Abstract
The aspartate receptor of bacterial chemotaxis is representative of a large family of taxis receptors widespread in prokaryotes. The homodimeric receptor associates with cytoplasmic components to form a receptor–kinase signaling complex. Within this complex the receptor is known to directly contact the histidine kinase CheA, the coupling protein CheW, and other receptor dimers. However, the locations and extents of the contact regions on the receptor surface remain ambiguous. The present study applies the protein-interactions-by-cysteine-modification (PICM) method to map out surfaces on the aspartate receptor that are essential for kinase stimulation in the assembled receptor–kinase complex. The approach utilizes 52 engineered cysteine positions scattered over the surface of the receptor periplasmic and cytoplasmic domains. When the bulky, anionic probe 5-fluorescein-maleimide is coupled to these positions, large effects on receptor-mediated kinase stimulation are observed at eight cytoplasmic locations. By contrast, no large effects are observed for probe attachment at exposed positions in the periplasmic domain. The results indicate that essential receptor surface regions are located near the hairpin turn at the distal end of the cytoplasmic domain and in the cytoplasmic adaptation site region. These surface regions include the docking sites for CheA, CheW, and other receptor dimers, as well as surfaces that transmit information from the receptor adaptation sites to the kinase. Smaller effects observed in the cytoplasmic linker or HAMP region suggest this region may also play a role in kinase regulation. A comparison of the activity perturbations caused by a dianionic, bulky probe (5-fluorescein-maleimide), a zwitterionic, bulky probe (5-tetramethyl-rhodamine-maleimide), and a nonionic, smaller probe (N-ethyl-maleimide) reveals the roles of probe size and charge in generating the observed effects on kinase activity. Overall, the results indicate that interactions between the periplasmic domains of different receptor dimers are not required for kinase activation in the signaling complex. By contrast, the observed spatial distribution of protein contact surfaces on the cytoplasmic domain is consistent with both (i) distinct docking sites for cytoplasmic proteins and (ii) interactions between the cytoplasmic domains of different dimers to form a trimer-of-dimers.
The transmembrane chemoreceptors of the Escherichia coli and Salmonella typhimurium chemotaxis pathway are members of a large receptor superfamily that modulates two-component signaling systems ubiquitous in prokaryotes and lower eukaryotes (1–4). These chemoreceptors span the bacterial inner membrane where they form a signaling complex with a cytoplasmic histidine kinase (CheA) and a coupling protein (CheW). In the absence of chemoattractant, each chemoreceptor stimulates the histidine kinase, which autophosphorylates itself on a specific His residue. Subsequently, the same phosphoryl group is transferred to an aspartate residue in the active site of a response regulator protein (CheY or CheB). The phosphorylated response regulator then diffuses to its site of action where it modulates either the switching of the motor complex between different swimming states (Pi-CheY) or the covalent modification of the receptor adaptation sites (Pi-CheB). When the periplasmic domain of the receptor binds an attractant molecule, typically a sugar, amino acid, or peptide, this attractant binding event triggers a transmembrane conformational change that inhibits the cytoplasmic histidine kinase.
Chemoreceptors exist as homodimers, even in the absence of ligand, and these dimers are the most stable unit of receptor structure since detergent solubilization yields a pure dimer population (2, 5). In the native membrane environment, receptor homodimers assemble into a higher order oligomer (6, 7) believed to contain three dimers (8–12). The crystal structure of the isolated cytoplasmic domain reveals a trimer-of-dimers stabilized by packing contacts between adjacent dimers near the cytoplasmic hairpin turn, and genetic studies of this region provide further evidence for such contacts (8, 10). These studies do not rule out, however, additional strong receptor contacts within the trimer-of-dimers that may lie outside the cytoplasmic regions targeted by the crystallographic and genetic studies. Moreover, it is possible that multiple trimers-of-dimers further assemble into a higher order structure containing large numbers of receptors since receptors and the other pathway components are found in extensive arrays or clusters at the poles of the cell (13–15). The protein–protein contacts that stabilize cluster formation are largely unknown. Some models propose that large regions of the receptor periplasmic or cytoplasmic domain serve as direct receptor–receptor contact surfaces that stabilize receptor clusters. By contrast, other models propose that another protein, such as the dimeric CheA molecule, serves as a bridge between receptors during cluster formation (8, 14, 16, 17). To fully understand the molecular mechanism by which the receptor regulates kinase activity, it is important to map out the key contact surfaces on the receptor that are essential for kinase activation in the functional receptor–kinase signaling complex. These essential packing surfaces will include the CheA and CheW docking sites as well as any other docking surfaces required for kinase activation. Such additional surfaces could include the contact sites utilized in trimer-of-dimers formation or higher-order receptor clustering, if such interactions are essential for stimulation of kinase activity.
The present study focuses on the aspartate receptor, which generates a transmembrane signal in response to the chemo-attractant asparate. Figure 1 illustrates the periplasmic, transmembrane, and cytoplasmic domains of this receptor, which is typical of E. coli and S. typhimurium chemotaxis receptors (2, 8, 18). The periplasmic domain is a pair of four-helix bundles, one from each subunit. Two helices from each periplasmic domain extend across the membrane, forming a membrane-spanning four-helix bundle that serves as the transmembrane region. Current evidence indicates that the transmembrane signal within this receptor, and other chemotaxis receptors, is carried by a small piston-type displacement of a specific transmembrane helix, termed the signaling helix, toward the cytoplasm (2, 19, 20). Because of the 2-fold symmetry of the homodimer, each subunit possesses one signaling helix, which is the C-terminal membrane-spanning helix. In the cytoplasm, the signaling helix is coupled to the cytoplasmic domain via a conserved motif termed the linker, or HAMP motif. The cytoplasmic domain is another four-helix bundle, formed from the association of two helical hairpins provided by the two receptor subunits. Thus, the receptor dimer is a highly helical structure in which four-helix bundles play a central architectural role.
