Abstract
The orphan nuclear receptor small heterodimer partner (SHP; NR0B2) is an unusual orphan nuclear receptor that lacks a conventional DNA-binding domain and acts as a modulator of transcriptional activities of a number of nuclear receptors. We have previously reported that the orphan nuclear receptor ERRγ activates the SHP promoter. In this study, we have found that basic helix–loop–helix (bHLH) transcription factors, the E2A proteins (E47, E12 and E2/5), activated the human but not the mouse SHP promoter. In contrast, the tissue-specific E47 heterodimer partner BETA2 repressed the E47- mediated transactivation of the human SHP (hSHP) promoter. Using serial deletions and E-box mutant constructs of the hSHP promoter, we identified two E-boxes (E6 and E7) as E47-responsive E-boxes, which are not conserved in the mouse SHP promoter. Moreover, gel shift, chromatin immunoprecipitation (ChIP) and northern blot assays demonstrated that E47 directly binds to the hSHP promoter in vivo and in vitro and that Id proteins inhibited E47 binding to the hSHP promoter. Finally, we found that E47 and steroidogenic factor 1 (SF-1), a regulator of the SHP promoter, synergistically activate the human but not the mouse SHP promoter. Our findings suggest that the E2A proteins differentially regulate the human and mouse SHP promoters and cooperate with orphan nuclear receptor SF-1 for transcriptional activation of the hSHP promoter.
INTRODUCTION
The nuclear receptor superfamily is a group of ligand-dependent transcriptional factors that usually contain two functionally independent and conserved domains (1). A conserved DNA-binding domain (DBD) allows them to associate directly with specific DNA response elements. A ligand-binding domain (LBD) is required for the binding of small lipophilic molecules, ligands or hormones, and for the transmission of ligand signals to transcriptional responses. The superfamily includes both conventional receptors with known ligands and orphan nuclear receptors that lack specific ligands [for reviews see Evans (2) and Giguere (3)].
SHP is one of unique members of the orphan nuclear receptor superfamily that lacks a conventional DBD, but contains a putative LBD (4). Various studies have reported that SHP is a repressor of transcriptional activities of a number of nuclear receptors, including both ligand-responsive receptors, such as ER, GR, TR, AR, RAR and RXR, and orphan receptors such as CAR, HNF4α, LRH-1, ERRγ and LXRα (4–12). However, SHP was also found to be able to activate the transcriptional activity of PPARγ (13). The very broad range of receptors sensitive to inhibition by SHP suggests a central role for SHP in modulation of nuclear receptor signaling pathways. Recent results demonstrate that SHP can compete with coactivators for binding to the AF-2 surface of nuclear receptors, and a direct transcriptional repression domain contributes to the inhibitory function of SHP (8,9). SHP is expressed in a wide variety of tissues, including heart, brain, liver, spleen, adrenal gland, small intestine and pancreas (7,14). The SHP gene consists of two exons and one intron (15), and SHP gene transcription is regulated by several members of the nuclear receptor superfamily, including bile acid receptor FXR (16–18), steroidogenic factor-1 (SF-1) (19), HNF4α (20), LRH-1 (18) and ERRγ (11). Moreover, previous reports have suggested that SHP plays a pivotal role in the regulation of cholesterol homeostasis via a bile acid-activated regulatory cascade in the liver (16–18). The FXR-mediated SHP gene induction has been shown to inhibit the activity of orphan nuclear receptor LRH-1, a positive regulator of cholesterol 7α-hydroxylase (CYP7A1) gene expression, which catalyzes the rate-limiting step in bile acid biosynthesis. However, loss-of-function studies of SHP demonstrated that redundant pathways independent of the SHP-mediated pathway are implicated in the negative feedback regulation of bile acid production (21,22). It has also been reported that mutations in the SHP gene in human are associated with mild hyperinsulinemia and obesity (14).
Basic helix–loop–helix (bHLH) transcription factors comprise a large family of transcription factors, which bind to the specific DNA sequence CANNTG, known as the E-box [reviewed in Massari and Murre (23)]. The E2A proteins (E47, E12 and E2/5), encoded by the E2A gene, are the founder members of the vertebrate class I bHLH proteins (also called E proteins) (24). E2A proteins are all very similar; E12 and E47 proteins differ in their C-terminal bHLH sequences, whereas the differences between the E2/5 and E47 proteins are within their N-terminal sequences. A large body of evidence indicates that the E2A proteins play a central role in tissue-specific gene regulation, usually after their dimerization with tissue-specific class II bHLH proteins (25–28). Nevertheless, homodimers of E2A proteins are functionally active in B-cell lineages where they participate in the transcriptional regulation of several Ig genes. In fact, the products of the E2A gene (E12/E47) are required for B-cell differentiation and Ig gene rearrangements (29–31). In addition, targeted disruption of the E2A gene leads to thymic lymphomas, suggesting that E2A proteins can act as tumor suppressors (32). The transcriptional activity of E2A proteins can be negatively regulated by their dimerization with another subclass of HLH factors, the Id proteins (33). The four Id proteins, Id-1, Id-2, Id-3 and Id-4, identified thus far have only a HLH domain and lack the basic DNA-binding region. Dimerization of Id proteins with bHLH proteins results in the inhibition of DNA binding of bHLH proteins (34–38).
The SHP promoter contains several E-box (CANNTG) sequences that are predicted to be bHLH protein response elements, and we investigated whether the bHLH protein subfamily members can regulate the SHP promoter. In this study, we demonstrated a preferential effect of E2A proteins (E47, E12 and E2/5) on the human but not the mouse SHP promoter. Detailed mapping studies revealed that only two (E6 and E7) of 13 E-box (CANNTG) motifs on the human SHP (hSHP) promoter are sufficient and necessary for the E47-mediated activation. Gel mobility shift assays and mutation studies confirmed the binding of E47 homodimers to the two E-boxes. In addition, Id-1, Id-2 and Id-3, known as inhibitory proteins of bHLH proteins, repressed E47-mediated transactivation of the hSHP promoter. Chromatin immunoprecipitation (ChIP) assay and northern blot analysis demonstrated that Id-2 inhibited E47 binding to the hSHP promoter in vivo and decreased hSHP expression. Furthermore, E47-mediated transactivation synergized with SF-1 to activate the hSHP promoter. These results suggest that the E2A gene products play a key role in the regulation of hSHP gene expression and cooperate with orphan nuclear receptor SF-1 for transcriptional activation of the hSHP promoter.
