Abstract
Mature U3 snoRNA in yeast is generated from the 3′-extended precursors by endonucleolytic cleavage followed by exonucleolytic trimming. These precursors terminate in poly(U) tracts and are normally stabilised by binding of the yeast La homologue, Lhp1p. We report that normal 3′ processing of U3 requires the nuclear Lsm proteins. On depletion of any of the five essential proteins, Lsm2–5p or Lsm8p, the normal 3′-extended precursors to the U3 snoRNA were lost. Truncated fragments of both mature and pre-U3 accumulated in the Lsm-depleted strains, consistent with substantial RNA degradation. Pre-U3 species were co-precipitated with TAP-tagged Lsm3p, but the association with spliced pre-U3 was lost in strains lacking Lhp1p. The association of Lhp1p with pre-U3 was also reduced on depletion of Lsm3p or Lsm5p, indicating that binding of Lhp1p and the Lsm proteins is interdependent. In contrast, a tagged Sm-protein detectably co-precipitated spliced pre-U3 species only in strains lacking Lhp1p. We propose that the Lsm2–8p complex functions as a chaperone in conjunction with Lhp1p to stabilise pre-U3 RNA species during 3′ processing. The Sm complex may function as a back-up to stabilise 3′ ends that are not protected by Lhp1p.
INTRODUCTION
The 3′ maturation of several small RNAs shares common steps and common processing factors in the yeast Saccharomyces cerevisiae. Normal 3′ processing of the U1, U2, U4 and U5 small nuclear RNAs (snRNAs) and U3 small nucleolar RNA (snoRNA) involves cleavage by the endonuclease Rnt1p, the yeast homologue of Escherichia coli RNase III (1–5). The mature 3′ ends of these RNAs are generated by 3′→5′ exonucleases, including the exosome complex and the Rex1–3p proteins (3,5–7). The exosome also participates in the 3′ maturation of the 5.8S rRNA and many snoRNAs (3,8) and in the degradation of cytoplasmic and nuclear RNAs, including mRNAs, pre-mRNAs, pre-snRNAs and pre-snoRNAs (3,9–14). This raises the obvious question of how the precursors to the stable RNA species avoid degradation by the RNA processing machinery?
In the case of the U3 snoRNA and the snRNAs, precursors generated by Rnt1p cleavage are stabilised against continued degradation at least in part by binding of Lhp1p (La-homologous protein), the yeast homologue of the human La phosphoprotein (5,15). Lhp1p/La binds poly(U) tracts located at the 3′ ends of newly synthesised RNA polymerase III transcripts, including precursors to tRNAs, 5S rRNA, SRP RNA and the U6 snRNA (16–19). Binding of Lhp1p protects newly transcribed U6 RNA against degradation (20) and stimulates the cleavage of tRNA 3′ ends while suppressing maturation by exonucleases (21). In contrast, the poly(U) tracts in the pre-U3 snoRNA and pre-snRNAs are located between the Rnt1p cleavage site and mature 3′ end of the RNAs.
Mature U1, U2, U4 and U5 snRNPs contain seven core Sm proteins (22), which form a closed ring structure (23). In contrast, the U6 snRNP associates with seven related proteins, Lsm2–8p (like-Sm) (20,24–29), which also assemble in a heptameric ring (30–32). The Lsm2–8p complex is important for U6 snRNA stability and biogenesis of the U6 snRNP, and is therefore involved in pre-mRNA splicing. The related Lsm1–7p complex functions in mRNA decapping and 5′→3′ degradation via association with cytoplasmic mRNA decay factors (33–36).
During the initial analysis of the lsm mutant strains, it was observed that the levels of several small RNAs were affected, in addition to U6 snRNA (27). The nuclear Lsm2–8p complex was subsequently reported to be involved in the processing and degradation of tRNAs and rRNAs (37,38). In Xenopus, Lsm2–4p and Lsm6–8p bind to the U8 snoRNA (39), while over-expression of yeast Lsm5p suppressed a defect in accumulation of the box H/ACA snoRNAs (40), suggesting that an Lsm2–8p complex also participates in snoRNP biogenesis. Tagged Lsm3p was shown to transiently bind tRNA precursors, presumably as part of an Lsm2–8p complex, and stimulate their association with Lhp1p. Association of Lhp1p with other RNA substrates, pre-RNase P RNA and the SRP RNA (scR1), was also affected in the absence of Lsm proteins (37). As processing of U3 snoRNA involves binding of Lhp1p to a poly(U) tract in the 3′-extended precursors, we tested U3 processing in the lsm mutants. Here we show that Lsm proteins bind U3 precursors, affect their association with Lhp1p and are involved in U3 synthesis and degradation, consistent with roles as chaperones for RNA–protein assembly.
MATERIALS AND METHODS
Strains and plasmids
The transformation procedure was as described (41). Yeast strains used are listed in Table 1. Strain YCA56 was generated by PCR-based gene disruption of LHP1 in the BMA64 strain using plasmid pTL54 as PCR template (42). Disruption was confirmed by PCR analysis. Strain YJK55 was constructed by a PCR strategy as described (43) in the YCA56 strain; construction was confirmed by PCR analysis, and the expression of Lsm3-TAP was tested by western blotting. Strains YRB15 and YRB20 were kindly provided by Rémy Bordonné (CNRS, Montpellier, France). Plasmid pBS1360 carrying a C-terminal fusion between SmE1p and two IgG-binding domains of Staphylococcus aureus protein A (ProtA) was kindly provided by Bertrand Séraphin (CNRS, Gif sur Yvette, France).
Table 1. Yeast strains used in this work.