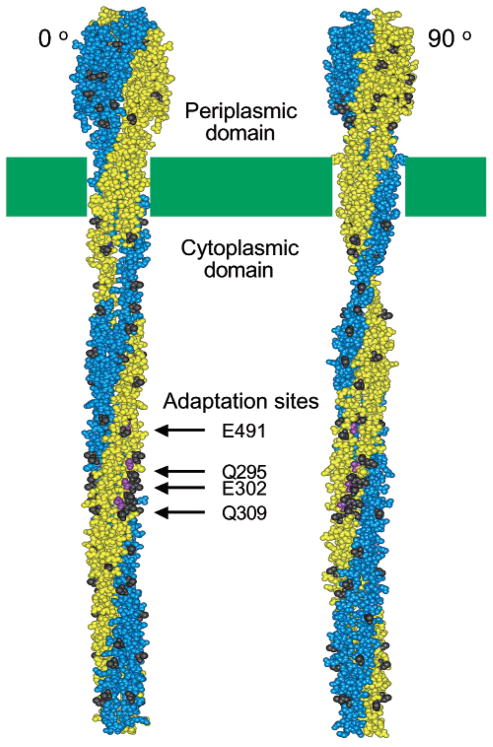
Figure 1.
Two orientations of the chemoreceptor structural model (2, 8) illustrating the positions of engineered cysteine positions. In each orientation, one subunit of the symmetric dimer is shown in gold, the other in blue. The two orientations differ by a 90° rotation about the long axis of the dimer. The dark side chains indicate the positions of the 52 engineered cysteine substitutions in the periplasmic and cytoplasmic domains. Also highlighted are the locations of the four adaptation sites on the cytoplasmic domain of the aspartate receptor.
The present study is designed to identify all of the protein–protein docking surfaces on the periplasmic and cytoplasmic domains of the aspartate receptor that are essential for kinase activation using the protein-interactions-by-cysteine-modification (PICM) method. Previous applications of the PICM method to the aspartate receptor scanned only a small region (21, 22), while the present study extends this analysis to all aqueous surfaces of the receptor. The results identify receptor surface regions essential for kinase activation in the distal half of the cytoplasmic domain and also suggest that the cytoplasmic linker or HAMP region could be coupled to kinase activation. By contrast, no surfaces on the periplasmic domain are found to be essential for kinase activation. Overall, the findings provide further evidence for functionally important contacts between the cytoplasmic domain and the other receptors, CheA or CheW, but disfavor models proposing essential functional contacts between the periplasmic domains of different receptor dimers.
MATERIALS AND METHODS
Materials
E. coli strains used for receptor expression and characterization were (a) RP3808 {Δ(cheA-cheZ)DE2209 tsr-1 leuB6 his-4 eda-50 rpsl136 [thi-1 Δ(gal-att)DE99 ara-14 lacY1 mtl-1 xyl-5 tonA31 tsx- 78]/mks/} and (b) RP8611 {ΔtsrDE7028 Δ(tar-tap)-DE5201 zbd::Tn5 Δ(trg)DE100 leuB6 his-4 rpsL136 [thi-1 ara-14 lacY1 mtl-1 xyl-5 tonA31 tsx-78]/CP326} kindly provided by John S. Parkinson, University of Utah (23). The expression plasmid pSCF6 used to generate membranes containing the S. typhimurium aspartate receptor has been previously described (24). Expression strains and plasmids used to produce CheA (HB101/pMO4) and CheW (HB101/pME5) were generously provided by Jeffrey Stock, Princeton University. The strain and plasmid used to generate CheY (RBB455/pRBB40) were kindly provided by Robert Bourret, University of North Carolina. The methods and materials used to express and isolate purified CheA, CheW, and CheY have been previously described (24). [γ-32P]-ATP was purchased from New England Nuclear. Deoxynucleotides were synthesized by Life Technologies Inc. Kunkel site-directed mutagenesis reagents (T7 DNA polymerase and deoxynucleoside triphosphates) were purchased from BioRad. MluI, EcoRI, BsmI, BclI, BsaI, and SacI were from New England Biolabs. PstI was from Roche, and Eco47 III was from Promega. The sulfhydryl-specific probes 5-fluorescein-maleimede (5FM) and 5-tetra-methyl-rhodamine-maleimide (5TRM) were purchased from Molecular Probes, Inc. Unless otherwise noted, all other reagents were purchased from Sigma and were reagent grade.
Mutagenesis
Site-directed mutagenesis was performed as previously described (21). Briefly, the Kunkel method (25) was used to engineer single cysteine substitutions into the S. typhimurium aspartate receptor in plasmid pSCF6. Silent restriction site mutations were designed to accompany the desired cysteine codons, allowing for pre-screening of mutagenesis reactions by restriction digestion. In all cases, DNA sequencing was carried out to identify plasmids containing the desired mutations.