MATERIALS AND METHODS
Plasmids and DNA construction
The plasmids of CMV4-E47, E12 and E2/5 were kind gifts from Dr Roland Stein. pCMV5-casein kinase (CK) II-α was donated by Dr Michael P. Czech. pcDNA3-human Id-1, Id-2 and Id-3 were kind gifts from Dr John D. Norton. pCR3.1-BETA2 and RIPE3 (–126 to –86 bp) reporters that contain E-box elements from the insulin promoter were donated by Dr Ming-Jer Tsai. (523)4 Reporter plasmid that contains multiple E-boxes upstream of the luciferase gene was provided by Dr John Kim Choi. The wild-type and mutant reporter genes of the hSHP promoter were cloned into the pGL2-basic plasmid (Promega) vector using restriction enzymes and specific primers. Wild-type and serial deletion constructs containing 5′-flanking sequences from the hSHP promoter were fused to the luciferase gene at +30 (–2.2 kb/Luc, –1.35 kb/Luc, –574 bp/Luc, –355 bp/Luc, –330 bp/Luc, –280 bp/Luc, –263 bp/Luc, –250 bp/Luc, –243 bp/Luc, and deletion –574 to –243 bp/Luc). E-box mutant reporters (mE7, mE6, mE5 and mE6,7) from the hSHP promoter –2.2 kb/Luc were constructed by site-directed mutagenesis using the primers containing CA to AG substitution mutations in the core E-box (CANNTG) of individual E5, E6 and E7 E-boxes, and both E6 and E7 E-boxes. Mutations (underlined) were introduced using PCR with the following pairs of primers: mE7, 5′-CCTGCTGATTGTGCACCT-3′ and 5′-AGGTGCACAATCAGCAGGCTTCCCCATTGGTGCCCG-3′; mE6, 5′-CCTGGGGCCTTGGTG-3′ and 5′-CACCAAGGCCCCAGGCTCACAATCAGCAGG-3′; mE5, 5′-CTTGAGTCATCTGATAAG-3′ and 5′-CTTATCAGATGACTCAAGCTGAT AAACAAGGTCATTAAC-3′. pcDNA3 hemagglutinin (HA)-E47, HA-SF-1, HA-LRH-1 and HA-ERRγ were constructed by inserting EcoRI-, SalI- and XhoI-digested PCR products encoding the open reading frame of E47, SF-1, LRH-1 and ERRγ into EcoRI- and XhoI-digested pcDNA3 HA vector.
Cell culture and transient transfection assay
HepG2, HeLa and CV-1 cells were maintained with Dulbecco’s modified Eagle’s medium (DMEM; Gibco-BRL), supplemented with 10% fetal bovine serum (FBS; Biowhittaker) and antibiotics (Life Technologies). Cells were split in 24-well plates at densities of 2–20 × 104 cells/well the day before transfection. Transient transfections were performed using the SuperFect transfection reagent (Qiagen Inc.) according to the manufacturer’s instructions. Cells were co-transfected with 0.2 µg of hSHP promoter wild-type, deletion constructs (–2.2 kb to –243 bp/Luc), E-box point mutant constructs (mE5/Luc, mE6/Luc, mE7/Luc and mE6,7), RIPE3/Luc and mouse SHP promoter (–2.2 kb/Luc) reporter plasmids together with 0.2 µg of pcDNA3 HA expression vectors for BETA2 and E47 and the indicated amounts of pCMV5-CK II-α, pcDNA3-hId-1, -2 and -3. Total DNA used in each transfection was adjusted to 1 µg/well by adding appropriate amount of pcDNA3 empty vector, and 0.2 µg of cytomegalovirus (CMV)-β-galactosidase plasmids were co-transfected as an internal control. Cells were harvested ∼40– 48 h after the transfection for luciferase and β-galactosidase assays. The luciferase activity was normalized by β-galactosidase activity.
Gel mobility shift assays
The probes used in gel shift assay cover the region –342 to –321 bp from the hSHP promoter. Mutated E-box sequences (CACCTG to agCCTG) in mE7, mE6 and mE6,7 are indicated in lower case in the following primers: E6,7 wild-type, 5′-GGGGACACCTGCTGATTGTGCACCTGGGGC-3′; mE7, 5′-GGGGAagCCTGCTGATTGTGCACCTGGGGC-3′; mE6, 5′-GGGGACACCTGCTGATTGTGagCCTGGGGC-3′, and mE6,7, 5′-GGGGAagCCTGCTGATTGTGagCCTGGGGC-3′. Double-stranded oligonucleotides were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. Bacterially expressed His-tagged E47 protein (His-E47) was purified as described previously (39), analyzed by SDS–PAGE and quantified by Coomassie blue staining. In vitro synthesized E47 and Id proteins were transcribed and translated using a coupled rabbit reticulocyte system (TNT, Promega) according to the manufacturer’s instructions. Gel shift assays contained 10 mM Tris pH 8.0, 40 mM KCl, 0.05% NP-40, 6% glycerol, 1 mM dithiothreitol (DTT) and 1 µg of poly(dI–dC). A 1 µg aliquot of His-tagged E47 was mixed with 10 000 c.p.m. of end-labeled oligonucleotide probes in 20 µl of each reaction. After 20 min incubation, DNA–protein complexes were analyzed on a 4% polyacrylamide gel in 1× TBE (90 mM Tris, 90 mM boric acid, 2 mM EDTA). Gels were dried and analyzed by autoradiography.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay described by Shang et al. (40) was adopted, with modifications. HepG2 cells were transfected with 5 µg of pcDNA3 HA-E47 and pcDNA3-Id-2. Total DNA used in each transfection was adjusted to 10 µg/60 mm dish by adding appropriate amounts of pcDNA3 HA empty vector. The cells were fixed with formaldehyde 30 h after transfection and harvested. For immunoprecipitation, anti-HA antibodies (Roche Molecular Biochemicals) and protein A–Sepharose beads CL-4B (Amersham Biosciences) were used. The final DNA extractions were amplified using pairs of primers that cover the E47 response region within the hSHP promoter (nucleotides –120 to –474) 5′-CCCCTGGCAGGAATG-3′ and 5′-AGGTTAGGCAAACAAGC-3′).
Northern blot analysis
HepG2 cells were maintained in DMEM supplemented with 0.5% FBS for 24 h and changed to medium containing 20% FBS. Total RNA was isolated at 0, 1, 3, 6 and 12 h after 20% serum stimulation. Northern blot analysis was carried out as described previously (15).
RESULTS
Identification of E2A-encoded gene products as transcription factors regulating the hSHP promoter
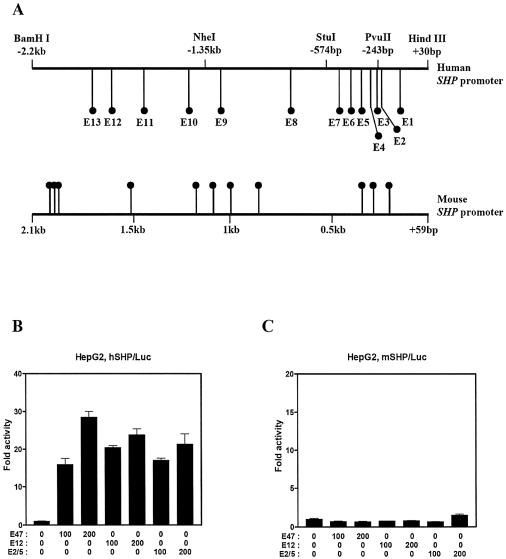
To identify the transcription factors that regulate the SHP promoter, we examined the nucleotide sequence of the hSHP promoter regions spanning from upstream –2.2 kb to downstream +10 bp of the putative transcription start site. Among the binding sites for various kinds of transcription factors in the hSHP promoter, we found 13 E-box (CANNTG) motifs (designated as E1–E13 in human), which are well known binding sites of bHLH transcription factors. We also aligned the mouse SHP (mSHP) promoter and found 11 E-boxes (Fig. 1A). To investigate whether bHLH protein members can regulate the SHP promoter, we performed a transient transfection assay using the reporter gene containing the –2.2 kb to +30 bp sequence of the hSHP promoter. We first examined the effects of E2A gene products (E2A proteins) including E47, E12 and E2/5, which belong to class I bHLH proteins (also called E proteins), on the hSHP promoter. Over-expression of E47, E12 and E2/5 in HepG2 cells strongly activated the hSHP promoter in a dose-dependent manner (Fig. 1B). In HeLa and CV-1 cells, a similar effect of E2A gene products on the hSHP promoter was observed (data not shown). Unexpectedly, over-expression of E47, E12 and E2/5 had no significant effect on the mSHP promoter, although the mSHP promoter also contains 11 potential E-boxes, indicating that E47-responsive regions may be present only in the hSHP promoter (Fig. 1C). Taken together, these results suggest that E2A proteins can be a positive regulator of the hSHP promoter and that E2A gene products can differentially regulate the hSHP and mSHP promoter.