| Strain | Genotype | Reference/note |
|---|---|---|
| AEMY19 | MATα ade2-1 his3Δ200 leu2-3,-112 trp1Δ1 ura3-1 LSM6::HIS3 | (27) |
| AEMY22 | MATα ade2-1 his3Δ200 leu2-3,-112 trp1Δ1 ura3-1 LSM7::HIS3 | (27) |
| AEMY24 | MATα ade2-1 his3-11,-15 leu2-3,-112 trp1Δ1 ura3-1 LSM1::TRP1 | (27) |
| AEMY31 | MATα ade2-1 his3-11,-15 leu2-3,-112 trp1Δ1 ura3-1 LSM3::TRP1 [pBM125-GAL1-HA-LSM3] | (27) |
| AEMY33 | MATα ade2-1 his3Δ200 leu2-3,-112 trp1Δ1 ura3-1 LSM2::HIS3 [pBM125-GAL1-LSM2-HA] | (27) |
| AEMY46 | MATα ade2-1 his3-11,-15 leu2-3,-112 trp1Δ1 ura3-1 LSM8::TRP1 [pBM125-GAL1-HA-LSM8] | (27) |
| AEMY47 | MATα ade2-1 his3-11,-15 leu2-3,-112 trp1Δ1 ura3-1 LSM5::TRP1 [pBM125-GAL1-HA-LSM5] | (27) |
| MCY4 | MATa ade1-101 his3Δ1 trp1-289 ura3-52 LEU2-GAL1-LSM4 | (24) |
| YDL401 | MATa his3Δ200 leu2Δ1 trp1 ura3-52 gal2 galΔ108 | (42) |
| YJK20 | as YDL401 but LHP1::ProtA-TADH1-HIS3MX6 | (37) |
| YJK21 | as AEMY31 but LHP1::ProtA-TADH1-HIS3MX6 | (37) |
| YJK22 | as AEMY47 but LHP1::ProtA-TADH1-HIS3MX6 | (37) |
| YJV140 | MATa ade2 his3 leu2 trp2 ura3 | (37) |
| YJK34 | as YJV140 but LSM3-TAP | (37) |
| BMA64 | MATα ade2-1 his3-11,-15 leu2-3,-112 trp1Δ ura3-1 | F. Lacroute |
| BMA38 | MATα ade2-1 his3Δ200 leu2-3,-112 trp1Δ1 ura3-1 | (27) |
| YCA53 | as AEMY46 + [pU3sub6 CBS1] | This work |
| YCA56 | as BMA64 but LHP1::Kl URA | This work |
| YJK55 | as YCA56 but Lsm3-TAP | This work |
| YJK59 | as YCA56 + [pBS1360] | This work |
| YJK60 | as BMA64 + [pBS1360] | This work |
| YRB15 | MATa ura3-52 lys2-801 ade2-110 trp1-Δ63 his3-Δ200 leu2-Δ1 SME::HIS3, pUN-SmE | (53) |
| YRB20 | MATa ura3-52 lys2-801 ade2-110 trp1-Δ63 his3-Δ200 leu2-Δ1 SME::HIS3, PGal1-SmE | (53) |
| YJK61 | as YRB20 + [pU3sub6 CBS1] | This work |
RNA extraction, northern hybridisation and primer extension
For depletion of the essential Lsm proteins, cells were harvested at intervals following the shift from RSG medium (2% galactose, 2% sucrose, 2% raffinose) or YPGal medium containing 2% galactose to YPD medium containing 2% glucose. Otherwise strains were grown in YPD medium. The lsm-Δ strains were pre-grown at 23°C and transferred to 37°C. RNA extraction and northern hybridisation were as described (44,45).
For RNA hybridisation, the following oligonucleotides were used: 031 (MRP), 5′-AATAGAGGTACCAGGTCAAGAAGC; 200 (U3), 5′-UUAUGGGACUUGUU; 203 (U3 boxA), 5′-CUAUAGAAAUGAUCCU; 230 (U3sub6), 5′-GATTCCTATAGAAACACAG; 250 (scR1), 5′-ATCCCGGCCGCCTCCATCAC; 251 (3′Ex-U3), 5′-GTGGTTAACTTGTCA; 254 (3′U3), 5′-CCAACTTGTCAGACTGCCATT; 261 (U6), 5′-AAAACGAAATAAATTCTTTGTAAAAC; 264 (5′U3), 5′-TCCTATGAAGTACGTCGAC; and 421 (RPL30), 5′-GGTTGATAGATTCTTGGGAT.
Expression of the U3 cDNA
The U3A sub6-CBS1 cDNA (46,47) was expressed from an ARS-CEN-ADE2 plasmid under the control of the natural promoter and terminator regions in the GAL::lsm8 and GAL::sme1 strains. U3 synthesised from the cDNA construct was detected by hybridisation with a probe specific for the U3sub6 mutation (oligo 230).
Immunoprecipitation
Whole-cell extracts from strains GAL::HA-lsm3, Lhp1p-ProtA, Lhp1p-ProtA/GAL::HA-lsm3 and Lhp1p-ProtA/GAL:: HA-lsm5 grown either in RSG medium or following the transfer to YPD medium for 4, 8.5 or 24 h were prepared as described (48). Extract from cells lhp1-Δ, SmE1-ProtA, SmE1-ProtA/lhp1-Δ and their respective isogenic wild-type strains grown in YPD medium were prepared in the same way. Immunoprecipitation of ProtA-tagged strains was performed with rabbit IgG–agarose beads (Sigma) as described (49). Immunoprecipitation using monoclonal A66 antibody against Nop1p (50) was performed in the same way except that antibodies were first bound to protein A–Sepharose (Pharmacia). The RNAs were extracted with GTC/phenol–chloroform and ethanol precipitated. Immunoprecipitation of TAP-tagged Lsm3p protein was performed as described (43) using extract equivalent to 400 OD600 of cells. Co-purified RNAs were recovered from the eluate of the IgG column by phenol–chloroform extraction and ethanol precipitation. Precursors and mature RNAs were identified by northern hybridisations. Untagged isogenic strains (YJV140 or YCA56) were utilised as controls.
RESULTS
The major 3′-extended U3 precursors are lost in lsm mutants
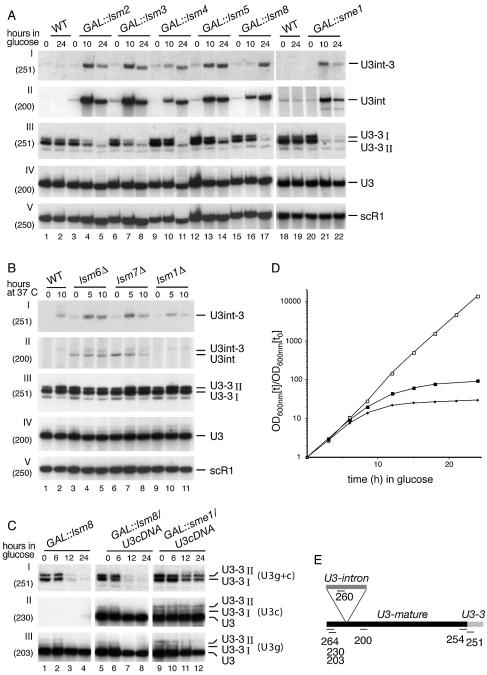
Yeast U3 snoRNA is synthesised from 3′-extended precursors, with major intermediates (U3-3′I and U3-3′II) that are stabilised by binding of the Lhp1p protein to 3′-terminal poly(U) tracts (5). In an lhp1-Δ strain, the normal U3 precursors are absent and are replaced by pre-U3 species that are shorter and more heterogeneous, but still terminate at poly(U) (5), indicating that other poly(U)-binding proteins also participate in pre-U3 processing. Moreover, the level of mature U3 was not affected in strains lacking Lhp1p, again implying the participation of additional cofactors. Since the Lsm2–8p complex is known to bind to poly(U) tracts, pre-U3 processing was analysed in strains in which expression of the essential Lsm proteins, Lsm2–5p and Lsm8p, was under GAL control (strains GAL::lsm2, GAL::lsm3, GAL::lsm4, GAL::lsm5 and GAL::lsm8). The genes encoding non- essential Lsm proteins, Lsm1p, Lsm6p and Lsm7p, were deleted, giving rise to temperature-sensitive (ts) strains (strains lsm1-Δ, lsm6-Δ and lsm7-Δ) (27). The GAL-regulated proteins were depleted by transferring the strains from permissive RSG medium (0 h samples) to repressive glucose medium. The strains deleted for Lsm proteins were grown in glucose medium at 23°C (0 h samples) and transferred to the non-permissive temperature of 37°C for up to 10 h, by which time growth had ceased.