Protein Expression and Isolation
The mutated pSCF6 plasmid coding for a given single-cysteine receptor was transformed and expressed E. coli strain RP3808, which lacks the major receptors and adaptation enzymes to ensure isolation of a homogeneous, overexpressed aspartate receptor population (adaptation state QEQE). Native E. coli membranes containing the overexpressed receptor were isolated as described previously (22). Membrane samples were assayed for total protein using the BCA assay (Pierce) calibrated against bovine serum albumin standards (Pierce) using a microplate reader (Molecular Devices). Protein purity was determined by comparing the integrals of receptor and nonreceptor bands on a Coomassie-stained 10% Laemmli SDS–PAGE gel (acrylamide/bisacrylamide ratio of 40:0.2) using a digital camera (Alpha Innotech). The soluble chemotaxis proteins CheA, CheW, and CheY were over-expressed in E. coli and purified as described previously (24, 26).
Receptor Alkylation Reactions
Prior to PICM analysis, receptor-containing membranes were labeled with 5-fluorescein-maleimide (5FM), 5-tetramethylrhodamine-maleimide (5TRM), or N-ethyl-maleimide (NEM). Receptors at a final concentration of 12 μM monomer were labeled with a 30-fold molar excess of probe added in a small volume (2%) of solvent (dimethylformamide for 5FM and NEM, dimethyl sulfoxide for 5TRM). At the same time, nonlabeled controls were generated by adding the same volume of solvent lacking probe. All samples were incubated for 10 min at 37 °C and then quenched by the addition of DTT to 35 mM and held on ice until reconstitution with CheA, CheW, and CheY. The extent of labeling of each receptor with the fluorescent probes (5FM, 5TRM) was determined as previously described (26) and ranged between 80 and 100%. Since NEM is smaller and more reactive than the fluorescent probes, it was assumed that NEM labeling under identical conditions proceeded to a similarly high extent.
PICM Assay
PICM analysis was carried out essentially as previously detailed (21) using the reconstituted receptor–kinase signaling complex (27, 28). Briefly, the receptor-coupled kinase reaction was initiated by the addition of [γ-32P]-ATP to a reaction mixture containing the reconstituted signaling complex of labeled or unlabeled receptor with CheW, CheA, and CheY. The reaction included sufficient CheY to ensure that the rate-limiting step in phospho-CheY formation is the autophosphorylation of CheA in the signaling complex. After 10 s, the reaction was quenched by the addition of an equal volume of 2X Laemmli sample buffer containing 25 mM EDTA and heated to 95 °C for 3 min prior to loading onto 15% Laemmli SDS–PAGE gels (acrylamide/bisacrylamide ratio of 40:1.25). The resulting gels were dried and phosphorimaged (Molecular Dynamics) to quantitate the level of [γ-32P]-phospho-CheY, which is proportional to the initial rate of the receptor-regulated kinase reaction. All reactions were performed in triplicate, and the PICM parameter was calculated as the ratio of kinase rates in the labeled/unlabeled samples.
Error Analysis
Indicated error ranges represent ± 1 standard deviation for n ≥ 3.
RESULTS
Experimental Strategy
The PICM approach is a generalizable scanning method that can be carried out in the native, membrane-bound receptor–kinase complex (21). The strategy begins by introducing cysteine substitutions at solvent-exposed positions scattered over the surface of the receptor dimer, in both the periplasmic and the cytoplasmic domains. Only those positions at which the cysteine substitution retains receptor-stimulated kinase activation are utilized, thereby eliminating engineered receptors that fold improperly or are otherwise damaged. E. coli membranes containing each engineered receptor are isolated and labeled with a bulky probe, 5-fluorescein-maleimide (5FM), which specifically alkylates the sulfhydryl residue of the exposed cysteine side chain. Next, the effect of probe attachment on receptor stimulation of kinase activity is quantitated in the reconstituted receptor–kinase signaling complex (27, 28) generated by combining the receptor-containing membranes with purified histidine kinase (CheA), coupling protein (CheW), and response regulator (CheY). Finally, the PICM parameter of a given mutant receptor is determined from the ratio of the histidine kinase activities obtained in the labeled and unlabeled states (21). This PICM parameter ranges from zero at positions where the bulky probe completely blocks kinase activation, to unity at positions where the probe has no effect, to greater-than-unity at positions where the probe super-activates the kinase.
Generation of the Cysteine Library
To investigate all exposed receptor surfaces on both the periplasmic and the cytoplasmic domains, a group of 52 surface positions were selected for cysteine substitution and PICM analysis as illustrated in Figure 1 (black side chains). These positions are fully solvent-exposed on the surface of an isolated receptor dimer and included a subset of positions implicated in contacts between dimers in the trimer-of-dimers. The 52 selected positions were approximately evenly spaced throughout the two domains as determined by examination of the crystal structures of the isolated periplasmic and cytoplasmic domains (8, 29) and of the current structural model of the full-length receptor dimer (2, 8, 16). For the periplasmic domain, the crystal structure utilized was that of the aspartate receptor periplasmic domain (29). For the cytoplasmic domain, the only crystal structure available is that of the serine receptor (8); however, this domain is highly homologous to the cytoplasmic domain of the aspartate receptor (30); thus, it was straightforward to identify the aspartate receptor positions corresponding to the selected positions in the serine receptor cytoplasmic domain.
