Abstract
The 26S proteasome is a large multi-protein complex involved in the regulated degradation of ubiquitinated proteins in the cell. The 26S proteasome has been shown to control an increasing number of essential biochemical mechanisms of the cellular lifecycle including DNA synthesis, repair, transcription, translation and cell signal transduction. Concurrently, it is increasingly seen that malfunction of the ubiquitin proteasome system contributes to the pathogenesis of disease. The recent identification of four molecular chaperones, in addition to five previously identified chaperones, have provided mechanistic insight into how this cellular megastructure is assembled in the cell. These data, together with new insights into the structure and function of the proteasome, provide a much better understanding of this complex protease.
Introduction
Eukaryotic protein homeostasis, proteostasis (see Glossary), is central to cell development, ameliorates the rigors of cellular ageing and protects cells against disease. Deficiencies in proteostasis can lead to metabolic, oncogenic, neurodegenerative, and cardiovascular disorders1. The 26S proteasome is at the heart of proteostatic mechanisms in the cell, because it is the major cellular protease. Therefore, the assembly, structure, and function of this regulated proteolytic machine is fundamental to the life process and when deranged contributes to disease.
The 26S proteasome (see text box 1) can be divided in two sub-complexes, the core particle (CP, 20S) and the regulatory particle (RP, 19S). The RP receives, assists in deubiquitination, and unfolds ubiquitinated protein substrates, that are subsequently translocated into an enclosed cavity formed by the CP. Here a variety of catalytic sites degrade the substrate into short peptides that are subsequently broken down to amino acids by peptidases and recycled by the cell. Recent developments in understanding proteasome assembly, structure and function will be highlighted in this review.
Assembly of the proteasome
Assembly of the 26S proteasome is not a straightforward process; numerous proteins (at least sixty six2) have to assemble into a functional complex, whilst proteolytic active sites have to mature and be properly controlled during the assembly process. Additionally, three ring structures consisting of 6 or 7 unique, but homologous, subunits (ATPase subunit ring in RP and α-subunit ring and β-subunit ring in CP, see figure 1) need to form without errors. This is not a trivial supra-macromolecular task. Recent studies have shown that at least nine dedicated chaperones assist in the formation of the 26S proteasome (see table 1). These chaperones bind to proteasome sub-complexes and facilitate the formation of the 26S proteasomes, but (like other chaperones) do not form part of the final biological functional complex. Five of these proteins are dedicated to the assembly of the CP; these have been excellently reviewed recently and will only be briefly discussed here3–5. The four remaining chaperones have been described only recently and will be discussed in more detail.
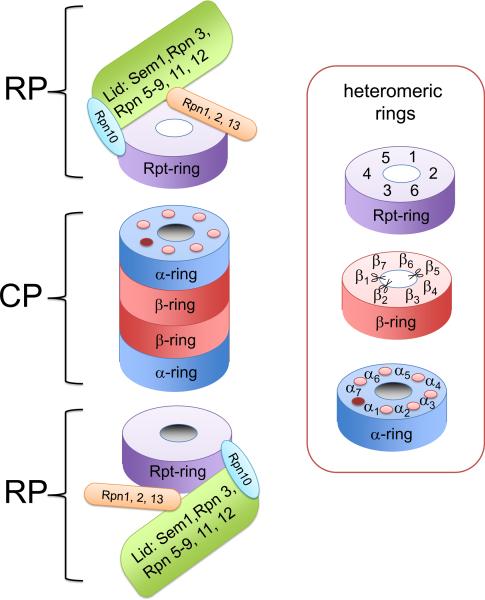
Figure 1.
Composition of the 26S proteasome. Proteasome is formed by two regulatory particles abutting cylindrically shaped core particle. Core particle is formed by two α-rings and two β-rings. Regulatory particle consists of a base complex and a lid complex. Rpn10 is at the interface between these complexes (shown in light blue). The lid (green) contains contains indicated subunits and its function is not well understood, with the exception of rpn11, which functions as a de-ubiquitinating enzyme. The base contains a ring formed by six AAA-ATPases, Rpt1–6 (purple ring), and the subunits Rpn1, Rpn2 and Rpn13 (shown as orange box; see text for more details). The proteasome contains three heteromeric ring structures, each present twice. The α and β-ring are formed by seven subunits; scissors indicate the active sites and the dots in the a ring show the binding pockets for the Rpt tails (see text). The pocket between α7 and α1 lacks a Lys typically found in the pocket and might not harbour an Rpt C-terminus41. The Rpt-ring is formed by six AAA-ATPases.