On depletion of any essential Lsm protein (Fig. 1A, lanes 3–17 and C, lanes 1–4), the two major 3′-extended species, U3-3′I and U3-3′II, were largely lost (Fig. 1A, III), whereas the level of mature U3 was not affected (Fig. 1A, IV). The level of these precursors was clearly reduced at 10 h of depletion and significantly lowered at 24 h. In contrast, the absence of Lsm1p, Lsm6p or Lsm7p had no effect on the levels of U3-3′I, U3-3′II or mature U3 snoRNA (Fig. 1B, III, lanes 3–11). Processing of U3 was also assessed in lsm2ts and lsm5ts strains (35,51,52), which are partially ts for growth and cytoplasmic mRNA degradation, but no clear differences were observed (data not shown).
Figure 1.
Normal 3′ processing of U3 snoRNA requires Lsm proteins. (A) Northern analysis of Lsm2–5p and Lsm8p. Strains carrying GAL-regulated constructs (GAL::lsm, lanes 3–17; and GAL::sme1, lanes 20–22) and the BMA64 and YRB15 wild-type strains (WT, lanes 1 and 2, and 18 and 19) were grown in permissive RSG medium (0 h) and transferred to repressive, glucose medium at 30°C for the times indicated. Probe names are in parentheses and RNA species are shown on the right. U3int-3′ is intron containing and 3′ unprocessed. U3int is intron containing and 3′ mature. U3-3′I and U3-3′II are spliced and 3′ unprocessed. (B) Analysis of non-essential Lsm proteins. Strains deleted for Lsm1p (lanes 9–11), Lsm6p (lanes 3–5) and Lsm7p (lanes 6–8) and the wild-type strain (WT, lanes 1 and 2) were pre-grown at 23°C (0 h) and transferred to 37°C for the times indicated. RNA was separated on a 6% polyacrylamide gel and hybridised with oligonucleotide probes. (C) Analysis of processing of intron-less U3 snoRNA. RNA was extracted from a GAL::lsm8 strain (lanes 1–4) and GAL::lsm8 (lanes 5–8) and GAL::sme1 (lanes 9–12) strains expressing a tagged U3 cDNA, and transferred to glucose medium for the times indicated. Probe names are in parentheses. Oligo 251 is specific for 3′-extended forms of both genomic U3 (U3g) and cDNA (U3c). Oligo 230 is specific for U3c. Oligo 203 is specific for mature U3g. (D) Growth curves of the wild-type (open square), GAL::sme1 (filled square) and GAL::HA-lsm3 (filled diamond) strains pre-grown in permissive, RSG medium and transferred to repressive, glucose medium for the times indicated. Strains were maintained in exponential growth by dilution with pre-warmed medium. Cell densities measured by OD600 are shown corrected for dilution. (E) U3 RNA species. Locations of the oligos are shown schematically.
Yeast U3 is unusual in containing an intron, and depletion of Lsm2–8p, but not Lsm1p, caused some accumulation of the intron-containing pre-U3 species, the 3′-processed species U3int and the 3′-extended species U3int-3′ (Fig. 1A, I and II, and B, I and II) (27). A low level of U3int-3′ is also visible in the wild-type and lsm1-Δ strains at 37°C but not at 23°C (Fig. 1B, I), probably reflecting differences in the relative order of 3′ processing versus splicing at elevated temperatures. Precursors to several tRNAs are similarly elevated at 37°C (37). Notably, the unspliced pre-U3 species were strongly detected even after 24 h of depletion, clearly showing that pre-U3 synthesis was still ongoing at this time.
To determine whether the defects in U3 processing are a consequence of impaired pre-mRNA splicing, we analysed a GAL-sme1 strain (Fig. 1A, lanes 20–22) (53), and the ts-lethal prp2-1 strain (data not shown) (54). Growth inhibition was comparable in the GAL-lsm3 and GAL-sme1 strains following transfer to glucose medium, with a clear growth defect observed after 8.5 h of depletion (Fig. 1D). Following SmE1p depletion, the level of the spliced U3-3′I and U3-3′II species was significantly decreased, whereas intron-containing U3 precursors (U3int and U3int-3′) accumulated (Fig. 1A, I–III, lanes 20–22 and data not shown). The level of mature U3 snoRNA was not clearly reduced by depletion of SmE1p. This was also seen for depletion of the Lsm proteins and may arise because even a strongly reduced U3 synthesis rate is able to maintain snoRNA levels at the low growth rate seen in these strains.
To confirm that the depletion of U3-3′I and II RNAs in the lsm mutants does not result from the inhibition of pre-U3 splicing, a tagged U3 cDNA (U3c) was expressed in the GAL-lsm8 and GAL-sme1 strains under the control of the U3 promoter on a low copy number plasmid (see Materials and Methods). Synthesis of U3c (Fig. 1C, II) in both strains was similar to the genomic U3 (U3g) (Fig. 1C, III). U3c was processed via U3-3′I and U3-3′II precursors, and these were lost on depletion of Lsm8p with similar kinetics to the U3g precursors. In contrast, while U3g precursors were reduced following depletion of Sme1p (Fig. 1C, I and III, lanes 9–12), the U3c precursors remained unchanged (Fig. 1C, I and II, lanes 9–12).
We conclude that the essential Lsm2–5p and 8p proteins are involved in the pre-U3 processing pathway, and their presence is required for accumulation of the major 3′-extended precursors.
Lsm3p binds to U3 precursors
To test whether the nuclear Lsm complex interacts directly with U3 precursors, immunoprecipitation was performed using C-terminal TAP-tagged Lsm3p (37,43) and haemagglutinin (HA)-tagged Lsm1p (27) (Fig. 2). As previously reported (25,27,28,30), Lsm3-TAP but not HA-Lsm1p co-precipitated U6 (Fig. 2F). In contrast, there was no clear precipitation of the RNA component of RNase MRP (Fig. 2G), P RNA or mature tRNAs (data not shown), which were recovered at the background level seen for the isogenic non-tagged control. Both Lsm3p and Lsm1p precipitated shortened deadenylated RPL30 mRNA (Fig. 2E), in agreement with the preferential binding of Lsm1–7p complex to deadenylated mRNAs (35,36).
Figure 2.