Figure 1.
E2A gene products of bHLH transcription factor activate the hSHP promoter. (A) Schematic diagrams of the human and mouse SHP promoter showing E-boxes. HepG2 cells were transfected with 200 ng of –2.2 kb human (B) and mouse (C) SHP promoter fused to a luciferase reporter, together with the indicated amount of expression vectors for E47, E12 or E2/5. Approximately 40 h after transfections, the cells were harvested and luciferase activity was measured and normalized against β-galactosidase activity. One representative experiment is shown. All values represent the mean of duplicate samples, and similar results were obtained in at least three independent experiments. The representative results were expressed as fold activation (n-fold) over the value obtained with vector alone, with the error bars as indicated.
E2A gene products are sufficient for the transactivation of the hSHP promoter
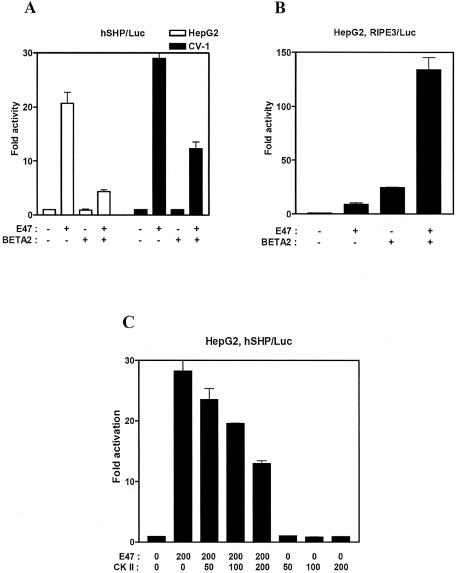
It has been reported that E2A proteins are potent transcriptional activators as either homodimers or heterodimers (25–31). However, the transcriptional activity of E2A proteins as homodimers or heterodimers is highly specific for the enhancers they stimulate. To investigate whether E2A proteins function as either homodimers or heterodimers on the hSHP promoter, we tested the effect of class II bHLH proteins such as BETA2/NeuroD, MyoD and myogenin, which are heterodimer partners of E2A proteins. Interestingly, co-expression of BETA2 with E47 reduced the E47-mediated transactivation of the hSHP promoter, whereas overexpression of BETA2 alone had little effect on the basal activity in HepG2 or CV-1 cells (Fig. 2A). In contrast, BETA2 and E47 strongly activated the RIPE3 reporter that contains E-box elements from the insulin gene promoter, as previously reported (Fig. 2B). Other heterodimer partners of E47, such as MyoD and myogenin, also repressed the E47-mediated transactivation of the hSHP promoter (data not shown). These results suggest that E2A gene products alone may be sufficient for the transactivation of hSHP promoter, and heterodimerizatin of BETA2 with E2A may interfere with E47 homodimer formation or perhaps may even be repressive on the hSHP promoter.
Figure 2.
E47 is sufficient for the transactivation of the hSHP promoter. (A) HepG2 (white bar) and CV-1 (black bar) cells were transfected with 200 ng of hSHP promoter reporter and expression vectors for E47 and BETA2 as indicated. (B) HepG2 cells were transfected with 200 ng of RIPE3 reporter gene and expression vectors for E47 and BETA2 as indicated. (C) HepG2 cells were transfected with 200 ng of hSHP/Luc and the indicated amount of the expression vectors for E47 and CK II. The luciferase activity was measured and normalized against β-galactosidase activity. All values represent the mean of duplicate samples, and similar results were obtained in at least three independent experiments. The representative results were expressed as fold activation (n-fold) over the value obtained with vector alone, with the error bars as indicated.
It has been reported that CK II repressed the transcriptional activity of E47 homodimer by inhibiting DNA binding, while the transcriptional activity of E47 heterodimer is enhanced by CK II (41). To further confirm whether the activation of the hSHP promoter by E47 is due to E2A homodimers or heterodimers, we transiently transfected the CK II expression vector with or without E47 expression vector in HepG2 cells. Over-expression of CK II repressed the E47-dependent transactivation of the reporter gene in a dose-dependent manner, whereas CK II had no significant effect on the basal promoter activity, indicating that E47-dependent transactivation of the hSHP promoter may be mediated by E47 homodimers (Fig. 2C).
Identification of two E-boxes (E6 and E7) as E47-responsive E-boxes on the hSHP promoter
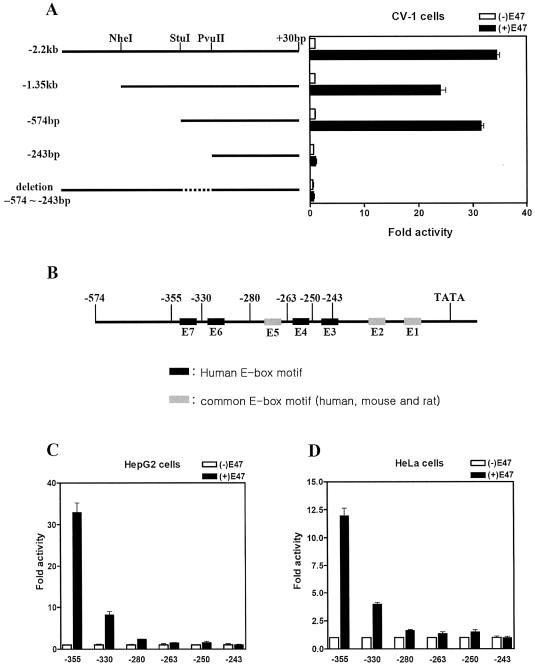
In order to map the sequences required for E47-mediated activation, a series of 5′ deletions of the hSHP promoter were used in a transient transfection assay. As shown in Figure 3A, E47 responsiveness was retained with a deletion up to –574 bp, but was lost with the –243 bp construct. In addition, a construct deleted from –574 to –243 within –2.2 kb of the hSHP promoter completely lost E47 responsiveness. These results indicate that the sequences required for E47 response lie in the region between –574 and –243, which contains five E-boxes (from E3 to E7). Notably, four (E3, E4, E6 and E7) out of the five E-boxes are unique to the hSHP promoter, while the E5 box is present in human, mouse and rat SHP promoters. To further identify E47-responsive E-boxes within this region (–574 to –243 bp), we made a series of E-box deletion constructs of the hSHP promoter as indicated in Figure 3B. In HepG2 and HeLa cells, E47 expression vectors were over-expressed together with the reporter genes driven by a series of E-box deletion constructs of the hSHP promoter. As shown in Figure 3C and D, E47-dependent activity of the hSHP promoter was still detected with further deletion up to –355 bp, which contains all seven E-boxes (E1–E7). However, the E47-mediated transactivity was markedly decreased by E7 deletion (–330 bp) and completely abolished by E6 deletion (–280 bp) from the hSHP promoter, although the –280 bp/Luc reporter contains five intact E-boxes (E1–E5). We could not observe E47-dependent activation of the further deletions of the SHP promoter, –263 bp, –250 bp and –243 bp/Luc. These results suggest that E47-dependent activation of the hSHP promoter is mediated through the –355 bp to –280 bp region containing the E6 and E7 E-boxes.
Figure 3.