Of the 52 single-cysteine receptors targetted for study, 31 already existed in the available library of receptor mutants and were known to retain significant kinase stimulation (21, 22, 26, 31), while the remaining 21 were created by site-directed mutagenesis. Subsequently, each of the 52 mutant receptors was expressed in E. coli, and cell membranes containing the overexpressed receptor were isolated. Reactions of mutant receptors with the bulky, aqueous 5FM and 5TRM probes were found to proceed to 80–100% completion, confirming aqueous exposure of their engineered cysteines (see Table 1 for representative examples). In addition, all 52 mutant receptors were successfully reconstituted into the receptor–kinase signaling complex and exhibited kinase stimulation within 3-fold of that measured for the wild type receptor, as well as normal aspartate-triggered kinase inhibition. Thus, each of the 52 mutant aspartate receptors possessed a unique surface-exposed cysteine suitable for alkylation and retained approximately native function in the reconstituted signaling complex.
Table 1.
Effect of Labeling with Covalent Probes on Receptor-Regulated Kinase Activity
| position | extent of labeling | labeled/unlabeled activity (PICM) |
|---|---|---|
| 5-Fluorescein-maleimide (5FM) | ||
| E294C | 79.1% | 1.8 ± 0.2 |
| E308C | 100% | 0.17 ± 0.02 |
| H350C | 100% | 0.14 ± 0.01a |
| S364C | 100% | 0.20 ± 0.01a |
| L378C | 100% | 0.03 ± 0.01 |
| V396C | 100% | 0.07 ± 0.01 |
| A399C | 100% | 0.06 ± 0.01 |
| R449C | 99.3% | 2.1 ± 0.3 |
| 5-Tetramethyl-rhodamine-maleimide (5TRM) | ||
| E294C | 92.1% | 3.3 ± 0.7 |
| E308C | 94.4% | 0.55 ± 0.03 |
| H350C | 95.3% | 0.9 ± 0.3 |
| S364C | 100% | 0.6 ± 0.1 |
| L378C | 100% | 0.01 ± 0.01 |
| V396C | 100% | 0.14 ± 0.02 |
| A399C | 100% | 0.03 ± 0.01 |
| R449C | 100% | 0.33 ± 0.04 |
| N-ethyl-maleimide (NEM) | ||
| E294C | NDb | 1.5 ± 0.3 |
| E308C | ND | 0.04 ± 0.01 |
| H350C | ND | 0.96 ± 0.05 |
| S364C | ND | 0.9 ± 0.1 |
| L378C | ND | 0.15 ± 0.01 |
| V396C | ND | 0.17 ± 0.01 |
| A399C | ND | 1.30 ± 0.02 |
| R449C | ND | 0.36 ± 0.04 |
Previous measurements (ref 21) of 5FM-PICM values for the two indicated mutants are in good agreement for S364C but significantly closer to 1.0 for H350C, suggesting that 5FM labeling of H350C was not complete in the earlier study.
Although labeling extents were not directly determined for NEM, they are expected to match or exceed the labeling extents of the bulkier fluorescent probes that typically exhibit lower reaction rates.
Effect of 5-Fluorescein-Maleimide Coupling on Receptor Stimulation of Kinase Activity
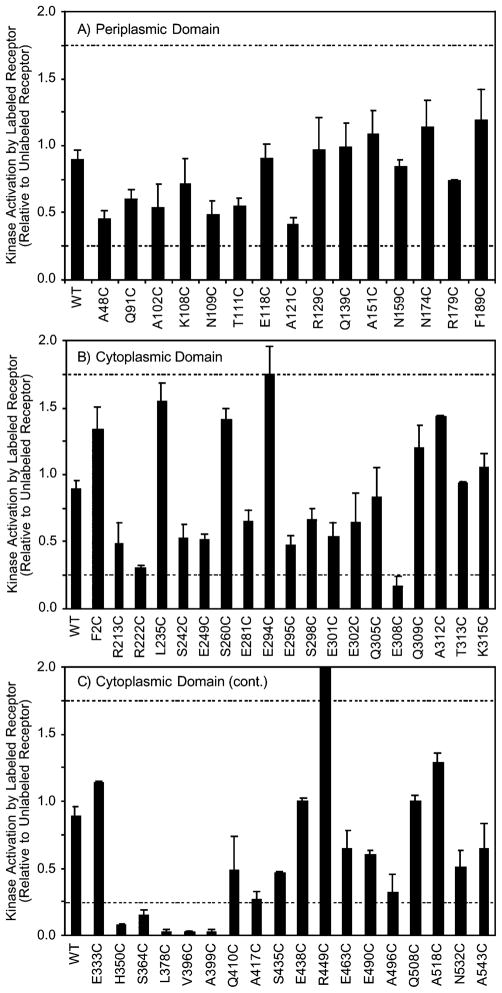
To identify engineered cysteine positions at which bulky probe attachment prevented the native receptor stimulation of kinase activity, the PICM procedure was carried out. Two identical aliquots of membranes containing a given mutant receptor were incubated with buffer containing or lacking 5-fluorescein-maleimide (5FM) for a sufficient time to drive the labeling reaction essentially to completion, then excess probe was quenched, and the receptor was reconstituted into the receptor–kinase signaling complex. Finally, receptor stimulation of kinase activity was quantitated in the in vitro assay, and the ratio of kinase activity in the labeled/unlabeled samples was determined in triplicate. The resulting 5FM–PICM parameters are summarized in Figure 2. As expected, the PICM parameter for the wild type receptor, which lacks cysteine and thus is not labeled by the probe, is nearly 1.0 indicating that incubation with the probe has little effect on kinase activation. The PICM parameters of the 52 single-cysteine receptors range from 0.03 to 2.1, representing nearly total probe-induced inhibition to significant superactivation, respectively. Examination of the data indicates that 5FM attachment slightly inhibits kinase stimulation at most of the 52 cysteine positions, yielding an average PICM value of 0.74 over the entire set of 52 sites. Of most interest are the eight positions at which probe attachment causes the largest effects. Here we operationally define inhibitory positions as those with PICM values below 0.25 of which there are six examples (E308C, H350C, S364C, L378C, V396C, and A399C). At the other extreme, superactivating positions are defined as those with PICM values exceeding 1.75, of which there are two examples (E294C and R449C). Notably, all eight of the highly perturbing sites lie in the cytoplasmic domain. To more carefully examine the molecular features of 5FM responsible for the largest inhibitory and super-activating effects, the eight highly perturbing positions were also labeled with a zwitterionic bulky probe and a small nonionic probe.