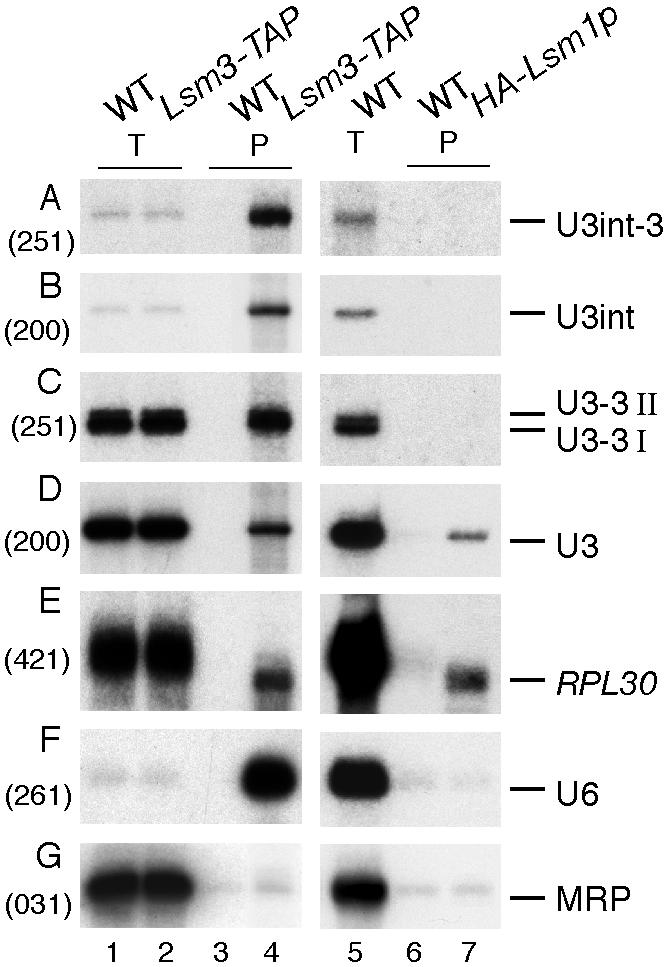
Lsm3 protein binds to U3 precursors. Immunoprecipitation of RNAs from strains expressing TAP-tagged Lsm3p and HA-tagged Lsm1p. Lysates from the Lsm3-TAP strain and the isogenic wild-type strain (YJV140) were immunoprecipitated with rabbit IgG–agarose beads (Sigma). Lysates from HA-Lsm1 and isogenic wild-type (BMA64) strains were immunoprecipitated with rat monoclonal anti-HA antibody bound to protein G–agarose. RNA was recovered from the lysate (T) and the immunoprecipitate (P) and analysed by northern hybridisation (A–G). Probe names are indicated in parentheses. RNA species are shown on the right. Approximately 30-fold more cell equivalents of RNA were loaded for the pellet fractions compared with the total fraction. Probe names are in parentheses and RNA species are shown on the right.
The U3-3′ extended precursors (U3int-3′, U3int, U3-3′I and U3-3′II) that were affected by depletion of Lsm3p were also detectably co-precipitated with Lsm3-TAP (Fig. 2A–C, lane 4), but not with HA-Lsm1p (Fig. 2A–C, lane 7). Recovery of the intron-containing U3int-3′ and U3int species may be due to their association with the splicing machinery, since they were also co-precipitated with an SmE1-ProtA C-terminal fusion (see Fig. 3H, lane 2). However, the spliced U3-3′I and U3-3′II RNAs were not recovered in the SmE1-ProtA precipitate (Fig. 3I, lane 2). Mature U3 was weakly co-precipitated with both Lsm3-TAP and HA-Lsm1p (Fig. 2D). The basis of this association is not known, but it might represent a low level of U3 that fails to dissociate from pre-ribosomal particles prior to their exit from the nucleus to the cytoplasm.
Figure 3.
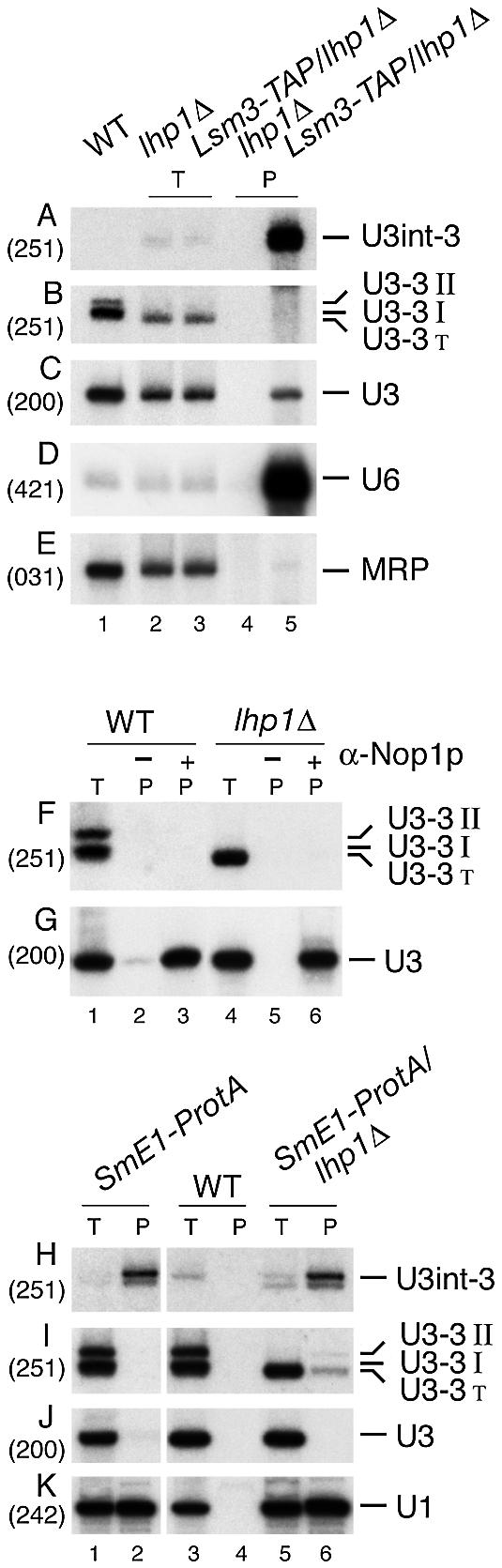
In the absence of Lhp1p, U3-3′ is not associated with Lsm3p or Nop1p but is co-precipitated with SmE1p. (A–E) Immunoprecipitation of RNAs from the lhp1-Δ strain expressing Lsm3-TAP. Lysates from lhp1-Δ and Lsm3-TAP/lhp1-Δ strains were immunoprecipitated as described in Figure 2. Total RNA from the wild-type strain (BMA64) in lane 1 was used as a control. U3-3′T is spliced and 3′ extended, but shorter than the wild-type U3-3′I species (see legend to Fig. 1 for other species). (F and G) Immunoprecipitation of RNAs with antibodies against Nop1p. Lysate from lhp1-Δ and isogenic wild-type (BMA64) strains was immunoprecipitated with monoclonal A66 antibody against Nop1p bound to protein A–Sepharose (Pharmacia). RNA was recovered from the lysate (T) and the immunoprecipitate (P) and analysed by northern hybridisation. (H–K) Immunoprecipitation of RNAs from the lhp1-Δ strain expressing SmE1-ProtA. Lysates from SmE-ProtA, SmE-ProtA/lhp1-Δ strains and the isogenic wild-type strain (BMA64) were immunoprecipitated as described in Figure 2. Probe names are in parentheses and RNA species are shown on the right.
We conclude that Lsm3p associates with 3′-extended U3 precursors, presumably as a component of an Lsm complex. The efficiency of precipitation of U3 precursors with Lsm3-TAP was lower than that of U6, but comparable with that of pre-tRNAs or pre-rRNAs (37) and significantly higher than that of RPL30 mRNA [Fig. 2E and Tharun et al. (35)]. This probably reflects the transient nature of these associations in vivo.