Determination of the E47-responsive region on the hSHP promoter. (A) Schematic diagram of the hSHP promoter deletion constructs subcloned in pGL2-basic vector. The restriction enzymes used to make the deletions are indicated on the top. CV-1 cells were transfected with 200 ng of reporter plasmids containing serial deletion of the hSHP promoter and 200 ng of E47 expression vector or empty vector. (B) Illustration of serial E-box deletion constructs of the hSHP promoter. E-boxes (E7 to E3) in the hSHP promoter from –355 bp to +30 bp were serially deleted and subcloned into pGL2-basic vector by inserting the PCR fragments using various primers. Primers used to make the deletions are indicated on the top. E-boxes found in only the hSHP promoter are indicated in black boxes, and common E-boxes are indicated in gray boxes. HepG2 (C) and HeLa (D) cells were transfected with 200 ng of reporter plasmids and 200 ng of E47 expression vector or empty vector.
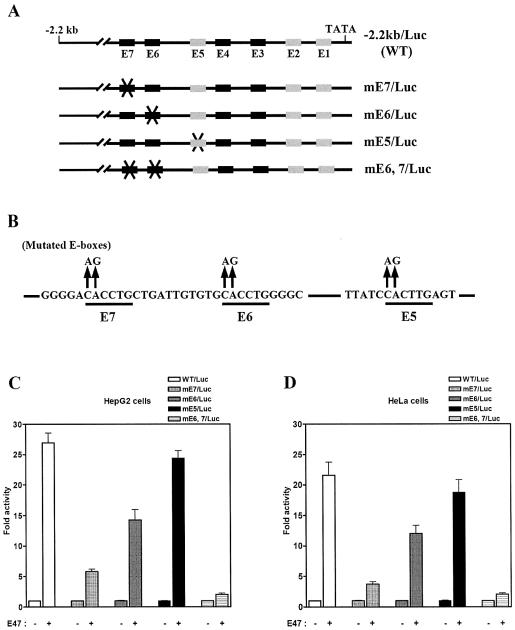
The results from Figure 3C and D suggested that E47-dependent transactivation of the hSHP promoter may be directly mediated by E6 and E7 E-boxes. To address this, a serious of E-box mutants within the –2.2 kb hSHP promoter was constructed and transiently transfected with E47 expression vectors in HepG2 and HeLa cells. E-box mutant reporters were made by introducing CA to AG substitution mutations into the core E-box (CANNTG) of individual boxes E5, E6 and E7, and double mutation of both E6 and E7 E-boxes (Fig. 4A and B). As shown in Figure 4C and D, E47-mediated activity was markedly decreased with E7 mutation reporter (mE7/Luc), by ∼75% of the wild-type hSHP promoter activity. However, we observed a slight decrease in E47-mediated activity with mE6/Luc, by ∼30% of wild-type reporter activity. Notably, E47-mediated activity was completely abolished with the double E6 and E7 mutant reporter (mE6,7/Luc), although the –2.2 kb hSHP promoter still contains other intact E-boxes. This observation is consistent with the results from Figure 3C showing that E47-mediated transactivity was completely abolished by E6 and E7 deletion, although five E-boxes (E1–E5) were intact. As expected, the E47-mediated activity of the E5 box mutant (mE5/Luc) was similar to that of the wild-type hSHP promoter, in good agreement with the results showing that the mSHP promoter was not activated by E47 even though it contains the E5 E-box. Taken together, these results suggest that E47 transactivates the hSHP promoter through E-boxes E6 and E7, which are present only in the hSHP promoter.
Figure 4.
E6 and E7 E-boxes are responsible for E47-mediated activation of the hSHP promoter. (A) Schematic of mutated E-boxes within the hSHP (–2.2 kb) promoter. (B) Illustration of mutated E-box sequences, in each of which the sequence CANNTG was replaced by AGNNTG. HepG2 (C) and HeLa (D) cells were transfected with 200 ng of each mutated E-box, reporter plasmids and E47 expression vector or empty vector.
E47 directly binds to the E6 and E7 E-boxes on the hSHP promoter
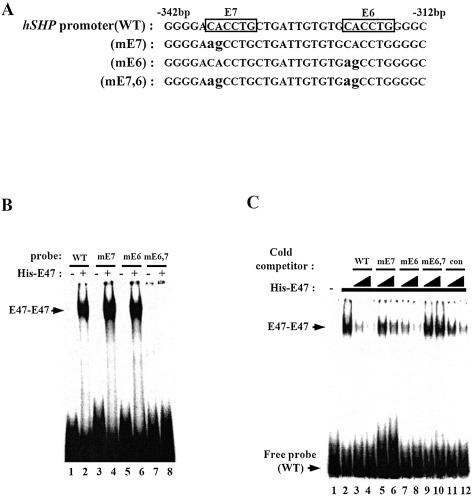
To further examine whether E47 directly binds to E6 and E7 E-boxes within the hSHP promoter, a series of gel mobility shift assays were conducted using oligonucleotides (–342 to –312 bp) containing wild-type, individual E6 and E7 E-box mutations (mE6, mE7) and a double E-box mutation (mE6,7) (Fig. 5A). Unexpectedly, only one band of E47–DNA complex was observed, although wild-type probes contain two E-boxes (E6 and E7). In addition, E47 bound to mE6 and mE7, but not mE6,7 double mutant probes, indicating that E47 may not bind to both E6 and E7 E-boxes simultaneously and either the E6 or the E7 E-box is required for E47 binding to the hSHP promoter (Fig. 5B). To determine to which E-box E47 binds with higher affinity, a series of competition assays were performed using 50- and 100-fold molar excess of unlabeled oligonucleotides of wild-type, mE6, mE7, mE6,7 and control probes from the insulin promoter E-box (RIPE3). As shown in Figure 5C, excess unlabeled oligonucleotides corresponding to wild type and mE6 greatly reduced the E47–DNA complex, while excess unlabeled mE7 oligonucleotides only marginally reduced the E47–DNA complex, indicating that E47 binds to the E7 E-box with higher affinity than to E6. As expected, mE6,7 had no significant effect on the E47–DNA complex, and RIPE3 control inhibited E47–DNA complex formation. These results are consistent with our previous observations showing that most of the transactivity of the hSHP promoter was decreased by E7 deletion or E7 mutation (Figs 3, 4C and D). Taken together, these data suggest that the E7 E-box is preferentially required for E47 binding, although both E-boxes are required for transactivation of the hSHP promoter and E47 can bind to either E-box (E6 or E7).
Figure 5.
E47 binds to E6 and E7 boxes in the hSHP promoter. (A) Wild-type or mutated E-box probe sequences (–342 to –312 bp from the hSHP promoter) used in gel mobility shift assays. Mutated nucleotides (CACCTG to agCCTG) in mE6, mE7 and mE6,7 are indicated in lower case. (B) Purified His-E47 protein was incubated with end-labeled wild-type (WT) or mutated E-box (mE6, mE7 and mE6,7) probes as indicated. (C) Cold competition assay. Purified His-E47 protein (lanes 2–12) was incubated with end-labeled wild-type probe as indicated. In the cold competition assay, black right-angled triangles indicate 50- to 100-fold molar excess of each unlabeled probe as indicated, and con indicates the E-box probe from RIPE3 (rat insulin promoter element 3).