Figure 2.
Summary of the effects of 5-fluorescein-maleimide (5FM) labeling on receptor-stimulated kinase activity in the reconstituted signaling complex. Each bar indicates the ratio of receptor-stimulated kinase activity in the labeled and unlabeled states of a given single-cysteine receptor, defined as the PICM parameter (see text). Dashed lines indicate the thresholds used for the operational definition of cysteine positions where coupling of the probe yields a superactivated receptor (PICM > 1.75) or inhibited receptor (PICM < 0.25).
Effect of 5-Tetramethyl-Rhodamine-Maleimide and N-Ethyl-Maleimide Coupling on Receptor Stimulation of Kinase Activity
While 5FM is a bulky dianion at neutral pH (32), 5-tetramethyl-rhodamine-maleimide (5TRM) is a bulky zwitterion, and N-ethyl-maleimide (NEM) is a smaller nonionic probe, as illustrated in Figure 3. Comparison of these three probes in the PICM assay provides information about the contribution of probe size and charge to activity perturbations. Table 1 summarizes the PICM effects of all three probes at the eight most highly sensitive positions identified in the 5FM–PICM analysis. At two positions (L378C and V396C) all three probes are perturbing, suggesting that these positions are at highly critical docking locations where any probe is disruptive. Notably, these two positions are located near the cytoplasmic hairpin turn in the most highly conserved region of the receptor where both CheA and other receptor dimers are proposed to dock (8, 14, 22, 33). At three positions (H350C, S364C, and R449C), the large kinase activity changes observed for the 5FM probe are significantly smaller for both the 5TRM and the NEM probes, suggesting that the negative charge of 5FM plays a role in perturbation. At two positions (E294C and A399C), similar kinase activity changes are observed for both 5FM and 5TRM but not NEM, suggesting that the probe size is responsible. Finally, at the remaining position (E308C) the 5FM and NEM probes are more perturbing than 5TRM, suggesting that a more complex dependence on probe charge, size, and shape may control PICM effects at this location. Overall, these findings indicate that comparison of different probes can generally provide insight into the molecular mechanisms of perturbations because of probe attachment.
Figure 3.
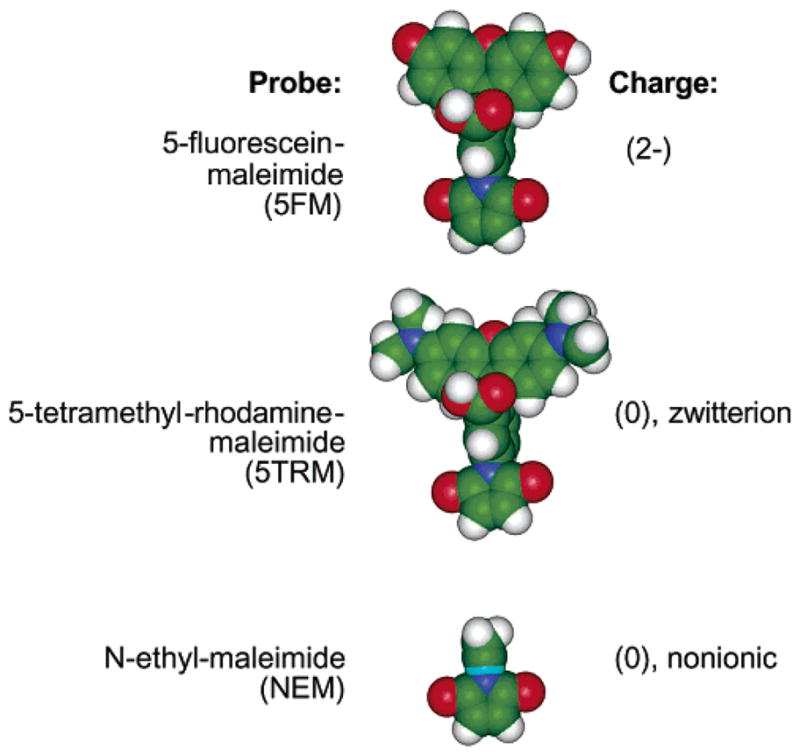
Comparison of the three cysteine-specific probes used to resolve the effects of probe size and charge on receptor function. The 5-fluorescein-maleimide (5FM) probe is a bulky dianion, while the 5-tetramethylrhodamine-maleimide (5TRM) probe is a bulky zwitterion with no net charge, and the N-ethyl-maleimide (NEM) probe is smaller and nonionic.
