Abstract
Rationale: Amplification of distal 3q is the most common genomic aberration in squamous lung cancer (SQC). SQC develops in a multistage progression from normal bronchial epithelium through dysplasia to invasive disease. Identifying the key driver events in the early pathogenesis of SQC will facilitate the search for predictive molecular biomarkers and the identification of novel molecular targets for chemoprevention and therapeutic strategies. For technical reasons, previous attempts to analyze 3q amplification in preinvasive lesions have focused on small numbers of predetermined candidate loci rather than an unbiased survey of copy-number variation.
Objectives: To perform a detailed analysis of the 3q amplicon in bronchial dysplasia of different histological grades.
Methods: We use molecular copy-number counting (MCC) to analyze the structure of chromosome 3 in 19 preinvasive bronchial biopsy specimens from 15 patients and sequential biopsy specimens from 3 individuals.
Measurements and Main Results: We demonstrate that no low-grade lesions, but all high-grade lesions, have 3q amplification. None of seven low-grade lesions progressed clinically, whereas 8 of 10 patients with high-grade disease progressed to cancer. We identify a minimum commonly amplified region on chromosome 3 consisting of 17 genes, including 2 known oncogenes, SOX2 and PIK3CA. We confirm that both genes are amplified in all high-grade dysplastic lesions tested. We further demonstrate, in three individuals, that the clinical progression of high-grade preinvasive disease is associated with incremental amplification of SOX2, suggesting this promotes malignant progression.
Conclusions: These findings demonstrate progressive 3q amplification in the evolution of preinvasive SQC and implicate SOX2 as a key target of this dynamic process.
Keywords: bronchial dysplasia, squamous lung cancer, gene amplification, molecular copy-number counting, SOX2
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Amplification of 3q is extremely common in squamous lung cancer, suggesting the presence of a key driver oncogene within the amplified part of the genome. A number of candidate 3q oncogenes have been proposed, including SOX2. The amplicon structure has been difficult to study in detail in preinvasive disease because bronchial biopsies are generally small, heterogeneous, and fixed in formalin to preserve histological appearance.
What This Study Adds to the Field
This work demonstrates that a novel single-molecule digital polymerase chain reaction–based technique can be used to analyze regional amplicons in archived preinvasive bronchial biopsies. We show that 3q amplification effectively segregates high-grade dysplasia from low-grade dysplasia and identify SOX2 as the most likely focus of the amplicon. We also show that clinical progression is associated with progressive amplification of SOX2.
Squamous lung cancer (SQC) is believed to develop through a series of preinvasive stages before invasion of the basement membrane. A theoretical advantage of studying preinvasive lesions is that key early events in the pathogenesis of cancer may be identified, providing molecular biomarkers predictive of future invasive disease and novel targets for therapeutics. A separate goal is to gain an understanding of the natural history of carcinoma development in vivo rather than in model systems.
The cancer genome is characterized by multiple genomic rearrangements, including deletions, translocations, and amplification (1, 2), which may be driver events (i.e., involved in the pathogenesis of disease) or so-called passenger events that are present but do not contribute to the cancer phenotype. Regional amplification of driver oncogenes is well-described in many cancers, including lung adenocarcinoma (3, 4). In squamous lung cancer 3q amplification is one of the most common and significant genomic aberrations (5) and is a recurrent finding in head and neck (6), esophageal (7), cervical (8), and other cancers. Many groups have sought to define the driver oncogenes on 3q with a view to exploiting these as molecular biomarkers or novel therapeutic targets. Array-based studies of SQC have identified a broad region encompassing many hundreds of genes on 3q as a recurrent region of amplification (5). Many candidate genes have been identified, including TP73L (9), ECT2 (7), PIK3CA (10), DCUND1 (11), and SOX2 (12). There are functional data implicating the latter three genes (10–12), and a recent publication reported SOX2 amplification was present in 23% of SQCs and proposed it as a lineage-specific oncogene (12, 13). SOX2 is a nuclear transcription factor with pleiotropic roles in key biological pathways; as well as being implicated in oncogenesis (14), it has critical roles in lung embryogenesis (15) and in the reprogramming of adult somatic cells to pluripotency (16).
Fewer studies have examined the timing and role of regional amplification in the preinvasive development of cancer. This is because archives of preinvasive lesions are often limited and studying them is technically challenging. Successful reports have largely used fluorescence in situ hybridization (FISH) to define aneusomy (17) or to examine specific loci on 3q (9, 18–21). The results vary, but 3q amplification was detected in at least 27% of higher-grade lesions in one study assessing eight 3q loci (21), whereas others suggested a higher incidence. FISH is a useful technique, but may be limited by the subjective interpretation of results (22) as well as the inevitable focus on a small number of predetermined target loci.
An alternative is an array-based approach. Customized arrays have been successfully used by one group to examine 1p (23) and 5p (24) in high-grade dysplastic lesions from archived surgical resection specimens. However, in general, it is difficult to retrieve sufficient amounts or quality of DNA from very small heterogeneous formalin-fixed paraffin-embedded bronchial biopsies to reliably perform such studies without performing a whole-genome amplification step (25). This measure inevitably introduces bias into results (26). A technique that addresses these issues is molecular copy-number counting (MCC) (27) and a modified protocol microdissection-MCC (μMCC) (28). This is a digital polymerase chain reaction (PCR) approach that facilitates the accurate analysis of large numbers of loci (hundreds) using the small amounts of degraded DNA available from archived clinical biopsies. It means that an unbiased assessment of copy-number variation across a genomic region of interest can be undertaken to any resolution. In this study we describe the use of μMCC to perform the first high-resolution analysis of 3q amplification in preinvasive bronchial lesions.