Table 1.
Names of the chaperones
| Saccharomyces cerevisiae | Homo Sapiens | ||||
|---|---|---|---|---|---|
| name | Systematic | alternatives | name | HGNC1 | alternatives |
| Pba1 | YLR199C | Poc1 | PAC1 | PSMG1 | DSCR2, c21-LRP, LRPC21, |
| Pba2 | YKL206C | Add66, Poc2 | PAC2 | PSMG2 | HCCA3, MDS003, MGC15092, CLAST3, HsT1707 TNFSF5IP1 |
| Pba3 | YLR021W | Poc3, Dmp2, Irc25 | PAC3 | PSMG3 | C7orf48, MGC10911, MGC10911 |
| Pba4 | YPL144W | Dmp1, Poc4 | PAC4 | PSMG4 | C6orf86 |
| Ump1 | YBR173C | Rns2 | UMP1 | POMP | HSPC014 |
| Hsm3 | YBR272C | S5b | PSMD5 | KIAA0072 | |
| Nas2 | YIL007C | P27 | PSMD9 | ||
| Nas6 | YGR232W | Gankyrin | PSMD10 | P28 | |
| Rpn14 | YGL004C | PAAF1 | PAAF1 | FLJ11848, Rpn14, WDR71 | |
In Bold are the SGD standard names (http://www.yeastgenome.org/).
Names based on HUGO gene Nomenclature Committee name assignment (http://www.genenames.org/).
Core particle chaperones
Assembled CP has a cylindrical shape created by four seven subunit rings designated as follows: α1–7, β1–7, β1–7, α1–7 (figure 1). The CP by itself is a functional entity that can associate with RP to form 26S, but also with other regulators, such as PA28/11S6. Consistent with this CP can assemble independently of the RP.
The assembly of the CP starts with the formation of a ring of α-subunits. However, the formation of this ring might be error prone. e.g. after in vitro translation, non-neighboring α-subunits have been shown to interact with each other7. Also, expression of some α-subunits in E.coli results in homoheptameric rings, that are stacked8, 9. In yeast, deletion of the α3 subunit results in the incorporation of two α4 subunits10. Presumably, such promiscuity led to the evolution of chaperones to control ordered assembly of α-rings in the cell and to prevent aggregation of `sticky' α-ring proteins. Indeed, deletion of the CP-chaperones from yeast results in lower levels of 20S proteasome, the accumulation of dead-end complexes, and the incorporation of a second α4 subunit instead of α311–13, 14.
Four CP-chaperones play a role in assembly of the α-ring; yeast Pba (proteasome biogenesis associated protein) 1–4 or their likely orthologs in human PAC (proteasome assembly chaperone)1–4. For simplicity we use only the human nomenclature here, but the assembly process seems largely conserved between yeast and humans. These proteins are only functional as heterodimers: PAC1-PAC2 and PAC3-PAC411, 15. The PAC3-PAC4 heterodimer is probably important early in assembly, when it attaches to α5 and assists in the recruitment and ordering of additional α-subunits14, 16.
Upon completion of α-ring assembly, β–subunits are then incorporated. To allow incorporation of all β–subunits, the PAC3-PAC4 heterodimer needs to dissociate. Structural studies show that PAC3-PAC4 cannot bind to α-rings that contain β4, because of steric hindrance between β4 and PAC316.
The PAC1-PAC2 heterodimer also assists in the formation of the α-ring, but unlike PAC3-PAC4 it remains associated with the complex upon incorporation of the β-subunits. PAC1-PAC2 can interact with α5 and α7 seems to bind the α-ring on the opposite side compared to PAC3-PAC43, 13.
The fifth CP-chaperone Ump1/POMP, which has no role in α-ring formation, seems to function as a quality control agent. The Ump1 protein prevents stable dimerization of two half CPs until all seven β–subunits are assembled on the α-ring (creating the half CP). The N-terminal pro-peptides of the β-subunits are crucial for dimerization of the half CPs. The formation of CP by two half CPs is accompanied by the maturation of the β-subunits and the degradation of Ump1 and PAC 1-PAC23. In summary, the formation of CP is assisted by five chaperones.