Id family proteins repress the E47-dependent transactivation of the hSHP promoter
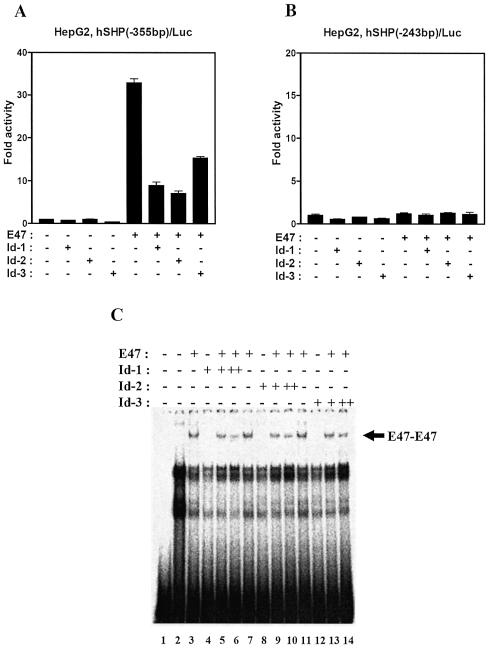
Id family proteins have been well documented to reduce E-box-mediated transactivation. Since Id proteins lack a DBD, dimerization of Id proteins with bHLH proteins results in the inhibition of DNA binding of bHLH proteins (30–34). To examine whether Id proteins can downregulate the hSHP promoter by repressing E47 activity, a transient transfection assay was performed with Id-1, Id-2, Id-3 and E47 expression vectors in HepG2 cells. As shown in Figure 6A, Id proteins inhibited E47-mediated transactivation of –355 bp hSHP reporter gene containing E6 and E7 E-boxes. However, we could not observe any specific inhibitory effect of Id proteins on the –243 bp reporter gene deleting E6 and E7 E-boxes (Fig. 6B). To investigate the effect of Id proteins on the DNA-binding activity of E47 to the hSHP promoter, a gel mobility shift assay was performed using the wild-type probe containing E6 and E7. As expected, a molar excess of Id proteins (Id-1, Id-2 and Id-3) reduced the E-box binding affinity of the E47 homodimer, suggesting that Id proteins repress E47-mediated hSHP promoter activity by inhibiting E47 binding to E-boxes within the hSHP promoter (Fig. 6C).
Figure 6.
Id proteins inhibit E47-mediated transactivation of the hSHP promoter. HepG2 cells were transfected with 200 ng of hSHP (–355 bp)/Luc (A) and hSHP (–243 bp)/Luc (B), and 200 ng of expression vectors for E47, Id-1, Id-2 and Id-3 as indicated. The luciferase activity was measured and normalized against β-galactosidase activity. (C) 32P-labeled DNA fragments containing E-boxes (E6,7) in the hSHP promoter were combined with in vitro transcribed and translated E47 and Id proteins as indicated. DNA–protein complexes were analyzed on a 4% polyacrylamide gel by autoradiography.
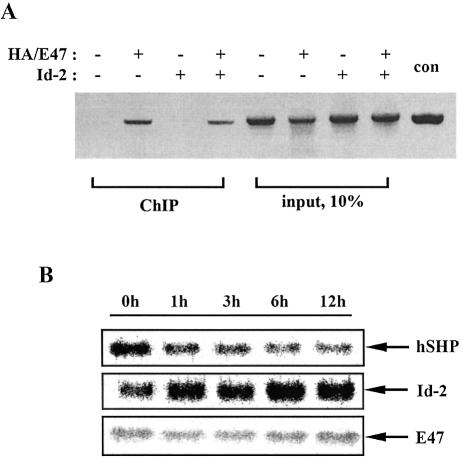
To further investigate whether E47 binds to the hSHP promoter and Id proteins inhibit its binding in vivo, we performed a ChIP assay using the PCR primers specific for the –474 to –120 bp region of the hSHP promoter. In HepG2 cells, expression vector encoding HA epitope-tagged E47 was transfected alone or together with expression vector for Id-2, and cell lysates were immunoprecipitated with the specific antibody against the HA epitope. As shown in Figure 7A, HA-E47 formed a specific complex with the hSHP promoter in vivo and this interaction was diminished when Id-2 was co-expressed with HA-E47. No bands were observed from cells transfected with either empty vector or Id-2 alone. These results demonstrated that E47 binds to the SHP promoter in vivo and Id-2 reduced the E47 binding affinity for the hSHP promoter (Fig. 7A). Previously, it has been reported that Id gene expression is induced by serum stimulation and Id proteins are involved in cell cycle progression (42). Given that Id-2 can interfere with E47-mediated activation of the hSHP promoter, we surmized that inducing expression of endogenous Id-2 might reduce the expression of hSHP mRNA. To this end, total RNA was isolated from HepG2 cells at 0, 1, 3, 6 and 12 h after 20% serum stimulation, and northern blot analysis was performed using the cDNA of SHP, Id-2 and E47 as a probe. As shown in Figure 7B, the SHP mRNA level was reduced while Id-2 gene expression was increased by serum stimulation in a time-dependent manner. However, we could not detect any significant difference in E47 mRNA level by serum stimulation. This inverse relationship might be explained at least partly by the activation of SHP expression by E2A proteins, whose activities are inhibited by Id proteins, although SHP gene expression can be affected by serum stimulation through other factors. Taken together, these results suggest that Id proteins repress hSHP gene expression by interfering with E47 binding to E-boxes.
Figure 7.
Id-2 downregulates hSHP gene expression via repression of E47 DNA binding in vivo. (A) ChIP assay. HepG2 cells were transfected with HA/E47 and Id-2 expression vectors. Soluble chromatin from these cells was prepared and immunoprecipitated with monoclonal antibody against HA as described previously (40). The final DNA extractions were amplified using pairs of primers that cover the E47 response region of the hSHP promoter (forward primer, 5′-AGGTTAGGCAAACAA-3′; reverse primer, 5′-CCCCTGGCAGGAATG-3′). (B) Northern blot analysis of SHP, Id-2 and E47 after serum stimulation in HepG2 cells. Total RNA was isolated from HepG2 cells at 0, 1, 3, 6 and 12 h after 20% serum stimulation. An RNA blot containing 20 µg of total RNA was hybridized with 32P-labeled probes for SHP, Id-2 and E47 as indicated.
Synergistic effects of SF-1 on the E47-medated transactivation of the hSHP promoter
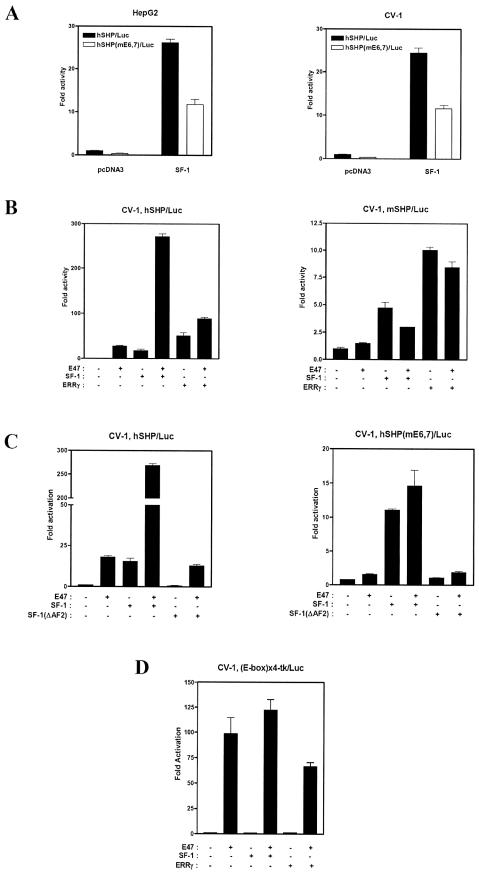
Based on the ChIP assay results showing that over-expressed E47 can bind to the hSHP promoter in vivo, we investigated whether endogenous E47 transactivates the hSHP promoter by comparing the basal promoter activities of wild-type hSHP promoter and mutant mE6,7/Luc reporter genes in HepG2 and CV-1 cells. As shown in Figure 8A, the basal promoter activity of mE6,7/Luc was drastically reduced to 35% of wild-type hSHP promoter activity, indicating that endogenous E47 may activate the hSHP promoter. It has been reported that the SHP promoter is regulated by several orphan nuclear receptors such as SF-1, FXR, LXR, LRH-1, ERRγ and HNF4 (11,16–18,43). In addition, we found that SF-1-mediated transactivation of the hSHP promoter was also significantly decreased by E6 and E7 E-box mutation, suggesting that E47 might have a significant effect on the SF-1-dependent activation of the hSHP promoter (Fig. 8A).