Some of the results of these studies have been previously reported in the form of abstracts (29–31).
METHODS
Patients and Samples
All samples were from patients enrolled in the University College London Hospital Early Lung Cancer Project (32). This is a bronchoscopic surveillance study in which patients undergo repeated assessment under a protocol that includes autofluorescence bronchoscopy, computed tomography, and fluorodeoxyglucose–positron emission tomography scanning. Patients are enrolled on the basis of having a biopsy-proven dysplastic lesion of the bronchial tree. At the time of enrollment none of the patients have an active diagnosis of lung cancer, although they may have a prior history of lung cancer. Local Regional Ethical Committee approval was obtained (01/0148). The patients included in this report had undergone an average of 7.4 bronchoscopies (range 1–19) in the surveillance study up to May 2007. The analyzed biopsies were obtained over a period between 1998 and 2007. Research biopsies were taken during surveillance bronchoscopies and fixed immediately for 4 hours in a solution of 4% formaldehyde in phosphate-buffered saline.
Biopsies were chosen from the research archive on the basis of the grade of lesion recorded on the paired clinical biopsy. Seven biopsies with low-grade dysplasia (LGD; mild or moderate dysplasia) and 10 with high-grade dysplasia (HGD; severe dysplasia or carcinoma in situ) were selected. Sections were then taken from the corresponding research biopsy. A team of three consultant pathologists, including the reference thoracic pathologist, read the clinical biopsies. The corresponding research biopsies were read “blind” by the reference thoracic pathologist (M.R.F.). In all except three lesions the paired clinical and research biopsies were read as the same grade, and in the three discordant readings the opinion of the reference thoracic pathologist was accepted. Further demographic and biopsy-related details are in Table 1.
TABLE 1.
DETAILS OF LESIONS ANALYZED AND CORRESPONDING STUDY PATIENTS
| Lesion Number | Patient Code | Sex | Age at Enrollment, yr | Pack-years* | Smoking Status at Enrollment in Study | Prior Lung Cancer | Duration Follow-up | Contemporaneous or Subsequent Lung Cancer | Same or Remote Anatomical Site | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| LG1 | 006† | M | 60 | 141 | Ex | N | 18 | No | N/A | Alive, free from lung cancer† |
| LG2 | 008 | M | 61 | 96 | Ex | N | 18 | No | N/A | Alive, free from lung cancer |
| LG3 | 019 | M | 70 | 60 | Current | N | 46 | No | N/A | Alive, free from lung cancer |
| LG4 | 020 | M | 53 | 45 | Current | N | 39 | No | N/A | Alive, free from lung cancer |
| LG5 | 064 | M | 52 | 30 | Current | N | 49 | No | N/A | Alive, free from lung cancer |
| LG6,7‡ | 073 | M | 53 | 33 | Ex | N | 25 | No | N/A | Alive, free from lung cancer |
| HG1 | 002 | F | 71 | 100 | Ex | N | 66 | Yes | Same | “Curative” surgical resection at month 13. Disease free for >5 yr post resection. Died year 9 from lung cancer |
| HG2 | 006† | M | 60 | 141 | Ex | N | 53 | Yes | Same | †Surgical resection. Alive, free from lung cancer |
| HG3 | 076 | M | 58 | NR | Ex | N | 15 | Yes | Same and remote | Recent diagnosis (2009) lung cancer |
| HG4 | 012 | M | 65 | 60 | Ex | Y | 46 | No | N/A | Alive, free from lung cancer |
| HG5 | 017 | M | 66 | NR | Ex | Y | 17 | Yes | Remote | Died, complications of treatment for lung cancer (PDT) |
| HG6 | 024 | M | 73 | 96 | Ex | Y | 17 | Yes, contemporary | Remote | PDT to cancer and high-grade lesion. Alive, free from lung cancer |
| HG7 | 026 | M | 74 | 40 | Ex | Y | 42 | Yes | Remote | Dead, lung cancer |
| HG8 | 056 | M | 70 | 65 | Ex | N | 48 | Yes | Not known | Dead, lung cancer |
| HG9 | 060 | M | 69 | 26 | Ex | N | 51 | Yes, contemporary | Adjacent | Treated with PDT. Alive, recurrent disease treated endobronchially |
| HG10 | 061 | M | 78 | 80 | Ex | Y | 9 | No | N/A | Unknown |
Definition of abbreviations: N/A = not applicable; NR = not recorded; PDT = photodynamic therapy.
Pack-year: 20 cigarettes per day for 1 year.
Patient 006 had a left upper lobe lesion (HG2) and subsequently developed a squamous carcinoma of the left upper lobe. LG1 is a biopsy taken from a lesion subsequently detected at a surveillance bronchoscopy in the contralateral lung (right lower lobe).
Patient had multifocal low-grade disease. Lesion LG7 was read as squamous metaplasia by the reference thoracic pathologist.