Lsm3p does not associate with spliced U3 precursors in the absence of Lhp1p
In strains lacking Lhp1p, U3-3′I and U3-3′II are lost and replaced by shorter, more heterogeneous species (labelled U3-3′T in Fig. 3B; compare lane 1 with lanes 2 and 3). We tested whether these RNAs bind the Lsm complex by performing immunoprecipitation with Lsm3-TAP expressed in the lhp1-Δ strain (Fig. 3A–E). A non-tagged lhp1-Δ strain was used as a control. As in the wild-type, Lsm3-TAP strongly co-precipitated U6 and moderately precipitated mature U3, but did not precipitate MRP RNA from the lhp1-Δ strain (Fig. 3C–E). The intron-containing U3int-3′ precursor was clearly co-precipitated with Lsm3-TAP (Fig. 3A), whereas the truncated U3-3′T species were not detectably co-precipitated with Lsm3-TAP from the lhp1-Δ strain (Fig. 3A and B). These U3 precursors remain stable in cell extracts and are not processed to mature U3 during immunoprecipitation, as they were efficiently recovered from the supernatants (data not shown). These observations confirm the specificity of pre-U3 co-precipitation with Lsm3-TAP in the otherwise wild-type strain.
We conclude that U3 precursors generated in the absence of Lhp1p are not stabilised by binding of the Lsm complex to the terminal poly(U) tract. Moreover, binding of 3′-extended pre-U3 by the Lsm proteins apparently requires the presence of Lhp1p.
Mature U3, but not pre-U3, associates with the core snoRNP proteins, Nop1p, Nop56p and Nop58p (5,55–57). These were proposed to displace Lhp1p from the 3′-flanking sequence of U3 during 3′ processing (5). Immunoprecipitation was performed using antibodies against Nop1p (50) with wild-type and lhp1-Δ cell extracts to assess whether Nop1p binds to the truncated U3-3′T precursors in the absence of Lhp1p (Fig. 3F and G). In the wild-type and lhp1-Δ cells, Nop1p associated strongly with mature U3 (Fig. 3G, lanes 3 and 6) but not with U3 precursors (Fig. 3F, lanes 3 and 6).
The seven-member Sm protein complex has a high affinity for uridine-rich RNA and binds to a U9 oligonucleotide in vitro with only 3-fold lower affinity than the consensus Sm-site (58). To test the association of Sm proteins with pre-U3, an SmE1-ProtA C-terminal fusion was expressed from a low copy CEN plasmid pBS1360 (28) in the wild-type and lhp1-Δ strains (Fig. 3H–K). SmE1-ProtA efficiently co-precipitated the U1 snRNA from both strains and also precipitated the intron-containing U3int-3′ precursor, probably due to its association with the spliceosome, whereas mature U3 was not recovered (Fig. 3H, J and K, lanes 2 and 6). The spliced U3-3′I and U3-3′II precursors were not co-precipitated with SmE1-ProtA from the wild-type strain (Fig. 3I, lane 2), but the U3-3′T species was co-precipitated with SmE1-ProtA from the lhp1-Δ strain (Fig. 3I, lane 5). The efficiency of co-precipitation of spliced pre-U3 with SmE1-ProtA in the lhp1-Δ strain and with Lsm3-TAP in the LHP1 strain is similar (6-fold more cell equivalents were loaded in the precipitate lane relative to the total in Fig. 3 and 30-fold more in Fig. 2).
We conclude that the pre-U3-3′T species present in lhp1-Δ cells are not detectably associated with an Lsm complex or the snoRNP protein Nop1p, but do associate with Sm proteins. The association of the Sm ring with a poly(U) sequence is more labile during incubation in vitro than with a consensus Sm-binding site (58), and the complex may well have undergone some dissociation during immunoprecipitation.
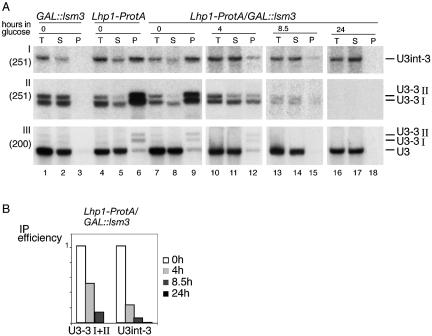
Binding of Lhp1p to U3 precursors is reduced by depletion of Lsm3p or Lsm5p
Lhp1p is required for the association of Lsm3p (and presumably an Lsm complex) with U3-3′I and II. To assess whether the association of Lhp1p with pre-U3 also requires an Lsm complex, we expressed an Lhp1-ProtA C-terminal fusion (37) in GAL::lsm3 and GAL::lsm5 strains. Very similar results were obtained for each strain, and data are shown only for Lsm3p depletion in Figure 4. Lhp1p-ProtA immunoprecipitation was performed with extracts from strains grown in permissive RSG medium (0 h samples) and following transfer to glucose medium for 4, 8.5 and 24 h to deplete Lsm3p or Lsm5p. Approximately 4-fold more cell equivalents were loaded for the pellet fraction in Figure 4. Data from the northern analyses in Figure 4A were quantified using a PhosphorImager and are presented graphically in Figure 4B.
Figure 4.
Lhp1p binds less efficiently to pre-U3 in the absence of Lsm proteins. (A) The strains GAL::lsm3 (lanes 1–3), Lhp1p-ProtA (lanes 4–6) and Lhp1p-ProtA/GAL::HA-lsm3 (lanes 7–18) were grown at 30°C either in RSG medium (lanes 1–9) or transferred to glucose medium for 4, 8.5 and 24 h (lanes 10–18). Lysates were immunoprecipitated using IgG–agarose, and RNA was recovered from the lysate (T), immune supernatant (S) and the immunoprecipitate (P) and analysed by northern hybridisation. Approximately 4-fold more cell equivalents are loaded for the bound material. Probe names are in parentheses and RNA species are shown on the right. (B) Graphic representation of immunoprecipitation efficiency by Lhp1p of RNAs from (A) in the Lhp1-ProtA/GAL::HA-lsm3 strain before depletion of Lsm3p (0 h, white bars) and after transfer to glucose for 4 (light grey bars), 8.5 (dark grsy bars) and 24 h (black bars). Immunoprecipitation efficiency was calculated from the ratio between S + P and P. Values after depletion are expressed relative to the value before depletion, which was arbitrarily set at 1.
Lhp1p-ProtA efficiently co-precipitated U3-3′I, U3-3′II and U3int-3′ from the LSM3+ strain (Fig. 4, lane 6) and from the Lhp1p-ProtA/GAL::lsm3 strain in permissive medium (Fig. 4, lane 9). The co-precipitation of U3-3′I, U3-3′II and U3int-3′ was markedly reduced after depletion of Lsm3p or Lsm5p for only 4 h (2- and 3.8-fold, respectively; Fig. 4A, lane 12), before the appearance of a growth defect (Fig. 1D) and also prior to any clear defects in pre-U3 processing. Immunoprecipitation of U3-3′I, U3′3-II and U3int-3′ was further reduced following depletion for 8.5 h (8.4- and 16.2-fold, respectively, Fig. 4A, lane 15). After 24 h of depletion, U3-3′I and II were not detectable, whereas the intron-containing U3int-3′ persists, indicating that pre-U3 is transcribed, but very poorly recovered in the Lhp1p-ProtA precipitate (∼50- to 60-fold reduction, Fig. 4A, lane 18). The affinity of Lhp1p for newly synthesised U6 snRNA was not affected by Lsm protein depletion [data not shown; (37)].