DISCUSSION
PICM analysis using 5-fluorescein-maleimide (5FM) as a probe was carried out for 52 engineered cysteine locations on the surface of the aspartate receptor. At all 15 sites in the periplasmic domain, coupling of this bulky, anionic probe has little or no effect on the ability of the receptor to stimulate kinase activity in the reconstituted receptor–kinase complex. It is expected that any protein–protein docking surface critical for kinase activation would be significantly disrupted by attachment of a probe as large as 5FM (see Figure 3) since many receptor point mutations that completely eliminate kinase activation have been described (21, 22, 26, 31). Thus, the lack of PICM effects in the periplasmic domain argues that this region does not participate in protein–protein contacts essential to kinase activation. This finding disfavors models proposing that assembly of a functional signaling complex requires specific contacts between the dimeric periplasmic domains of adjacent receptors, either in the trimer-of-dimers or larger receptor arrays. Additional evidence disfavoring specific contacts between periplasmic domains is provided by the failure to observe a higher order oligomer in the crystal structure of the isolated periplasmic domain (8) and by the sharp NMR resonances observed for the isolated domain (34). Thus, the available evidence strongly supports the conclusion that the dimeric periplasmic domain is independent and has no functionally significant contacts with the periplasmic domains of nearby receptors.
By contrast, large effects on kinase activation are observed when 5FM is coupled to eight of the 37 selected sites in the cytoplasmic domain. At six locations (E308C, H350C, S364C, L378C, V396C, and A399C) coupling of 5FM inhibits kinase activity over 4-fold. At two locations (E294C and R449C), coupling of the probe superactivates the kinase over 1.75-fold. All eight of these probe-sensitive positions are located in the half of the cytoplasmic domain distal from the membrane as summarized in Figure 4. Three of the highly perturbing positions (E294C, E308C, and R449C) are located in or near the region containing the receptor adaptation sites, consisting of specific glutamate side chains that are methyl esterified and demethylated by the adaptation enzymes of the chemotaxis pathway (2). Covalent modification of the receptor adaptation sites is known to modulate kinase activity (9, 28, 35, 36); thus, it is quite possible that covalent 5FM coupling in this region perturbs a native mechanism that regulates the bound kinase. Interestingly, 5FM coupling in this region can both increase and decrease kinase activity, indicating that the perturbations can be activating as well as inhibitory. Similarly, covalent modification of the receptor adaptation sites can both activate and inhibit kinase activity (9, 28, 35, 36). The nature of the protein–protein interactions essential for coupling the adaptation sites to kinase regulation is not yet known but could involve electrostatic or steric interactions between helices in the same receptor subunit, between different subunits in the same dimer, between dimers in the same trimer-of-dimers, or between different trimer-of-dimers. The present approach does not ascertain whether the effects because of probe attachment in this region are transmitted via a conformational or dynamical change within a single dimer, or rather via packing changes between dimers.
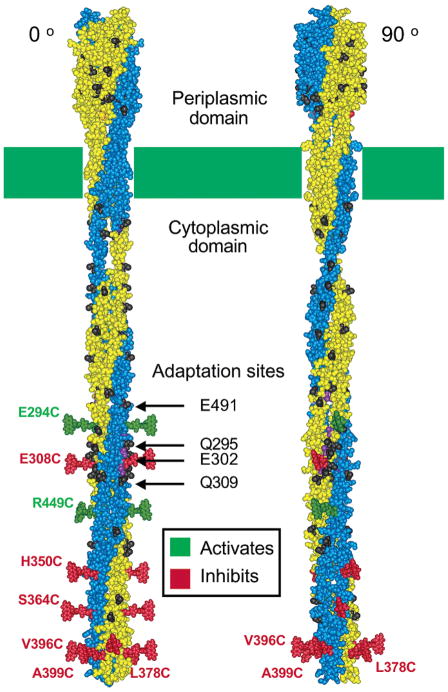
Figure 4.
Two orientations of the receptor structural model (2, 8) illustrating the eight positions at which 5-fluorescein-maleimide (5FM) probe attachment yields the largest perturbations of receptor-stimulated kinase activity. The two orientations differ by a 90° rotation about the long axis of the dimer. Shown at each perturbing position is a plausible orientation for a 5FM probe coupled to that position. Green and red probes indicate positions at which 5FM attachment strongly superactivates or inhibits the kinase, respectively. Prominent in the left orientation are the two symmetric linear interaction faces that contain the adaptation sites, while in the right orientation the two symmetric radial interaction faces are more prominent.