We obtained DNA from laser-capture microdissected dysplastic epithelium and from peripheral blood as described. For some experiments, pooled normal DNA was ultrasonicated to a median fragment size of 600 bp to mimic the effect of formalin fixation.
μMMC
This method was recently described in detail (28). In brief, test DNA is diluted and dispensed across a microtiter plate to less than a haploid genome per aliquot. Each aliquot is tested for the presence or absence of a target sequence in a two-phase hemi-nested digital PCR assay. The relative copy number of individual markers is derived by comparing the number of aliquots positive for each marker. In this study data were normalized to the mean value of three to five reference markers from regions of the genome previously shown to generally be at normal copy in SQC (28). Primer design was as previously described (28). All primers were supplied by Operon GmBH (Germany) or Sigma (Dorset, UK). The genomic position of markers was taken from the Ensembl database, National Center for Biotechnology Information reference human genome sequence release 36 – NCBI36 (www.ensembl.org). All oligonucleotide sequences are freely available on request.
FISH
FISH was performed on metaphase spreads and tissue sections. For metaphase spreads a standard protocol was followed to confirm Bacterial Artificial Chromosome BAC position. For paraffin-embedded archived biopsy specimens, 3-μm sections were freshly cut onto polylysine-coated microscope slides (VWR, Lutterworth, UK) and heated overnight at 58°C. Previously described pretreatment and hybridization protocols were followed (33, 34). Slides were visualized with a Nikon E800 microscope mounted with a 100 W mercury lamp light source. Composite raw images were pseudocolored and enhanced using CytoVision software (Genetix, New Milton, UK). Images presented were exported from CytoVision and processed with Adobe Photoshop and Adobe Illustrator.
Immunohistochemistry
Immunohistochemistry was performed using antibodies to human SOX2 (R&D Systems, Minneapolois, USA clone 245610) and PI3Kα (Sigma; catalog number HPA009985). Sections (2 μm) were cut onto polylysine-coated slides and incubated overnight at room temperature. Sections were dewaxed, and antigen retrieval was performed by microwave treatment in citrate buffer. Antigen detection was performed using the primary antibodies (both 1:200), biotinylated secondary antibodies, and streptavidin-horse radish peroxidase (DAKO, Glostrup, Denmark)/3,3′-diaminobenzidine (Vector Labs, Burlingame, USA). Slides were counterstained with hematoxylin.
Statistical Methods
P values are two-sided t tests.
RESULTS
Seven low-grade (including 1 squamous metaplasia) and 10 high-grade bronchial lesions were assessed (Table 1) and a comparison made of sequence copy-number in the two groups of lesions, of which representative examples are shown (Figure 1). For each LGD, there was no difference in copy number between 3p and 3q. However, in each case, the high-grade lesions had amplification of 3q relative to both 3p and reference autosomal markers. Importantly, as this is a longitudinal bronchoscopic surveillance study, the histological and clinical outcome for each patient and/or lesion is known and can be compared with the genomic signature (Table 1). In this series, none of the low-grade lesions progressed, and none of those individuals has subsequently gone on to develop lung cancer. Of 10 individuals with HGD and 3q amplification, 2 had contemporary invasive cancer, and 6 others later developed cancer. Of the eight patients who developed cancer, four had tumors detected at sites remote from the HGD analyzed, an observation consistent with “field cancerization.” One of the patients, Patient 006, had a low-grade lesion diagnosed at surveillance bronchoscopy after having a HGD and subsequent cancer diagnosed and resected from the contralateral lung. He has not had a recurrence of cancer since his low-grade lesion was diagnosed.
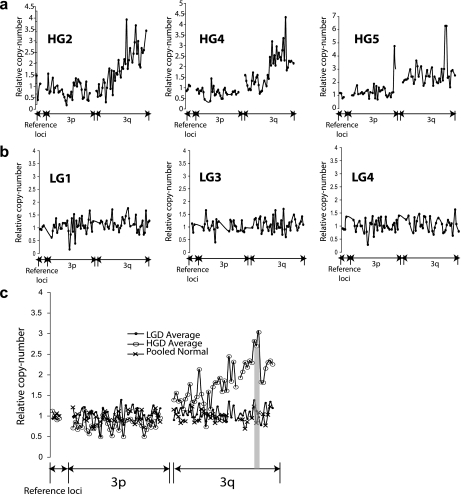
Figure 1.
Comparison of chromosome 3 profile in high-grade (HG) and low-grade (LG) bronchial dysplasia. Markers are located at approximately 2.2-Mb intervals along chromosome 3 and have previously been validated (28). Results are normalized to the average of three to four reference autosomal loci (see Methods). Representative results are shown for (A) three HG lesions or (B) three LG lesions. The HG dysplasias (HGDs) show marked amplification of 3q relative to 3p when compared with the same markers tested against low-grade dysplasias (LGDs). This was true for each LG and HG lesion assessed. (C) The average result for each marker is plotted comparing HGDs (n = 10) with LGDs (n = 7) and pooled normal DNA from peripheral blood leukocytes. Iterative microdissection molecular copy-number counting experiments (see Figure 2) identified a minimal common amplified region shaded in gray, which corresponded to the region of peak amplification.