We conclude that binding or stable association between Lhp1p and 3′-extended pre-U3 is facilitated by the Lsm proteins.
Lsm proteins function in U3 and pre-U3 degradation
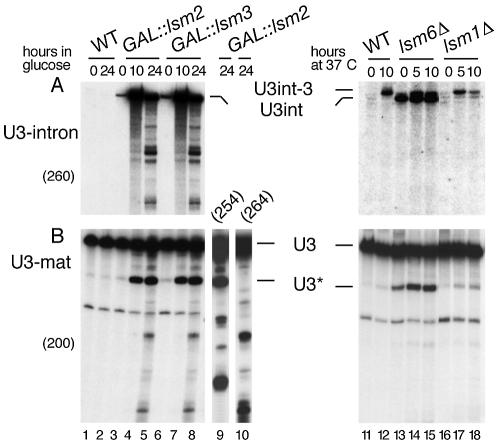
Truncated forms of several stable RNAs, including tRNAs and rRNAs, accumulate in lsm mutants and are assumed to represent degradation intermediates (37,38). Faster migrating U3 species were seen in Lsm-depleted strains at later time points (Fig. 1A). The appearance of truncated U3 and pre-U3 species was assessed in strains carrying GAL::lsm2–5 or 8 and lsm1-, 6- or 7-Δ (Fig. 5; data shown for GAL::lsm2, GAL::lsm3, lsm6-Δ and lsm1-Δ).
Figure 5.
Lsm proteins participate in degradation of U3 species. Northern hybridisation of U3 snoRNA in GAL::lsm2, GAL::lsm3 (lanes 3–10) and lsm6-Δ and lsm1-Δ (lanes 11–18) mutant strains. Strains were grown and RNA was prepared as described for Figure 1. Probe names are indicated in parentheses. RNA species are shown between the two columns. The major truncated species is indicated as U3*. Oligo 200 (lanes 1–8) hybridises downstream of position 82; oligo 254 (lane 9) hybridises downstream of position 316; oligo 264 (lane 10) hybridises downstream of position 1. Hybridisation with oligos 254 and 264 is shown as separate panels (lanes 9–10) for the sample from the GAL::lsm2 strain at 24 h after depletion on glucose medium. For location of the probes against mature U3, see Figure 1E.
Hybridisation with a probe against the U3A intron showed that intron-containing U3 precursors initially accumulate in lsm mutants and are subsequently degraded with substantial accumulation of intermediates (Fig. 5A). Such degradation intermediates are not observed when splicing is inhibited by the prp2-1 mutation or by depleting SmE1p (data not shown), indicating that they are not due to a general splicing defect. The 5′-truncated U3* RNA clearly accumulated in strains depleted of Lsm2p–Lsm8p, whereas little accumulation was seen on depletion of the predominantly cytoplasmic Lsm1p (Fig. 5B and data not shown). This species was also accumulated in strains with defects in the exosome (5). Comparison of hybridisation with probes specific for the 5′ region (Fig. 5B, lane 9) and 3′ region of mature U3 (Fig. 5B, lane 10) showed that both 3′- and 5′-truncated U3 species were accumulated.
We propose that an Lsm2–8p complex participates in degradation of mature and precursor U3 molecules. Both 3′-truncated and 5′-truncated RNAs were accumulated, suggesting that Lsm2–8p participates in both 5′→3′ and 3′→5′ degradation. Intermediates in the degradation of stable RNA species are not normally observed, and our interpretation of these data is that Lsm2–8p complexes enhance the processivity of the exonucleases during RNA degradation. However, an alternative explanation is also possible: that Lsm complex, by stabilising pre-U3 molecules, enhances the specific assembly of U3 RNPs, preventing fast degradation of unincorporated RNAs.
DISCUSSION
Lsm complexes have been proposed to act as chaperones that modify the structure of RNP complexes (27,35). This function would be consistent with the many and diverse consequences of the depletion of Lsm proteins. Here we report that the Lsm2–8p proteins act in the processing and degradation of U3 snoRNA, showing transient or weak interactions with U3 precursors that would be consistent with roles as chaperones.
In the absence of the essential proteins Lsm2–5p and 8p, the major 3′-extended precursors U3-3′I and U3-3′II were lost, suggesting that the Lsm2–8p complex plays an important role in their processing or stability. In contrast, lack of the non-essential Lsm6p and Lsm7p had no dramatic effects. It may be that non-essential proteins can be replaced by other Lsm proteins or even by related Sm proteins in complexes that retain partial activity. Alternatively, active hexameric complexes may form in the absence of a non-essential protein.
While we cannot exclude the possibility that the effects of depletion of the Lsm proteins are secondary effects due to defective pre-mRNA splicing or mRNA degradation, we feel that this is unlikely. Other pre-mRNA splicing defects cause accumulation of intron-containing pre-U3 but have no effect on the processing of an intron-less U3 expressed from a cDNA construct. Mutations in the components of the cytoplasmic mRNA degradation pathway, including the decapping enzyme Dcp1p, the exosome and 5′→3′ exonuclease Xrn1p, do not result in similar U3 processing defects [(5); and data not shown]. Even at late times of Lsm depletion, U3 transcription remains robust, as judged by the strong accumulation of the normally unstable, intron-containing pre-U3 species. This appears to exclude the possibility that the loss of the 3′-extended pre-U3 species is due to the absence of pre-U3 transcription.
In wild-type strains, the 3′-extended pre-U3 species terminate at poly(U) tracts that normally bind the Lhp1p protein. However, the poly(U) tracts form the 3′ end of the transcript only when the exonucleases, which process the pre-U3 from a downstream Rnt1p cleavage site, are arrested at this position (5). This indicates that some factor(s) can bind these regions as internal poly(U) tracts. Lhp1p is not reported to bind internal poly(U) tracts in vitro, and its binding at these sites in vivo may be facilitated by the Lsm2–8p complex. The mode of association of the Lsm proteins with RNA is not known, but the related Sm complex binds to internal Sm sites that include poly(U), assembling the intact ring structure from pre-assembled subcomplexes (58,59). Consistent with this model, depletion of Lsm3p or Lsm5p strongly reduced the association of Lhp1p with pre-U3-3′I and U3-3′II as well as the intron-containing precursors. The association of pre-U3 with Lhp1p was reduced after only 4 h of depletion of Lsm3p or Lsm5p, when the level of the U3-3′I and U3-3′II precursors was little affected and it is most unlikely that transcription was strongly impaired. Depletion of Lsm proteins also reduced the association of Lhp1p with several other RNA precursors (5,15,21,37).