The remaining five highly perturbing positions (H350C, S364C, L378C, V396C, and A399C), which exhibit the largest effects because of probe attachment, are all located in the region of the cytoplasmic domain most distal to the membrane and near the helical hairpin turn (Figure 4). This region is the most highly conserved among chemotaxis receptor structures (30) and is believed to contain the conserved docking surface for the histidine kinase (CheA) and the coupling protein (CheW) essential for the formation of the receptor–kinase signaling complex based on previous genetic and PICM studies (37, 21, 22, 33). Moreover, this same region provides the bulk of receptor–receptor contacts that stabilize the trimer-of-dimers (8, 10). In this region, 5FM modification of any of the five highly sensitive sites inhibits kinase activity by a factor of 5-fold or more, and the effect becomes more dramatic as the probe moves closer to the hairpin turn. The strongest inhibition, over 10-fold, is observed for labeling at the three positions (L378C, V396C, and A399C) closest to the turn. Thus, the protein–protein interaction surfaces most critical for kinase stimulation are located in the cytoplasmic hairpin region. The protein–protein contacts that give rise to these PICM effects presumably include both dimer–dimer contacts within the trimer-of-dimers and contacts between the receptor and the cytoplasmic proteins CheA and CheW. Because of the importance of the region in both the assembly of the trimer and the kinase signaling complex, it is not possible to determine whether the inhibitory effects of probe attachment stem from disruption of trimer formation, kinase docking, or both.
The spatial distribution of the eight highly perturbing positions in the cytoplasmic domain reveals a striking bimodal spatial pattern illustrated in Figure 4. Three positions (L378C, V396C, and A399C) form a radial interaction face near the hairpin turn. By contrast, seven highly perturbing positions (E294C, E308C, H350C, S364C, V396C, A399C, and R449C) are located on a linear interaction face parallel to the long axis of the dimer. One position, R449C, is located where the radial and linear interaction faces overlap; thus, this position is included in both faces. Because of the C2 symmetry of the receptor dimer, both the radial and the linear interaction faces are duplicated at symmetric locations on the dimer surface (Figure 4). The radial interaction face includes residues believed to participate in the CheA/CheW docking surface (33, 37) and in the dimer–dimer packing interactions that stabilize the trimer-of-dimers (8, 10). The two symmetric linear faces include the receptor adaptation sites. In the trimer-of-dimers crystal structure, one of the two symmetric linear faces is proposed to be oriented toward solvent where it can provide docking sites for the cytoplasmic proteins CheA, CheW, CheR, and CheB (8, 26, 33, 37). The crystal structure indicates that the other symmetric linear face is oriented toward the center of the trimer where it participates in the receptor–receptor interactions that stabilize trimer-of-dimers formation (8). It follows that the two symmetric, linear interaction faces detected by PICM analysis are identical in an isolated receptor dimer, but their symmetry is lost in the trimer-of-dimers. Moreover, their functional roles are different in the trimer-of-dimers. Thus, some residues on these symmetric surfaces must be bifunctional since they contact different protein partners when facing the interior and exterior of the trimer-of-dimers. Notably, the linear interaction surface identified by PICM is found precisely at the location where dimer–dimer contacts are observed in the trimer-of-dimers crystal structure; thus, the PICM data strongly support the trimer-of-dimers model that has emerged from crystallographic and genetic studies and from studies of receptor cooperativity (8–11, 41).
Comparison of the effects of the 5FM, 5TRM, and NEM probes at the eight highly sensitive positions provides information about the molecular features of a probe that are responsible for perturbing receptor function at a given probe position. At two positions (L378C and V396C), all three probes greatly inhibit kinase activity, suggesting that the native side chains at these positions are especially critical for kinase docking, trimer-of-dimers formation, or both. At three positions (H350C, S364C, and R449C), the dianionic 5FM probe generates a large degree of kinase inhibition or superactivation, while the zwitterionic 5TRM and nonionic NEM probes yield much smaller effects, suggesting that the negative charge of 5FM plays a role in modulating kinase activity at these locations. At two positions (E294C and A399C), the bulky 5FM and 5TRM probes both generate significantly more kinase inhibition or superactivation than the smaller NEM probe, suggesting that probe size is coupled to kinase regulation at these locations.
In addition to the largest perturbations observed at the eight positions discussed above, more subtle perturbations that may be significant are observed at other locations as well. In the periplasmic domain, there appears to be a general trend of increasing kinase inhibition as the 5FM labeling position moves toward the N-terminal end of the domain. This trend suggests that probe coupling causes greater perturbation at positions near the subunit interface of the dimer, which is dominated by the N-terminal α-helix of the domain (α 1), and to a lesser extent, the second helix of the domain (α 2) (2, 29, 33, 37–39). Probe attachment near the subunit interface, even at a solvent exposed position, could destabilize the static subunit–subunit interaction that is important for receptor stability and function. In the cytoplasmic domain, intermediate levels of kinase inhibition and superactivation are observed for multiple sites in the linker or HAMP region (40) located at the N-terminal end of the cytoplasmic domain where it emerges from the membrane (R213C, R222C, L235C, S242C, E249C, S260C, and the nearby F2C). These perturbations suggest that the linker region may participate in an as yet unidentified protein–protein interaction in the receptor–kinase signaling complex, perhaps involving contacts within the tertiary structure of the folded linker region for which a structure is not yet available. If this is the case, then the continuous helix model for the structure of the linker (8, 16) would need revision. Alternatively, the perturbations may arise from contacts between adjacent dimers in the trimer-of-dimers or higher order oligomers.
In summary, the PICM analysis of the receptor periplasmic and cytoplasmic domains yields two principal conclusions. First, protein–protein interactions between dimeric periplasmic domains are not essential for receptor stimulation of kinase activity, suggesting that contacts between periplasmic domains are not required for kinase activation in the trimer-of-dimers or higher order receptor clusters. Second, the dimeric cytoplasmic domain participates in numerous protein–protein contacts, and the highest density of critical contact sites lies near the cytoplasmic hairpin turn. These contact sites include packing surfaces involved in the stable docking of CheA and CheW and in the stabilization of the trimer-of-dimers. Contacts less critical for kinase activation are also likely to be present in the linker or HAMP region where the cytoplasmic domain couples to the transmembrane signaling helix.