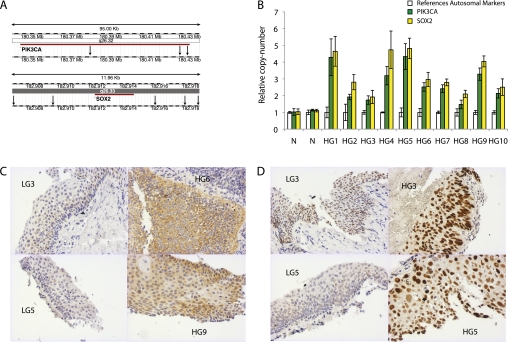
The 3q amplicon had a different structure in each HGD lesion analyzed (Figure 1). The minimum commonly amplified region (MCAR) across the cohort of HGDs was defined by using μMCC to iteratively resolve amplicon borders so that genes lying within the MCAR could be identified (Figure 2). The MCAR spanned approximately 4.3 Mb and encompassed 17 genes, 17 noncoding RNAs, and 6 pseudogenes (see Table E1 in the online supplement). It corresponded well to the region of peak amplification across the same cohort of high-grade lesions (Figure 1). Of note, previously suggested candidate 3q oncogenes, including TP73L (18), DCUND1/SCCRO (11), and ECT2 (7) did not lie within the MCAR. Using online databases and previously published reports, PIK3CA and SOX2 were identified as the most likely driver oncogenes within the MCAR. The fact that the regional amplification encompassed PIK3CA and SOX2 suggests, but does not specifically confirm, amplification of specific loci. Therefore, new μMCC markers were designed to specifically test the relative copy number of these genes, and increased copy of both PIK3CA and SOX2 was confirmed in each HGD (Figures 3A and 3B). Immunohistochemical analysis of the same low- and high-grade lesions was performed for both PI3Kα and SOX2 expression and demonstrated differential expression consistent with the genomic changes noted (Figures 3C and 3D).
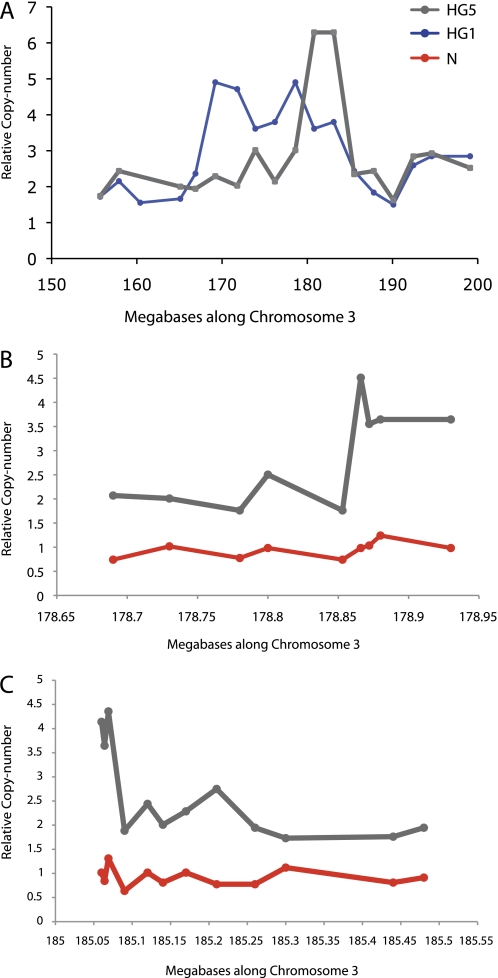
Figure 2.
Identifying the minimal commonly amplified region (MCAR). (A) Two lesions, high-grade (HG)1 and HG5, were informative for defining the MCAR. The markers from the original chromosome survey were at approximately 2-Mb intervals. Examples of the iterative higher-resolution analysis of HG5 are shown. This was performed to better define the (B) proximal and (C) distal amplicon boundaries and so identify genes lying within the amplicon. For HG5 the boundary was around 178.85 Mb and the distal boundary at around 185.05 Mb. Similar experiments on HG1 revealed the distal boundary for this lesion to be around 183.2 Mb. For this cohort of lesions, therefore, the MCAR lay between 178.85 and 183.2 Mb. In (B) and (C) microdissection molecular copy-number counting results on control DNA from peripheral blood from the same patients is used to demonstrate the somatic nature of the amplicon in the microdissected dysplastic tissue.
Figure 3.
(A) Multiple markers were designed to interrogate the copy number of two genes, PIK3CA (n = 3) and SOX2 (n = 5); the markers' location is indicated by black arrows in a modified screenshot from the Ensembl database. (B) The copy number of PIK3CA and SOX2 was analyzed for each high-grade dysplasia (HGD) and pooled normal genomic DNA (N). Results from individual experiments show the mean and standard deviation of multiple markers for each locus (reference autosomal, [n = 3–5]; PIK3CA and SOX2). (C) Immunohistochemistry for PI3Kα on low-grade dysplasia (LGD) and HGD biopsies. In seven of nine biopsies there was strong cytoplasmic staining in HGD compared with weak staining in five of seven LGD lesions. (D) Nuclear expression of SOX2 was increased in a number of high-grade lesions relative to low-grade lesions. However, for a number of lesions it was not possible to discriminate LGD and HGD on the basis of SOX2 immunostaining.