In strains lacking Lhp1p, the residual U3-3′T species are shorter and more heterogeneous than U3-3′I and II but still terminate within the poly(U) tract. Since the Lsm2–8p complex binds to poly(U) (30,31), we speculated that it might replace Lhp1p and stabilise the U3-3′T precursors in the lhp1-Δ cells. However, these species were not co-precipitated with tagged Lsm3p from an extract lacking Lhp1p. This suggests that mutual binding of Lsm2–8p and Lhp1p is required.
The seven-member Sm core also has substantial affinity for a poly(U) sequence in vitro (58). In vertebrates, specificity of snRNP and snoRNP assembly is likely to be greatly enhanced by association of Sm and Lsm proteins with the multimeric SMN complex, which acts as an assembly chaperone (60–65). How such specificity is achieved in S.cerevisiae, which lacks SMN complex components, is currently unclear. The yeast Sm complex is able to bind and stabilise RNA species with 3′-terminal poly(U) tracts in vivo, since the yeast U1, U4 and U5S snRNAs lack terminal stems beyond the Sm-binding sites (66–69). ProtA-tagged SmE1p failed to co-precipitate U3-3′I or U3-3′II from an otherwise wild-type strain, but was able to co-precipitate U3-3′T from the strain lacking Lhp1p. This indicates that the Sm complex binds to the 3′ poly(U) tracts only in the absence of Lhp1p. The efficiency of co-precipitation of U3-3′T with SmE1-ProtA was low. However, in vitro binding of the Sm heptamer to a poly(U) tract showed only limited thermodynamic stability, with dissociation in 10 min, in contrast to the stable binding seen with a consensus Sm-binding site (58). This suggests that substantial dissociation of the bound Sm complex from a terminal poly(U) tract may occur during the extended incubation required for immunoprecipitation. It is unclear why the Sm proteins apparently fail to bind pre-U3 in the presence of Lhp1p or in the absence of the Lsm complex, and it seems likely that additional factors are involved in these interactions.
It is notable that the level of the mature U3 snoRNA was little affected by the absence of any of the Lsm proteins or Lhp1p (5). Similarly, the levels of mature tRNAs or the U1, U4 and U5 snRNAs were not changed in cells lacking Lhp1p or the Lsm2–8p proteins (3,21,37). It appears that 3′-processing pathways in yeast have substantial redundancy, and we predict that still other pathways and factors remain to be identified.
Processing of U3 shares common steps with processing of spliceosomal snRNAs and some snoRNAs. It would be interesting to establish whether the role of the Lsm complex is specific for U3 or more general in maturation of other stable RNPs. However, analysis of their processing is more problematic than for U3. Synthesis of snRNAs is affected in splicing mutants [(70–72); and our unpublished observations] and it is not trivial to discriminate between the effects of Lsm proteins due to splicing against processing. On the other hand, 3′-extended precursors of snoRNAs are not readily detectable in wild-type cells and their fate in lsm mutants or their association with Lsm complex is difficult to assess. At least some snoRNAs of boxC+D and boxH+ACA classes contain poly(U) tracts 3′ of their mature ends, and they may associate with Lhp1p and require both Lhp1p and Lsm proteins for processing; however, their 3′ end formation pathway is not well characterised at present.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Bertrand Séraphin and Rémy Bordonné for generously providing the plasmid pBS1360 and strains YRB15 and YRB20, respectively. This work was supported by the Wellcome Trust.
REFERENCES
- 1.Chanfreau G., Abou Elela,S., Ares,M.,Jr and Guthrie,C. (1997) Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev., 11, 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou Elela S. and Ares,M.J. (1998) Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J., 17, 3738–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allmang C., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seipelt R.L., Zheng,B., Asuru,A. and Rymond,B.C. (1999) U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res., 27, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kufel J., Allmang,C., Chanfreau,G., Petfalski,E., Lafontaine,D.L.J. and Tollervey,D. (2000) Precursors to the U3 snoRNA lack snoRNP proteins but are stabilized by La binding. Mol. Cell. Biol., 20, 5415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Hoof A., Lennertz,P. and Parker,R. (2000) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol., 20, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Hoof A., Lennertz,P. and Parker,R. (2000) Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J., 19, 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- 9.Anderson J.S. and Parker,R. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allmang C., Mitchell,P., Petfalski,E. and Tollervey,D. (2000) Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res., 28, 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bousquet-Antonelli C., Presutti,C. and Tollervey,D. (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell, 102, 765–775. [DOI] [PubMed] [Google Scholar]
- 12.Burkard K.T. and Butler,J.S. (2000) A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol., 20, 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torchet C., Bousquet-Antonelli,C., Milligan,L., Thompson,E., Kufel,J. and Tollervey,D. (2002) Processing of 3′ extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell, 9, 1285–1296. [DOI] [PubMed] [Google Scholar]
- 14.Libri D., Dower,K., Boulay,J., Thomsen,R., Rosbash,M. and Jensen,T.H. (2002) Interactions between mRNA export commitment, 3′-end quality control and nuclear degradation. Mol. Cell. Biol., 22, 8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue D., Rubinson,D., Pannone,B.K., Yoo,C.J. and Wolin,S.L. (2000) U snRNP assembly in yeast involves the La protein. EMBO J., 19, 1650–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinke J. and Steitz,J.A. (1982) Precursor molecules of both human 5S ribosomal RNA and transfer RNAs are bound by a cellular protein reactive with anti-La lupus antibodies. Cell, 29, 149–159. [DOI] [PubMed] [Google Scholar]
- 17.Chambers J.C., Kurilla,M.G. and Keene,J.D. (1983) Association between the 7S RNA and the lupus La protein varies among cell types. J. Biol. Chem., 258, 11438–11441. [PubMed] [Google Scholar]
- 18.Stefano J.E. (1984) Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell, 36, 145–154. [DOI] [PubMed] [Google Scholar]
- 19.Rinke J. and Steitz,J.A. (1985) Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res., 13, 2617–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pannone B.K., Xue,D. and Wolin,S.L. (1998) A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J., 17, 7442–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo C.J. and Wolin,S.L. (1997) The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell, 89, 393–402. [DOI] [PubMed] [Google Scholar]
- 22.Hermann H., Fabrizio,P., Raker,V.A., Foulaki,K., Horning,H., Brahms,H. and Lührmann,R. (1995) SnRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein–protein interaction. EMBO J., 14, 2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kambach C., Walke,S., Young,R., Avis,J.M., de la Fortelle,E., Raker,V.A., Lührmann,R., Li,J. and Nagai,K. (1999) Crystal structure of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell, 5, 375–387. [DOI] [PubMed] [Google Scholar]
