Scholar]
- 22.Rath V.L., Silvian,L.F., Beijer,B., Sproat,B.S. and Steitz,T.A. (1998) How glutaminyl-tRNA synthetase selects glutamine. Structure, 6, 439–449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.