Interestingly, 20S proteasome structures formed by only one or two α and βsubunits, such as in prokaryotes, do not require chaperones for their formation3. Thus, the challenges of proper positioning of the seven homologous α-subunits and β-subunits in the eukaryotic CP structure seems to invoke the requirement for chaperones.
Regulatory particle chaperones
A third ring structure present in 26S proteasomes is an AAA-ATPase ring formed by the proteins Rpt1 to Rpt6 (see figure 1). These six proteins together with Rpn1, Rpn2, Rpn10 and Rpn13 form the so-called base complex. The base subunits Rpn10 and Rpn13 function as ubiquitin receptors and are the only non-essential base subunit in yeast2. They are, therefore, expected to associate peripherally to the base. Rpn1 and 2 are homologous proteins, both forming a toroid structure17. They function as a scaffold as they bind several of the proteasome subunits as well as some proteasome associated proteins2. All other RP subunits are contained in the lid complex (Figure 1). The function of this complex remains elusive, but includes de-ubiquitination of substrates by the Rpn11 subunit. The CP together with base and lid complexes can be combined in vitro to form proteasomes18; thus it is intuitive to think that 26S assembly proceeds through these sub-complexes. However, mammalian proteasomes have been successfully reconstituted from a different set of stable subcomplexes19. So, it remains to be seen how de novo formation of 26S proteasomes actually occurs in vivo.
Recently, a series of papers has revealed important roles for RP chaperones in proteasome assembly20–25. These papers suggest that the lid complex and CP can form independently, as both readily accumulate in yeast mutant strains used in these and other studies14, 26. However, base complex formation is compromised in the chaperone mutants.
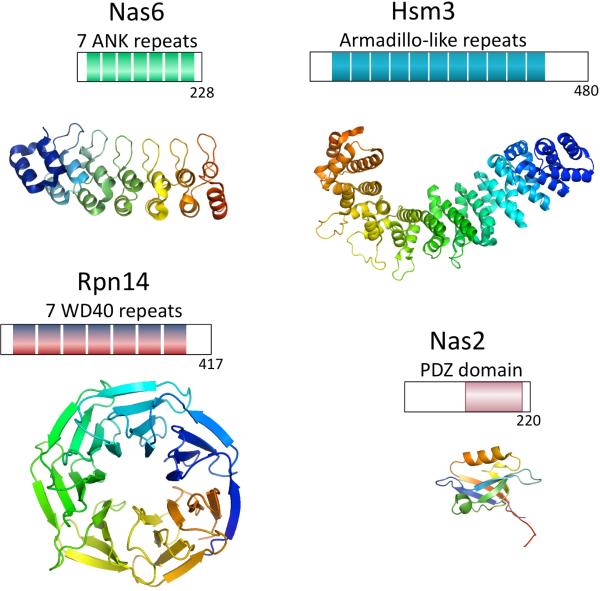
At least four chaperones are involved in the RP assembly: Nas2/p27, Nas6/gankyrin/p28, Rpn14/PAAF1 and Hsm3/S5b (Figure 2a). While all these proteins have been reported previously to associate with proteasome complexes27–33 their absence from mature 26S proteasomes in yeast and their chaperone function had not been appreciated until these recent papers shed light on their mechanisms of action. For PAAF1 and, especially, gankyrin other proteasome functions (including a role as proteasome inhibitor and intermediate in proteasome binding respectively) have been described33–35. The chaperones are evolutionary unrelated and structurally very different (see Figure 2A). Although they don't have recognizable enzyme activity, all four proteins contain domains known to be involved in protein-protein interactions36, 37. This suggests that these chaperones mainly act by binding to specific partners. Indeed, three chaperones bind to the small C-domain present in the Rpt proteins (Figure 2B): Nas6-Rpt3, Rpn14-Rpt6, Hsm3-Rpt1, while Nas2 binds either the C-domain or the C-terminus of Rpt520, 23, 25, 32. Thus the chaperones might be important in the formation of the AAA-ATPase ring containing the Rpt1–6 subunits.
Figure 2.