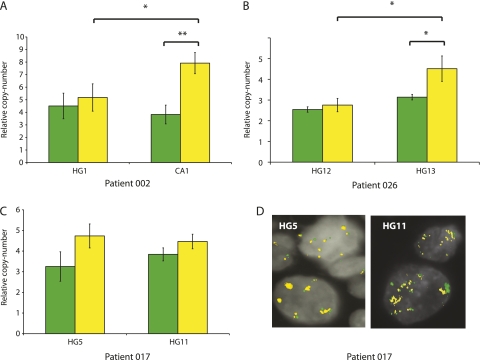
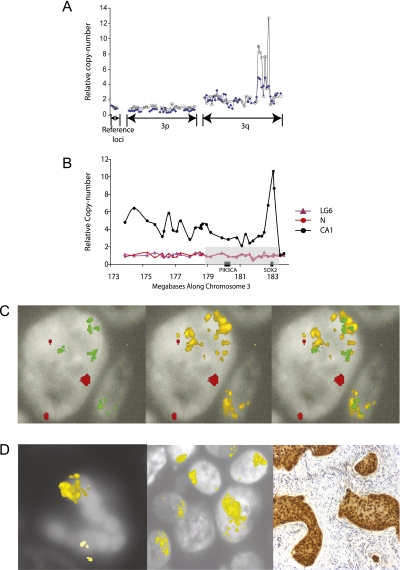
The samples derive from a longitudinal clinical study; therefore, for some participants serial bronchial biopsies are available over a follow-up period. The combination of this rare resource and the MCC technique provided a novel opportunity to precisely define changes in the amplitude of copy-number gain at specific loci in sequential biopsies from the same individual. A comparison of PIK3CA and SOX2 copy number was therefore performed on temporally separate biopsies from the same anatomic location in two cases (002 and 026), and in a third (017) from a left upper lobe biopsy and a later tracheal biopsy (Figure 4). Patient 002 was previously reported in a comparative genomic hybridization analysis (35); further details of their clinical histories are available in Figure E1. The results showed that the relative copy number of SOX2, but not PIK3CA, increased between biopsies in two of the three pairs of lesions. In the third case (Patient 017), although the relative copy number of both SOX2 and PIK3CA did not significantly increase between the first and second biopsy, FISH analysis revealed an increase in the number of signals per nucleus in the later biopsy. μMCC measures relative copy number of analyzed loci, whereas FISH estimates the absolute number of copies per nucleus. One explanation for these results is that the relative copy number remained static but there was an increase in ploidy between the two lesions leading to an increase in absolute copies of PIK3CA and SOX2. The results from Patient 002 (Figure 4) suggested a dramatic preferential amplification of SOX2 relative to PIK3CA in the progression from HGD to cancer. This was corroborated by comparing the low-resolution chromosome 3 data from the high-grade lesion and a subsequent cancer (002-CA1) (Figure 5A). The chromosome 3 profile of the cancer resection specimen (002-CA1) was previously used to demonstrate the ability of the μMCC technique to analyze regional genome structure using picogram quantities of degraded DNA (28). A subsequent high-resolution μMCC scan across the 3q amplicon in 002-CA1 demonstrated an intraamplicon subpeak of super-amplification that spanned up to 1 Mb (Figure 5B) but contained only a single gene, SOX2. This result was confirmed by FISH data from the same lesion, which demonstrates paired amplification of PIK3CA and SOX2, as would be expected from loci that are only 2.6 Mb apart, but also discrete further amplification of SOX2. In addition to the SOX2 amplification, immunohistochemistry revealed a high nuclear expression of SOX2 in cancer cells compared with surrounding stroma (Figures 5C and 5D).
Figure 4.
PIK3CA and SOX2 copy number was analyzed as before, this time in sequential biopsies from three patients (002, 017, 026). (A–C) Results represent the mean ± SD of duplicate experiments for the two genes and three to five reference autosomal markers. In the case of Patient 017 (C) there was no significant increase in relative copy number in the later biopsy; however (D) there was an absolute gain in copy of both loci (PIK3CA = green; SOX2 = yellow) on the basis of fluorescence in situ hybridization experiments on the same biopsies.
Figure 5.
(A) Comparison of copy number along chromosome 3 in a high-grade lesion (HG1), and a cancer (CA1) diagnosed 21 months later at the same anatomical location. (B) High-resolution analysis of the 3q amplicon in CA1. The minimum commonly amplified region is again indicated in gray. There is regional amplification in the interval 173.9 Mb to 183.1 Mb, with an intraamplicon peak between 182.49 Mb and 183.47 Mb. A single annotated gene, SOX2, lies within this interval. Corresponding microdissection molecular copy-number counting results against DNA from pooled normal DNA and from LG6 are shown. (C) Representative images from a fluorescence in situ hybridization experiment on 002-CA1 are shown. Probes for chromosome 3 centromere (red), SOX2 (yellow), and PIK3CA (green) are shown; there are more copies of SOX2 than PIK3CA. (D) In a separate experiment on CA1, multiple SOX2 probe signals are clustered at the upper pole of a mitotic body and within interphase nuclei. SOX2 amplification is accompanied by a high level of SOX2 nuclear expression in tumor relative to stromal cells.