Figure 8.
E47-mediated transactivation of the hSHP promoter is enhanced synergistically by SF-1 in CV-1 cells. (A) HepG2 and CV-1 cells were transfected with 200 ng of wild-type hSHP promoter reporter (–2.2 kb/Luc) or mE6,7/Luc together with pcDNA3 or SF-1 expression vectors. (B) CV-1 cells were co-transfected with 200 ng of human or mouse SHP promoter reporter (–2.2 kb) and 100 ng of expression vectors for E47 alone or together with SF-1 and ERRγ as indicated. (C) Human wild-type or the E6,7 E-box mutant of the SHP promoter reporter (–2.2 kb) were co-transfected with 100 ng of expression vectors for E47, SF-1 wild type and AF2 deletion mutant (SF-1ΔAF2) as indicated. (D) (523)4 Reporter plasmids that contain multiple E-boxes upstream of the luciferase gene were co-transfected with 100 ng of expression vectors for E47, SF-1 and ERRγ as indicated. Two days after transfection, cells were harvested for luciferase and β-galactosidase assays. The representative results were expressed as fold activation (n-fold) over the value obtained with vector alone, with the error bars as indicated.
To further investigate whether E47 cross-talks with the transcription factors that regulate the SHP promoter, a transient transfection assay was performed with expression vectors for FXR, LXR, SF-1 and ERRγ together with E47 expression vectors in CV-1 cells. As shown in Figure 8B, E47-mediated transactivation of human but not mouse SHP promoter was enhanced synergistically by SF-1 and additively by ERRγ. However, FXR, LXR and LRH-1 had no significant effects on the E47-mediated transactivation (data not shown). Interestingly, cooperative activation of the hSHP promoter by SF-1 and E47 was completely abolished, although SF-1 activated the mE6,7/Luc reporter gene with E47-responsive E-box mutations within the hSHP promoter. In addition, SF-1(ΔAF2), an AF2 deletion mutant, neither activated nor synergistically augmented E47-mediated transactivation of the hSHP promoter, suggesting that the AF2 region of SF-1 is indispensable for both transactivation and synergistic activation with E47 of the hSHP promoter (Fig. 8C). To further investigate whether SF-1 binding to SF-1RE is required for the synergistic effect of E47 and SF-1 on the hSHP promoter, CV-1 cells were co-transfected with (523)4 luciferase reporter plasmids and expression vectors for E47, SF-1 and ERRγ. The (523)4 luciferase reporter contains multiple E-boxes upstream of the luciferase gene and is specific for active homodimers of E47. The result showed that SF-1 and ERRγ had no significant effects on the E47-mediated transactivation on the four-copy E-box luciferase reporter, indicating that SF-1 binding is essential for the synergy with E47 (Fig. 8D). Overall, these results demonstrated that binding to their responsive elements and the AF-2 domain of SF-1 are necessary for synergistic transactivation of the hSHP promoter by E47 and SF-1.
DISCUSSION
Recent reports have identified several nuclear receptors, such as SF-1, LRH-1, HNF4, FXR, ERRγ and LXRα, as potent regulators of SHP gene expression (11,16–18,43). However, it has not been fully characterized whether other families of transcription factors regulate SHP gene transcription. In this report, we have demonstrated that E2A gene products (E47, E12 and E2/5) are novel regulators of hSHP gene expression. Interestingly, E2A gene products regulate only the human, but not the mouse SHP promoter, and two E-boxes (E6 and E7) present only in the hSHP promoter are identified as E47-responsive E-boxes. Furthermore, Id proteins downregulate SHP gene expression in vivo by inhibiting E47 binding to the SHP promoter. This study may provide an important insight into SHP gene regulation and further into the cross-talk mechanisms of nuclear receptor function with bHLH and Id proteins.
Transcription factors belonging to the bHLH family are known to regulate a variety of developmental programs, including myogenesis, neurogenesis, sex determination and hematopoiesis (27–31). Many of these processes are controlled by E protein homodimer or heterodimer composed of E proteins and tissue-specific bHLH factors such as BETA2, MyoD and myogenin. In this report, we have provided potential evidence that the hSHP promoter may be activated by E2A protein homodimers. First, we have shown that over-expression of E2A proteins alone is sufficient to transactivate the hSHP promoter in a couple of cell lines such as CV-1, HepG2 and HeLa cells, which originate from different tissues, i.e. kidney, liver and epithelium, respectively. We also observed that class II bHLH proteins, such as BETA2, myogenin and MyoD, inhibited the E2A protein-mediated transactivation of the hSHP promoter. These results suggest that tissue-specific class II bHLH proteins may not be required for this activation. In addition, it is reported that CK II acts as a positive regulator of myogenesis and a negative regulator of E protein homodimers by preventing E protein homodimers from binding to E-box elements (41). In good agreement with a previous report, we demonstrated that CK II downregulated E47 transactivity on the hSHP promoter, indicating that E47 homodimers may be involved in transactivation of the hSHP promoter (Fig. 5C). Overall, this evidence suggests that E2A proteins alone are sufficient and necessary for the transactivation of the hSHP gene, although it is still possible that other tissue-restricted bHLH proteins may be involved in this activation.
It has been previously reported that ligands for nuclear receptor LXRα activate rat CYP7A gene expression via direct binding to LXRE on the rat CYP7A promoter. However, the LXRα-binding site is not conserved in the human CYP7A promoter and instead LXRα effectively represses the human CYP7A gene (44). Recently, it also has been demonstrated that the repression of human CYP7A gene expression by LXRα is due to LXRα-mediated human, but not rat, SHP gene induction, which is a negative regulator of CYP7A gene expression, suggesting that LXRα activates hSHP gene transcription by direct binding to LXRE that is not conserved in the rat SHP promoter (43). In this study, we showed that E2A gene products regulate human, but not mouse SHP gene expression via two E-boxes (E6 and E7) unique to the human gene promoter. These results suggest that E2A proteins may regulate SHP gene expression differentially among different species and, in turn, species-specific SHP expression may lead to its distinct functions in various species in an E2A protein-dependent manner. In addition, Id proteins downregulate the E protein-mediated transactivation of the human but not the mouse SHP gene. Therefore, we postulate that E2A proteins may indirectly regulate bile acid production, and bHLH proteins and Id proteins can be related to the function of nuclear receptors in vivo. It has been demonstrated that Id family proteins repress the transcriptional activity of bHLH proteins by blocking their binding to E-boxes (34–38). Therefore, we speculated that E47-mediated transactivation of the hSHP promoter might be reduced by Id proteins. Expectedly, Id-1, Id-2 and Id-3 repressed the E47-mediated transactivity of hSHP, but not mSHP, and reduced E47 binding to the hSHP promoter in vivo and in vitro (Figs 6 and 7). In addition, we demonstrated that SHP mRNA levels were decreased under conditions where Id-2 mRNA levels were increased by serum stimulation, although the E47 expression level showed no significant difference according to serum stimulation. These results suggested that Id proteins downregulate hSHP gene expression by repressing the E2A proteins. The notion that the ratio of E2A proteins to class II bHLH and Id proteins is a key regulator of hSHP gene expression is supported by the evidence that Id proteins and E2A heterodimer partners, such as class II bHLH proteins, are induced under specific physiological conditions or in a tissue-specific manner (25–28,34–38).