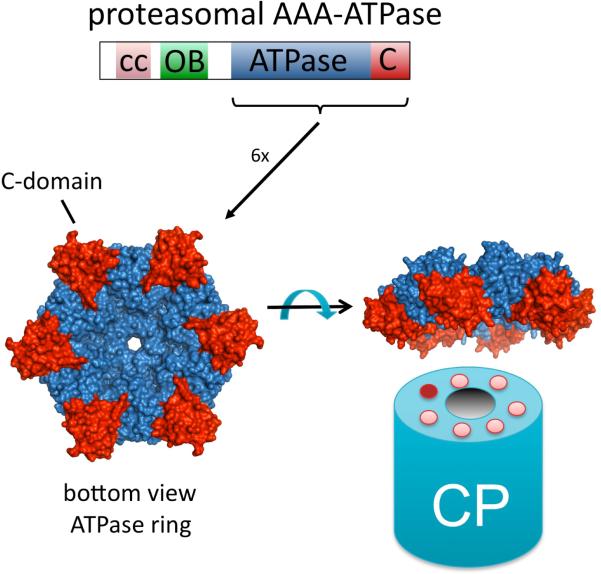
(A) Domain topology of Nas6, Rpn14, Hsm3 and Nas2. Below the topology is shown a 3D-structure of the domains to illustrate the structural difference. Only for Nas6 a structure been determined, for the others the structure of a similar domain from a different protein have been used (pdb coordinates used 1IXV for Nas6, 2H12 for WD40 repeat, 3GRL for Armadillo like repeats, 1G9O for PDZ domain). Hsm3 shows little conservation and detailed analysis of the domain can be found in Le Tallec et al.16. Surprisingly, while structurally unrelated, these chaperones bind to the same small C-domain in the AAA-ATPases. (B) The domain topology of the proteasomal AAA-ATPases. cc is coiled coil region, OB is the OB-domain, ATPase is the ATPase domain containing the walker A and walker B motifs and the C indicates the C-domain, typically found in AAA-ATPases behind the ATPase domain. The structure shown is a ring formed by the ATPase (Blue) and C-domains (red) of six proteasome-activating nucleotidase (PAN) AAA-ATPases from the archaea Methanocaldococcus jannaschii44. The CP cartoon is shown to illustrate the expected interface of the CP and the ATPase ring. Cartoons and structures are not drawn to scale.
Most AAA-ATPases form hexameric rings (Figure 2b) consisting of six identical ATPases. They appear to require no assistance in ring formation. The AAA-ATPase ring present in eukaryotic proteasomes, however, is formed by six different, but related, ATPases. This might suggest that the chaperones have evolved in order to regulate the formation of the hetero-hexameric ring. Other AAA-ATPase assemblies containing six different ATPases include dynein, midasin, and the MCM helicase complex. Dynein and midasin have all six ATPase domains in one polypeptide, providing physical restriction to the order of the ATPases38. The six MCM helicase proteins can assembly into a ring in vitro39 without the assistance of chaperones. However, a subset of subunits, e.g. MCM 4 and 6 or MCM 4, 6, and 7, can also form rings in vitro. This could indicate that there is a need for factors capable of directing ring assembly, although no chaperones have yet been identified. In summary, there is no precedent for the need of chaperones to assembly an AAA-ATPase hetero hexameric ring.
How the proteasomal chaperones assist in the ATPase ring formation is not fully understood and several potential mechanisms are discussed below. To assist directly in the formation of a complex each chaperone could either form a scaffold that binds multiple proteins or, alternatively, chaperone binding could induce a conformational change in a protein partner to create a new binding site: neither of these properties have been shown so far for these chaperones. Chaperones could also help assembly by stabilizing their binding partner(s), but RNAi studies in human tissue culture cell lines have shown that only depletion of Nas2 results in reduced levels of its binding partner24. Finally, chaperones could shield otherwise exposed surfaces. This property is important to avoid undesirable interactions (e.g. to avoid aggregation) or premature binding of other binding partners. The prevention of premature interactions has been observed (similar to Ump1 in CP assembly). The absence of human Nas2 causes a premature association of the Rpt4–5 complex with other base components24. Furthermore, the chaperones probably prevent premature binding of the Rpt subunits to the CP (Figure 3). Modelling studies suggest that binding of Nas6 to Rpt3 creates steric hindrance, so that the Rpt3 C-terminal tail cannot dock into its binding pocket on the CP21. Biochemical data support this and show a similar role for Rpn14 and Hsm321, 25. In a separate study it was found that human complexes containing p27 (Nas2) and S5b (Hsm3) did not bind CP. However, the role of the chaperones in this observation remains unexplored19. These processes are not mutually exclusive and each chaperone might employ a different set mechanisms.
Figure 3.