DISCUSSION
In this work, MCC has facilitated the most detailed genomic dissection to date of the critical 3q amplicon in preinvasive bronchial biopsies, material that heretofore was difficult to study. We have confirmed that μMCC can define regional genomic structure in detail using limited amounts of archived material. We have also shown that 3q amplification is a consistent finding in all HGD lesions analyzed and that it readily discriminates LGD from HGD in this cohort of samples. Therefore, 3q amplification may well represent a useful molecular prognostic biomarker for bronchial dysplasia, depending on the loci chosen for analysis (20). Our data are consistent with previous FISH data reporting 3q amplification as a biomarker of progression in cervical (36) and head and neck cancer (37) but suggest a different amplification target—SOX2.
The target(s) of the 3q amplicon have been a subject of much interest and debate, with particularly strong cases having previously being made for TP73L (18), SCCRO/DCUND1 (11), and PIK3CA (10). It remains possible that there are multiple targets for this regional amplicon. Coamplification of adjacent oncogenes can have a synergistic effect in assays designed to test the functional relevance of putative oncogenes. Such a mechanism has recently been shown to be important in lung adenocarcinoma (38).
The data presented here are consistent with recently published single nucleotide polymorphism array and functional data implicating SOX2 as a driver oncogene (12). These investigators reported SOX2 amplification (regarded as 3.6 copies per genome) in 23% of SQC (12). More recently a second group has published work corroborating this finding in invasive cancers and providing further functional data implicating SOX2 as an oncogene in SQC (39). Using a digital PCR approach, we demonstrate that PIK3CA and SOX2 are amplified in all HGDs examined. The consistent finding of SOX2 amplification differs from findings based on a single nucleotide polymorphism array analysis of invasive cancer and FISH surveys of preinvasive lesions (21). This probably reflects three factors: first, array-based studies tend to underestimate the degree of amplification; second, microdissection overcomes the problem of extracting DNA from heterogeneous biopsies comprising regions of dysplasia/cancer and cells; and third (22), the FISH probes used in previous studies did not encompass SOX2.
Bass and colleagues also proposed SOX2 as a lineage-survival oncogene (12). This refers to a recent model proposing that cell lineage–specific genes involved in development can become dysregulated and promote tumorigenesis (13). This is a similar concept to the master gene hypothesis, which proposed that key developmental genes, including some encoding transcription factors, are inappropriately activated by chromosomal translocations in the pathogenesis of leukemia (40). SOX2 encodes a key stem cell transcription factor that is one of a few factors required for the induction of pluripotency in adult fibroblasts (16). It has also been shown to have a critical role in the developing mouse trachea (15). The lineage-specific model would predict the dysregulation of SOX2 in the early stages of SQC development, as we have demonstrated in this work. We further show—in a few rare cases in which sequential biopsies are available from individual lesions—that 3q amplification is an evolving process that may be positively selected for in clinical progression and that the focus of the amplicon progression encompasses SOX2, and in one instance SOX2 alone.
A limitation of this study has been the number of lesions reported. This reflects the significant difficulties faced in studying preinvasive disease because of the nature of archived tissue and the scarcity of long-standing archives (17). This is reflected in the relatively small cohorts of preinvasive lesions studied in lung (21, 23, 24, 41) and other epithelial cancers (42, 43). Nevertheless, the study of preinvasive disease is key to understanding the pathogenesis of cancer; this work describes a much more detailed analysis of the critical 3q amplicon than has previously been attempted in bronchial preinvasive disease. The potential use of SOX2 amplification as a prognostic marker in preinvasive disease would require replication of these findings in a much larger cohort, ideally in the context of a multicenter prospective clinical trial.
There is much to be learned about the functional impact of SOX2 amplification in epithelial cancers, the relationship between its potential roles in cancer and in stem cell biology, and its potential synergism with other candidate 3q oncogenes. As discussed above, this work suggests that SOX2 amplification may be a useful molecular biomarker for clinical progression in bronchial dysplasia. Furthermore, when considered along with work from others (12, 39), SOX2 and its downstream effector targets may be targets for biological therapeutics for the treatment and chemoprevention of squamous carcinomas.
Supplementary Material
Acknowledgments
The authors thank S. Newman and P. Edwards (Department of Pathology, University of Cambridge) for advice on FISH, and P. Edwards for advice on the manuscript. They also thank Karl Storz, GmBH & Co (Tuttlingen, Germany) for the loan of an autofluorescence bronchoscope to University College London Hospitals for part of this study.
Supported by the UK Medical Research Council (F.M., P.H.D.), The Rosetrees and Bernard Coleman Trusts (F.M., P.J.G., B.C.), Yorkshire Cancer Research (T.H.R., P.H.R.), and Cancer Research UK grant C1023/A5977 (J.C.M.P.). This work was in part funded by the Department of Health's NIHR Biomedical Research Centres funding scheme.