The current study demonstrates that the SHP promoter is regulated by SF-1 and ERRγ (11,19). In addition, it has been reported that SF-1 regulates transcription of the FSHR gene via a mechanism that involves USF1 and USF2 binding to the proximal E-box element (45). Herein we demonstrated that E47 and SF-1 stimulate cooperatively the human, but not the mouse SHP promoter, supported by the results showing that mutations of the E6 and E7 E-boxes found only in the human promoter eliminated the synergistic effect by E47 and SF-1 (Fig. 8A and B). Interestingly, DNA binding by both E47 and SF-1 within the hSHP promoter is indispensable for the cooperative transactivation of the hSHP promoter. We also demonstrated the requirement for the AF2 region of SF-1 for both transactivation by SF-1 itself and synergistic activation with E47 of the hSHP promoter. This synergistic activation can be mediated by several mechanisms: (i) physical interaction between E47 and SF-1; (ii) recruitment of cofactors; and (iii) enhancement of their DNA binding. We observed that E47 directly interacted with SF-1 and that the A/B-DBDs of SF-1 are involved in the interaction with E47 in a GST pull-down assay (data not shown), suggesting that the synergistic activation can be mediated by direct interaction between them. However, the mechanism by which SF-1 and E47 cooperatively activate the hSHP promoter remains to be elucidated. The current results suggest that E47 and synergy with SF-1 could have important effects on a variety of nuclear receptor signaling pathways as a consequence of their effects on SHP.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Roland Stein, Michael P. Czech, John D. Norton and Ming-Jer Tsai for kind gifts of the plasmids, and Drs David D. Moore, Pyeung-Hyeun Kim and Kang-Yul Yoo for helpful advice and discussions and Dr Ming-Jer Tsai for critical reading. This work was supported by Korea Research Foundation Grant KRF-2000-015-DS0037 and in part by Hormone Research Center Grant HRC2001G0201.
REFERENCES
- 1.Mangelsdorf D.J. and Evans,R.M. (1995) The RXR heterodimers and orphan receptors. Cell, 83, 841–850. [DOI] [PubMed] [Google Scholar]
- 2.Evans R.M. (1988) The steroid and thyroid hormone receptor superfamily. Science, 240, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giguere V. (1999) Orphan nuclear receptors: from gene to function. Endocr. Rev., 20, 689–725. [DOI] [PubMed] [Google Scholar]
- 4.Seol W., Choi,H.S. and Moore,D.D. (1996) An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science, 272, 1336–1339. [DOI] [PubMed] [Google Scholar]
- 5.Seol W., Chung,M. and Moore,D.D. (1997) Novel receptor interaction and repression domains in the orphan receptor SHP. Mol. Cell. Biol., 17, 7126–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seol W., Hanstein,B., Brown,M. and Moore,D.D. (1998) Inhibition of estrogen receptor action by the orphan receptor SHP (short heterodimer partner). Mol. Endocrinol., 10, 1551–1557. [DOI] [PubMed] [Google Scholar]
- 7.Johansson L., Thomsen,J.S., Damdimopoulos,A.E., Spyrou,G., Gustafsson,J.A. and Treuter,E. (1999) The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERα and ERβ. J. Biol. Chem., 274, 345–353. [DOI] [PubMed] [Google Scholar]
- 8.Lee Y.K., Dell,H., Dowhan,D.H., Hadzopoulou-Cladaras,M. and Moore,D.D. (2000) The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol. Cell. Biol., 20, 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson L., Bavner,A., Thomsen,J.S., Farnegardh,M., Gustafsson,J.A. and Treuter,E. (2000) The orphan nuclear receptor SHP utilizes conserved LXXLL-related motifs for interactions with ligand-activated estrogen receptors. Mol. Cell. Biol., 20, 1124–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gobinet J., Auzou,G., Nicolas,J.C., Sultan,C. and Jalaguier,S. (2001) Characterization of the interaction between androgen receptor and a new transcriptional inhibitor, SHP. Biochemistry, 40, 15369–15377. [DOI] [PubMed] [Google Scholar]
- 11.Sanyal S., Kim,J.Y., Kim,H.J., Takeda,J., Lee,Y.K., Moore,D.D. and Choi,H.S. (2002) Differential regulation of the orphan nuclear receptor small heterodimer partner (SHP) gene promoter by orphan nuclear receptor ERR isoforms. J. Biol. Chem., 277, 1739–1748. [DOI] [PubMed] [Google Scholar]
- 12.Brendel C., Schoonjans,K., Botrugno,O.A., Treuter.E. and Auwerx,J. (2002) The small heterodimer partner interacts with the liver X receptor alpha and represses its transcriptional activity. Mol. Endocrinol., 16, 2065–2076. [DOI] [PubMed] [Google Scholar]
- 13.Nishizawa H., Yamagata,K., Shimomura,I., Takahashi,M., Kuriyama,H., Kishida,K., Hotta,K., Nagaretani,H., Maeda,N., Matsuda,M., Kihara,S., Nakamura,T., Nishigori,H., Tomura,H., Moore,D.D., Takeda,J., Funahashi,T. and Matsuzawa,Y. (2002) Small heterodimer partner, an orphan nuclear receptor, augments peroxisome proliferator-activated receptor-γ transactivation. J. Biol. Chem., 277, 1586–1592. [DOI] [PubMed] [Google Scholar]
- 14.Nishigori H., Tomura,H., Tonooka,N., Kanamori,M., Yamada,S., Sho,K., Inoue,I., Kikuchi,N., Onigata,K., Kojima,I., Kohama,T., Yamagata,K.,Yang,Q., Matsuzawa,Y., Miki,T., Seino,S., Kim,M.Y., Choi,H.S., Lee,Y.K., Moore,D.D. and Takeda,J. (2001) Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc. Natl Acad. Sci. USA, 98, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H.K., Lee,Y.K., Park,S.H., Kim,Y.S., Park,S.H., Lee,J.W., Kwon,H.B., Soh,J., Moore,D.D. and Choi,H.S. (1998) Structure and expression of the orphan nuclear receptor SHP gene. J. Biol. Chem., 273, 14398–14440. [DOI] [PubMed] [Google Scholar]
- 16.Goodwin B., Jones,S.A., Price,R.R., Watson,M.A., McKee,D.D., Moore,L.B., Galardi,C., Wilson,J.G., Lewis,M.C., Roth,M.E., Maloney,P.R., Willson,T.M. and Kliewer,S.A. (2000) A regulatory cascade of the nuclear receptors FXR, SHP-1 and LRH-1 represses bile acid biosynthesis. Mol. Cell, 6, 517–526. [DOI] [PubMed] [Google Scholar]
- 17.Sinal C.J., Tohkin,M., Miyata,M., Ward,J.M., Lambert,G. and Gonzalez,F.J. (2000) Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell, 102, 731–744. [DOI] [PubMed] [Google Scholar]
- 18.Lu T.T., Makishima,M., Repa,J.J., Schoonjans,K., Kerr,T.A., Auwerx,J. and Mangelsdorf,D.J. (2000) Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell, 6, 507–515. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y.K., Parker,K.L., Choi,H.S. and Moore,D.D. (1999) Activation of the promoter of the orphan receptor SHP by orphan receptors that bind DNA as monomers. J. Biol. Chem., 274, 20869–20873. [DOI] [PubMed] [Google Scholar]