Footnotes
Support provided by NIH Grant GM R01-40731 (to J.J.F.).
References
- 1.Bourret RB, Stock AM. J Biol Chem. 2002;277:9625–8. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- 2.Falke JJ, Hazelbauer GL. Trends Biochem Sci. 2001;26:257–65. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armitage JP. Adv Microbiol Physiol. 1999;41:229–89. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- 4.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milligan DL, Koshland DE., Jr J Biol Chem. 1988;263:6268–75. [PubMed] [Google Scholar]
- 6.Seeley SK, Weis RM, Thompson LK. Biochemistry. 1996;35:5199–206. doi: 10.1021/bi9524979. [DOI] [PubMed] [Google Scholar]
- 7.Murphy OJ, III, Yi X, Weis RM, Thompson LK. J Biol Chem. 2001;276:43262–9. doi: 10.1074/jbc.M105585200. [DOI] [PubMed] [Google Scholar]
- 8.Kim KK, Yokota H, Kim SH. Nature. 1999;400:787–92. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 9.Bornhorst JA, Falke JJ. Biochemistry. 2000;39:9486–93. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ames P, Studdert CA, Reiser RH, Parkinson JS. Proc Natl Acad Sci USA. 2002;99:7060–5. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falke JJ. Proc Natl Acad Sci USA. 2002;99:6530–2. doi: 10.1073/pnas.112214199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levit MN, Stock JB. J Biol Chem. 2002;277:36760–5. doi: 10.1074/jbc.M204325200. [DOI] [PubMed] [Google Scholar]
- 13.Maddock JR, Shapiro L. Science. 1993;259:1717–23. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu TS, Le Novere N, Levin MD, Beavil AJ, Sutton BJ, Bray D. Nat Cell Biol. 2000;2:792–6. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 15.Lybarger SR, Maddock JR. J Bacteriol. 2001;183:3261–7. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SH, Wang W, Kim KK. Proc Natl Acad Sci USA. 2002;99:11611–5. doi: 10.1073/pnas.132376499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levit MN, Grebe TW, Stock JB. J Biol Chem. 2002;277:36748–54. doi: 10.1074/jbc.M204317200. [DOI] [PubMed] [Google Scholar]
- 18.Falke JJ, Kim SH. Curr Opin Struct Biol. 2000;10:462–9. doi: 10.1016/s0959-440x(00)00115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chervitz SA, Falke JJ. Proc Natl Acad Sci USA. 1996;93:2545–50. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughson AG, Hazelbauer GL. Proc Natl Acad Sci USA. 1996;93:11546–51. doi: 10.1073/pnas.93.21.11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bass RB, Falke JJ. J Biol Chem. 1998;273:25006–14. doi: 10.1074/jbc.273.39.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bass RB, Coleman MD, Falke JJ. Biochemistry. 1999;38:9317–27. doi: 10.1021/bi9908179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu JD, Parkinson JS. Proc Natl Acad Sci USA. 1989;86:8703–7. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chervitz SA, Lin CM, Falke JJ. Biochemistry. 1995;34:9722–33. doi: 10.1021/bi00030a010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunkel TA, Bebenek K, McClary J. Methods Enzymol. 1991;204:125–39. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 26.Danielson MA, Bass RB, Falke JJ. J Biol Chem. 1997;272:32878–88. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borkovich KA, Kaplan N, Hess JF, Simon MI. Proc Natl Acad Sci USA. 1989;86:1208–12. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ninfa EG, Stock A, Mowbray S, Stock J. J Biol Chem. 1991;266:9764–70. [PubMed] [Google Scholar]
- 29.Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Jr, Kim SH. Science. 1991;254:1342–7. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 30.Le Moual H, Koshland DE., Jr J Mol Biol. 1996;261:568–85. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 31.Butler SL, Falke JJ. Biochemistry. 1998;37:10746–56. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sjoback R, Nygren J, Kubista M. Spectrochim Acta. 1995;A51:L7–L14. [Google Scholar]
- 33.Bass RB, Falke JJ. Structure Fold Des. 1999;7:829–40. doi: 10.1016/s0969-2126(99)80106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danielson MA, Biemann HP, Koshland DE, Jr, Falke JJ. Biochemistry. 1994;33:6100–9. doi: 10.1021/bi00186a009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borkovich KA, Alex LA, Simon MI. Proc Natl Acad Sci USA. 1992;89:6756–60. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li G, Weis RM. Cell. 2000;100:357–65. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 37.Ames P, Yu YA, Parkinson JS. Mol Microbiol. 1996;19:737–46. doi: 10.1046/j.1365-2958.1996.408930.x. [DOI] [PubMed] [Google Scholar]
- 38.Pakula AA, Simon MI. Proc Natl Acad Sci USA. 1992;89:4144–8. doi: 10.1073/pnas.89.9.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee GF, Burrows GG, Lebert MR, Dutton DP, Hazelbauer GL. J Biol Chem. 1994;269:29920–7. [PubMed] [Google Scholar]
- 40.Aravind L, Ponting CP. FEMS Microbiol Lett. 1999;176:111–6. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 41.Sourjik V, Berg HC. Proc Natl Acad Sci USA. 2002;99:123–7. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]