Current address for J.C.M.P. is BlueGnome Limited, Breaks House, Mill Court, Great Shelford, Cambridge, CB22 5LD, UK.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201001-0005OC on March 18, 2010
Conflict of Interest Statement: F.M. is employed by the UK Medical Research Council ($10,001–$50,000); the UK Medical Research Council holds the patent to MCC, the technique used in this article. J.C.M.P. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.T.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.A.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.F. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. T.H.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. P.J.G. has performed advisory board duties for Alveolus for which he has received no fees; he has received lecture fees from Glaxo Wellcome (up to $1,000); he has received expert witness fees from Blake Lapthorn Solicitors ($1,000–$5,000). P.H.D. is employed by UK Medical Research Council (more than $100,000); the UK Medical Research Council holds a patent on MCC, the technique used in this article, on which he is named as an inventor; he does not believe this has influenced his independence. P.H.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature 2009;458:719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabbitts TH. Commonality but diversity in cancer gene fusions. Cell 2009;137:391–395. [DOI] [PubMed] [Google Scholar]
- 3.Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med 2005;353:133–144. [DOI] [PubMed] [Google Scholar]
- 4.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039–1043. [DOI] [PubMed] [Google Scholar]
- 5.Qian J, Massion PP. Role of chromosome 3q amplification in lung cancer. J Thorac Oncol 2008;3:212–215. [DOI] [PubMed] [Google Scholar]
- 6.Redon R, Hussenet T, Bour G, Caulee K, Jost B, Muller D, Abecassis J, du Manoir S. Amplicon mapping and transcriptional analysis pinpoint cyclin l as a candidate oncogene in head and neck cancer. Cancer Res 2002;62:6211–6217. [PubMed] [Google Scholar]
- 7.Yen CC, Chen YJ, Pan CC, Lu KH, Chen PC, Hsia JY, Chen JT, Wu YC, Hsu WH, Wang LS, et al. Copy number changes of target genes in chromosome 3q25.3-qter of esophageal squamous cell carcinoma: TP63 is amplified in early carcinogenesis but down-regulated as disease progressed. World J Gastroenterol 2005;11:1267–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA 1996;93:479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massion PP, Taflan PM, Jamshedur Rahman SM, Yildiz P, Shyr Y, Edgerton ME, Westfall MD, Roberts JR, Pietenpol JA, Carbone DP, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res 2003;63:7113–7121. [PubMed] [Google Scholar]
- 10.Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, Soh J, Suzuki M, Wistuba II, Fong KM, et al. Pik3ca mutations and copy number gains in human lung cancers. Cancer Res 2008;68:6913–6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarkaria I, Oc P, Talbot SG, Reddy PG, Ngai I, Maghami E, Patel KN, Lee B, Yonekawa Y, Dudas M, et al. Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer Res 2006;66:9437–9444. [DOI] [PubMed] [Google Scholar]
- 12.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41:1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer 2006;6:593–602. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, Sun L, Yang X, Wang Y, Zhang Y, et al. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem 2008;283:17969–17978. [DOI] [PubMed] [Google Scholar]
- 15.Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for SOX2 in the developing and adult mouse trachea. Development 2009;136:1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–676. [DOI] [PubMed] [Google Scholar]
- 17.Jonsson S, Varella-Garcia M, Miller YE, Wolf HJ, Byers T, Braudrick S, Kiatsimkul P, Lewis M, Kennedy TC, Keith RL, et al. Chromosomal aneusomy in bronchial high-grade lesions is associated with invasive lung cancer. Am J Respir Crit Care Med 2008;177:342–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massion PP, Taflan PM, Rahman SM, Yildiz P, Shyr Y, Carbone DP, Gonzalez AL. Role of p63 amplification and overexpression in lung cancer development. Chest 2004;125:102S. [PubMed] [Google Scholar]
- 19.Massion PP, Taflan PM, Shyr Y, Rahman SM, Yildiz P, Shakthour B, Edgerton ME, Ninan M, Andersen JJ, Gonzalez AL. Early involvement of the phosphatidylinositol 3-kinase/akt pathway in lung cancer progression. Am J Respir Crit Care Med 2004;170:1088–1094. [DOI] [PubMed] [Google Scholar]
- 20.Massion PP, Zou Y, Uner H, Kiatsimkul P, Wolf HJ, Baron AE, Byers T, Jonsson S, Lam S, Hirsch FR, et al. Recurrent genomic gains in preinvasive lesions as a biomarker of risk for lung cancer. PLoS ONE 2009;4:e5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelosi G, Del Curto B, Trubia M, Nicholson AG, Manzotti M, Veronesi G, Spaggiari L, Maisonneuve P, Pasini F, Terzi A, et al. 3q26 amplification and polysomy of chromosome 3 in squamous cell lesions of the lung: a fluorescence in situ hybridization study. Clin Cancer Res 2007;13:1995–2004. [DOI] [PubMed] [Google Scholar]
- 22.Brown LA, Hoog J, Chin SF, Tao Y, Zayed AA, Chin K, Teschendorff AE, Quackenbush JF, Marioni JC, Leung S, et al. ESR1 gene amplification in breast cancer: a common phenomenon? Nat Genet 2008;40:806–807; author reply 810–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garnis C, Campbell J, Davies JJ, Macaulay C, Lam S, Lam WL. Involvement of multiple developmental genes on chromosome 1p in lung tumorigenesis. Hum Mol Genet 2005;14:475–482. [DOI] [PubMed] [Google Scholar]