- 20.Shih D.Q., Screenan,S., Munoz,K.N., Philipson,L., Pontoglio,M., Yaniv,M., Polonsky,K.S. and Stoffel,M. (2001) Loss of HNF-1α function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes, 50, 2472–2480. [DOI] [PubMed] [Google Scholar]
- 21.Kerr T.A., Saeki,S., Schneider,M., Schaefer,K., Berdy,S., Redder,T., Shan,B., Russell,D.W. and Schwarz,M. (2002) Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell, 2, 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L., Lee,Y.K., Bundman,D., Han,Y., Thevananther,S., Kim,C.S., Chua,S.S., Wei,P., Heyman,R.A., Karin,M. and Moore,D.D. (2002) Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell, 2, 721–731. [DOI] [PubMed] [Google Scholar]
- 23.Massari M.E. and Murre,C. (2000) Helix–loop–helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol., 20, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murre C., Bain,G., Dijk,M.A., Engel,V.I., Furnari,B.A., Massari,M.E., Matthews,J.R., Quong,M.W., Rivera,R.R. and Stuiver,M.H. (1994) Structure and function of helix–loop–helix proteins. Biochim. Biophys Acta, 1218, 129–135. [DOI] [PubMed] [Google Scholar]
- 25.Murre C. and Baltimore,D. (1992) The helix–loop–helix motif: structure and function, In McKnight,S.L. and Yamamoto,K.R. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 861–879. [Google Scholar]
- 26.Mutoh H., Fung,B.P., Naya,F., Tsai,M.J., Nishitani,J. and Leiter,A.B. (1997) The basic helix–loop–helix transcription factor BETA2/NeuroD is expressed in mammalian enteroendocrine cells and activates secretin gene expression. Proc. Natl Acad. Sci. USA, 94, 3560–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naya F.J., Huang,H.P., Qiu,Y., Mouth,H., DeMayo,F.J., Leiter,A.B. and Tsai,M.J. (1997) Diabetes, defective pancreatic morphogenesis and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev., 11, 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naya F.J., Stellrecht,C.M.M. and Tsai,M.J. (1995) Tissue-specific regulation of the insulin gene by a novel basic helix–loop–helix transcription factor. Genes Dev., 9, 1009–1019. [DOI] [PubMed] [Google Scholar]
- 29.Bain G., Maandag,E.C., Izon,D.J., Amsen,D., Kruisbeek,A.M., Weintraub,B.C., Krop,I., Schlissel,M.S., Feeney,A.J., van Roon,M., van der Valk,M., te Riele,H.P.J., Berns,A. and Murre,C. (1994) E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell, 79, 885–892. [DOI] [PubMed] [Google Scholar]
- 30.Bain G., Robanus,M.E., te Riele,H.P.J., Feeney,A.J., Sheehy,A., Schlissel,M., Shinton,S.A., Hardy,R.R. and Murre,C. (1997) Both E12 and E47 allow commitment to the B cell lineage. Immunity, 6, 145–154. [DOI] [PubMed] [Google Scholar]
- 31.Herblot S., Aplan,P.D. and Hoang,T. (2002) Gradient of E2A activity in B-cell development. Mol. Cell. Biol., 22, 886–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikkers H., Allen,J. and Berns,A. (2002) Proviral activation of the tumor suppressor E2a contributes to T cell lymphomagenesis in EmuMyc transgenic mice. Oncogene, 21, 6559–6566. [DOI] [PubMed] [Google Scholar]
- 33.Littlewood T.D. and Evan,G.J. (1994) Transcription factors 2: helix–loop–helix. Protein Profile, 1, 639–709. [PubMed] [Google Scholar]
- 34.Biggs J., Murphy,E.B. and Israel,M.A. (1992) A human Id-like helix–loop–helix protein expressed during early development. Proc. Natl Acad. Sci. USA, 89, 2512–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy J.J. and Norton,J.D. (1990) Cell-type-specific early response gene expression during plasmacytoid differentiation of human B lymphocytic leukemia cells. Biochim. Biophys Acta, 1049, 261–271. [DOI] [PubMed] [Google Scholar]
- 36.Deed R.W., Bianchi,S.M., Atherton,G.T., Johnston,D., Santibanez-Koref,M., Murphy,J.J. and Norton,J.D. (1993) An immediate early human gene encodes an Id-like helix–loop–helix protein and is regulated by protein kinase C activation in diverse cell types. Oncogene, 8, 599–607. [PubMed] [Google Scholar]
- 37.Hara E., Yamaguchi,T., Nojima,H., Ide,T., Campisi,J., Okayama,H. and Oda,K. (1994) Id-related genes encoding helix–loop–helix proteins are required for G1 progression and are repressed in senescent human fibroblasts. J. Biol. Chem., 269, 2139–2145. [PubMed] [Google Scholar]
- 38.Deed R.W., Jasiok,M. and Norton,J.D. (1998) Lymphoid-specific expression of the Id3 gene in hematopoietic cells. Selective antagonism of E2A basic helix–loop–helix protein associated with Id3-induced differentiation of erythroleukemia cells. J. Biol. Chem., 273, 8278–8286. [DOI] [PubMed] [Google Scholar]
- 39.Ohneda K., Mirmira,R.G., Wang,J.H., Johnson,J.D. and German,M.S. (2000) The homeodomain of PDX-1 mediates multiple protein–protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol. Cell. Biol., 20, 900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang Y., Hu,X., DiRenzo,J., Lazar,M.A. and Brown,M. (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- 41.Johnson S.E., Wang,X., Hardy,S., Taparowsky,E.J. and Konieczny,S.F. (1996) Casein kinase II increases the transcriptional activities of MRF4 and MyoD independently of their direct phosphorylation. Mol. Cell. Biol., 16, 1604–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barone M.V., Pepperkok,R., Peverali,F.A. and Philipson,L. (1994) Id proteins control growth induction in mammalian cells. Proc. Natl Acad. Sci. USA, 91, 4985–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodwin B., Watson,M.A., Kim,H., Miao,J., Kemper,J.K. and Kliewer,S.A. (2003) Differential regulation of rat and human CYP7A1 by the nuclear oxysterol receptor liver X receptor-α. Mol. Endocrinol., 17, 386–394. [DOI] [PubMed] [Google Scholar]
- 44.Chiang J.Y., Kimmel,R. and Stroup,D. (2001) Regulation of cholesterol 7α-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRα). Gene, 262, 257–265. [DOI] [PubMed] [Google Scholar]
- 45.Muschen M., Lee,S., Zhou,G., Feldhahn,N., Barath,V.S., Chen,J., Moers,C., Kronke,M., Rowley,J.D. and Wang,S.M. (2002) Activation of the rat follicle-stimulating hormone receptor promoter by steroidogenic factor 1 is blocked by protein kinase A and requires upstream stimulatory factor binding to a proximal E box element. Proc. Natl Acad. Sci. USA, 99, 10014–10019.12119411 [Google Scholar]