- 24.Cooper M., Johnston,L.H. and Beggs,J. (1995) Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J., 14, 2066–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Séraphin B. (1995) Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J., 14, 2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottschalk A., Neubauer,G., Banroques,J., Mann,M., Lührmann,R. and Fabrizio,P. (1999) Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6·U5] tri-snRNP. EMBO J., 18, 4535–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayes A.E., Verdone,L., Legrain,P. and Beggs,J.D. (1999) Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J., 18, 4321–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salgado-Garrido J., Bragado-Nilsson,E., Kandels-Lewis,S. and Séraphin,B. (1999) Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J., 18, 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens S.W. and Abelson,J. (1999) Purification of the yeast U4/U6·U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl Acad. Sci. USA, 96, 7226–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Achsel T., Brahms,H., Kastner,B., Bachi,A., Wilm,M. and Lührmann,R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J., 18, 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Achsel T., Stark,H. and Lührmann,R. (2001) The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl Acad. Sci. USA, 98, 3685–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins B.M., Harrop,S.J., Kornfeld,G.D., Dawes,I.W., Curmi,P.M. and Mabbutt,B.C. (2001) Crystal structure of a heptameric Sm-like protein complex from Archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol., 309, 915–923. [DOI] [PubMed] [Google Scholar]
- 33.Boeck R., Lapeyre,B., Brown,C.E. and Sachs,A.B. (1998) Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol., 18, 5062–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouveret E., Rigaut,G., Shevchenko,A., Wilm,M. and Séraphin,B. (2000) An Sm-like protein complex that participates in mRNA degradation. EMBO J., 19, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tharun S., He,W., Mayes,A.E., Lennertz,P., Beggs,J.D. and Parker,R. (2000) Yeast Sm-like proteins function in mRNA decapping and decay. Nature, 404, 515–518. [DOI] [PubMed] [Google Scholar]
- 36.Tharun S. and Parker,R. (2001) Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p–7p complex on deadenylated yeast mRNAs. Mol. Cell, 8, 1075–1083. [DOI] [PubMed] [Google Scholar]
- 37.Kufel J., Allmang,C., Verdone,L., Beggs,J. and Tollervey,D. (2002) Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol. Cell. Biol., 22, 5248–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kufel J., Allmang,C., Petfalski,E., Beggs,J. and Tollervey,D. (2003) Lsm proteins are required for normal processing and stability of ribosomal RNAs. J. Biol. Chem., 278, 2147–2156. [DOI] [PubMed] [Google Scholar]
- 39.Tomasevic N. and Peculis,B.A. (2002) Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol. Cell. Biol., 22, 4101–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y. and Meier,U.T. (2003) Genetic interaction between a chaperone of small nucleolar ribonucleoprotein particles and cytosolic serine hydroxymethyltransferase. J. Biol. Chem., 278, 23553–23560. [DOI] [PubMed] [Google Scholar]
- 41.Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Improved method for high efficient transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lafontaine D. and Tollervey,D. (1996) One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res., 24, 3469–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puig O., Caspary,F., Rigaut,G., Rutz,B., Bouveret,E., Bragado-Nilsson,E., Wilm,M. and Seraphin,B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods, 24, 218–229. [DOI] [PubMed] [Google Scholar]
- 44.Tollervey D. (1987) A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J., 6, 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beltrame M. and Tollervey,D. (1992) Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J., 11, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma K. and Tollervey,D. (1999) Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol., 19, 6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beltrame M. and Tollervey,D. (1995) Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J., 14, 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Séraphin B. and Rosbash,M. (1989) Identification of functional U1 snRNA–pre-mRNA complexes committed to spliceosome assembly and splicing. Cell, 59, 349–358. [DOI] [PubMed] [Google Scholar]
- 49.Lygerou Z., Mitchell,P., Petfalski,E., Séraphin,B. and Tollervey,D. (1994) The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev., 8, 1423–1433. [DOI] [PubMed] [Google Scholar]
- 50.Aris P. and B.,G. (1988) Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J. Cell Biol., 107, 17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fromont-Racine M., Mayes,A.E., Brunet-Simon,A., Rain,J.C., Colley,A., Dix,I., Decourty,L., Joly,N., Ricard,F., Beggs,J.D. and Legrain,P. (2000) Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast, 17, 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He W. and Parker,R. (2001) The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics, 158, 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bordonne R. and Tarassov,I. (1996) The yeast SME1 gene encodes the homologue of the human E core protein. Gene, 176, 111–117. [DOI] [PubMed] [Google Scholar]
- 54.Plumpton M., McGarvey,M. and Beggs,J.D. (1994) A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J., 13, 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schimmang T., Tollervey,D., Kern,H., Frank,R. and Hurt,E.C. (1989) A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J., 8, 4015–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lafontaine D.L.J. and Tollervey,D. (1999) Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA, 5, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lafontaine D.L.J. and Tollervey,D. (2000) Synthesis and assembly of the box C+D snoRNPs. Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raker V.A., Hartmuth,K., Kastner,B. and Lührmann,R. (1999) Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol., 19, 6554–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raker V.A., Plessel,G. and Lührmann,R. (1996) The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J., 15, 2256–2269. [PMC free article] [PubMed] [Google Scholar]
- 60.Fischer U., Liu,Q. and Dreyfuss,G. (1997) The SMN–SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell, 90, 1023–1029. [DOI] [PubMed] [Google Scholar]
- 61.Meister G., Buhler,D., Pillai,R., Lottspeich,F. and Fischer,U. (2001) A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nature Cell Biol., 3, 945–949. [DOI] [PubMed] [Google Scholar]
- 62.Meister G. and Fischer,U. (2002) Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J., 21, 5853–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jones K.W., Gorzynski,K., Hales,C.M., Fischer,U., Badbanchi,F., Terns,R.M. and Terns,M.P. (2001) Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem., 276, 38645–38651. [DOI] [PubMed] [Google Scholar]
- 64.Pellizzoni L., Baccon,J., Charroux,B. and Dreyfuss,G. (2001) The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol., 11, 1079–1088. [DOI] [PubMed] [Google Scholar]
- 65.Pellizzoni L., Yong,J. and Dreyfuss,G. (2002) Essential role for the SMN complex in the specificity of snRNP assembly. Science, 298, 1775–1779. [DOI] [PubMed] [Google Scholar]
- 66.Kretzner L., Krol,A. and Rosbash,M. (1990) Saccharomyces cerevisiae U1 small nuclear RNA secondary structure contains both universal and yeast-specific domains. Proc. Natl Acad. Sci. USA, 87, 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siliciano P.G., Jones,M.H. and Guthrie,C. (1987) Saccharomyces cerevisiae has a U1-like small nuclear RNA with unexpected properties. Science, 237, 1484–1487. [DOI] [PubMed] [Google Scholar]
- 68.Patterson B. and Guthrie,C. (1987) An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell, 49, 613–624. [DOI] [PubMed] [Google Scholar]
- 69.Brow D.A. and Guthrie,C. (1988) Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature, 334, 213–218. [DOI] [PubMed] [Google Scholar]
- 70.Noble S.M. and Guthrie,C. (1996) Transcriptional pulse–chase analysis reveals a role for a novel snRNP-associated protein in the manufacture of spliceosomal snRNPs. EMBO J., 15, 4368–4379. [PMC free article] [PubMed] [Google Scholar]
- 71.Roy J., Zheng,B., Rymond,B.C. and Woolford,J.L.,Jr (1995) Structurally related but functionally distinct yeast Sm D core small nuclear ribonucleoprotein particle proteins. Mol. Cell. Biol., 15, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rymond B.C. (1993) Convergent transcripts of the yeast PRP38-SMD1 locus encode two essential splicing factors, including the D1 core polypeptide of small nuclear ribonucleoprotein particles. Proc. Natl Acad. Sci. USA, 90, 848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]