- 24.Garnis C, Davies JJ, Buys TP, Tsao MS, MacAulay C, Lam S, Lam WL. Chromosome 5p aberrations are early events in lung cancer: Implication of glial cell line-derived neurotrophic factor in disease progression. Oncogene 2005;24:4806–4812. [DOI] [PubMed] [Google Scholar]
- 25.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet 2005;37:S11–S17. [DOI] [PubMed] [Google Scholar]
- 26.Little SE, Vuononvirta R, Reis-Filho JS, Natrajan R, Iravani M, Fenwick K, Mackay A, Ashworth A, Pritchard-Jones K, Jones C. Array CGH using whole genome amplification of fresh-frozen and formalin-fixed, paraffin-embedded tumor DNA. Genomics 2006;87:298–306. [DOI] [PubMed] [Google Scholar]
- 27.Daser A, Thangavelu M, Pannell R, Forster A, Sparrow L, Chung G, Dear PH, Rabbitts TH. Interrogation of genomes by molecular copy-number counting (MCC). Nat Methods 2006;3:447–453. [DOI] [PubMed] [Google Scholar]
- 28.McCaughan F, Darai-Ramqvist E, Bankier AT, Konfortov BA, Foster N, George PJ, Rabbitts TH, Kost-Alimova M, Rabbitts PH, Dear PH. Microdissection molecular copy-number counting (microMCC)–unlocking cancer archives with digital PCR. J Pathol 2008;216:307–316. [DOI] [PubMed] [Google Scholar]
- 29.McCaughan F, Bankier AT, Konfortov BA, Pole J, Carroll B, Falzon MR, Rabbitts TH, George PJ, Dear PH, Rabbitts PH. Chronology of 3q amplification in preinvasive bronchial lesions of increasing histological abnormality and identification of candidate oncogenes/molecular targets in the pathogenesis of squamous lung cancer [abstract]. Thorax 2008;63:A62. [Google Scholar]
- 30.McCaughan F, Bankier AT, Konfortov BA, Pole J, Carroll B, Falzon MR, Rabbitts TH, George PJ, Dear PH, Rabbitts PH. Exploring the precancer genome with microdissection molecular copy-number counting (μMCC) [abstract]. European Respiratory Society Congress, European Respiratory Society Lung Science Conference, 27-29 March, 2009, Estoril, Vienna, p. 247s.
- 31.McCaughan F, Bankier AT, Konfortov BA, Pole J, Carroll B, Falzon MR, Rabbitts TH, George PJ, Dear PH, Rabbitts PH. Progressive and clonal 3q amplification in preinvasive lesions of the bronchial epithelium: a detailed characterization [abstract]. National Cancer Research Institute Conference poster presentation, 7–10 November 2009, Birmingham, UK. p. LB67.
- 32.George PJ, Banerjee AK, Read CA, O'Sullivan C, Falzon M, Pezzella F, Nicholson AG, Shaw P, Laurent G, Rabbitts PH. Surveillance for the detection of early lung cancer in patients with bronchial dysplasia. Thorax 2007;62:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin SF, Daigo Y, Huang HE, Iyer NG, Callagy G, Kranjac T, Gonzalez M, Sangan T, Earl H, Caldas C. A simple and reliable pretreatment protocol facilitates fluorescent in situ hybridisation on tissue microarrays of paraffin wax embedded tumour samples. Mol Pathol 2003;56:275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia MJ, Pole JC, Chin SF, Teschendorff A, Naderi A, Ozdag H, Vias M, Kranjac T, Subkhankulova T, Paish C, et al. A 1 Mb minimal amplicon at 8p11–12 in breast cancer identifies new candidate oncogenes. Oncogene 2005;24:5235–5245. [DOI] [PubMed] [Google Scholar]
- 35.Foster NA, Banerjee AK, Xian J, Roberts I, Pezzella F, Coleman N, Nicholson AG, Goldstraw P, George JP, Rabbitts PH. Somatic genetic changes accompanying lung tumor development. Genes Chromosomes Cancer 2005;44:65–75. [DOI] [PubMed] [Google Scholar]
- 36.Heselmeyer-Haddad K, Sommerfeld K, White NM, Chaudhri N, Morrison LE, Palanisamy N, Wang ZY, Auer G, Steinberg W, Ried T. Genomic amplification of the human telomerase gene (TERC) in Pap smears predicts the development of cervical cancer. Am J Pathol 2005;166:1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh B, Stoffel A, Gogineni S, Poluri A, Pfister DG, Shaha AR, Pathak A, Bosl G, Cordon-Cardo C, Shah JP, et al. Amplification of the 3q26.3 locus is associated with progression to invasive cancer and is a negative prognostic factor in head and neck squamous cell carcinomas. Am J Pathol 2002;161:365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA 2007;104:16663–16668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hussenet T, Dali S, Exinger J, Monga B, Jost B, Dembele D, Martinet N, Thibault C, Huelsken J, Brambilla E, et al. Sox2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS ONE 2010;5:e8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabbitts TH. Translocations, master genes, and differences between the origins of acute and chronic leukemias. Cell 1991;67:641–644. [DOI] [PubMed] [Google Scholar]
- 41.Salaun M, Sesboue R, Moreno-Swirc S, Metayer J, Bota S, Bourguignon J, Thiberville L. Molecular predictive factors for progression of high-grade preinvasive bronchial lesions. Am J Respir Crit Care Med 2008;177:880–886. [DOI] [PubMed] [Google Scholar]
- 42.Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, Kuo WL, Ljung BM, Chew K, Myambo K, et al. In situ analyses of genome instability in breast cancer. Nat Genet 2004;36:984–988. [DOI] [PubMed] [Google Scholar]
- 43.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet 2007;39:41–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.