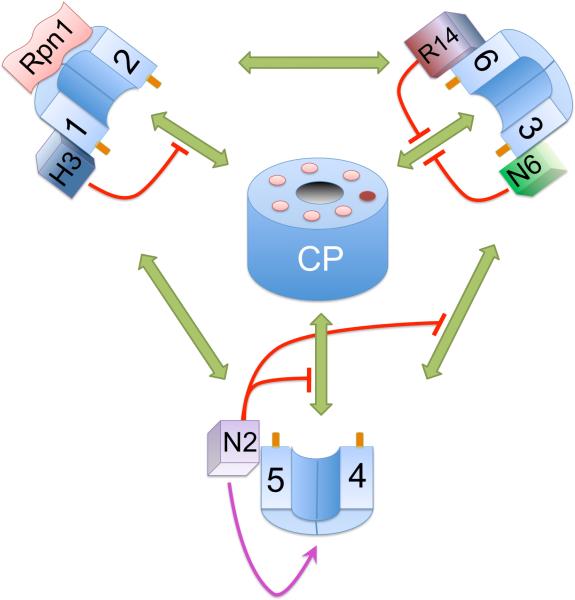
Cartoon showing the components known to be involved in the RP-assembly. The pathways and order of events are still unclear. Therefore an interaction map is displayed instead. The known potential interactions between the different complexes are shown in green. The chaperones are known to interfere with the interactions indicated in red, suggesting these proteins have a quality control role in assembly. Nas2 has also been shown to stabilize Rpt4 and 5 (purple arrow). In the middle of the cartoon are the seven pockets of the CP surrounding the gate (which is shown in an open conformation, although generally this is closed without activators bound to the CP). The dark pocket indicates the only pocket without a conserved positive charge expected not to be able to host a tail. Rpt tails that dock in the pockets are shown as dark orange extensions from the Rpt proteins. Numbers 1 to 6 indicate the different Rpt proteins, H3 Hsm3, R14 rpn14, N6 Nas6 and N2 Nas2.
Base assembly
The assembly of the base seems to occur by initial pairing of Rpt proteins (Figure 3). The Rpt1 and Rpt2 ATPases together with Rpn1 and Hsm3 have been repeatedly observed in cell lysates. The stability of this complex depends on Hsm321–25. Rpt5 might associate with this complex as well21. This entity, dubbed BP1, is a true assembly intermediate and not a dead-end product as it disappears in a chase experiment21. Most Rpt1-bound Hsm3 seems to associate with BP1 although some Hsm3 is found associated with base and RP23, 25. A second pair formed by Rpt4 and Rpt5 with the chaperone Nas2 has also been characterized in detail22, 23. Nas2 is important for the presence of this complex but dissociates upon further assembly. Rpt3 and Rpt6, the third pair of Rpt proteins, have been found in a complex with Nas6 and Rpn14, although these proteins have been more readily detected when associated with the base and/or RP23–25. Each pair interacts with at least three other subcomplexes in assembled proteasomes as indicated by the green arrows in Figure 3. In what order these interactions are established is not well understood.
Some models suggest that these complexes, together with Rpn2 and Rpn13, form the base22–24. In this process Hsm3 and Nas2 might dissociate. Upon association with the lid, RP is formed. When the RP associates with CP the Rpn14 and Nas6 proteins leave the RP, although Rpn14 dissociation might also occur at an earlier step. These models, however, do not address all observations. Mutants defective in CP assembly have shown defects in RP assembly14. Furthermore, deletion of the C-terminal amino acid of Rpt 4 and Rpt6 causes dramatic assembly defects21. The tails of the Rpt proteins dock into pockets in the CP40–42 and deleting the C-terminal amino acid disturbs this CP-Rpt interaction. Thus, there is a clear role for the CP in RP assembly (Figure 3). To ensure proper ordering of the Rpt proteins scaffolding proteins might play a role in assembly, however, such a role has not yet been shown for the chaperones. Rpn1 and Rpn2 are potential scaffolds21, 25. Rpn1 can bind to Rpt1 and Rpt2, thus it might act as a scaffold for the BP1. Rpn2, Rpt6 and Rpt3 interact14, 43, 44 but these proteins do not appear to form a precursor complex together23, 24. The CP itself might form another scaffold21, 25 with the pockets of the CP presumably having Rpt tail specificity40, 41 and thus directing Rpt proteins to distinct positions in the ring. With CP as a scaffold, the chaperones could regulate the incorporation of the different precursors. However, if the precursor complexes assemble on the CP, free base and free RP observed in cell lysates must have dissociated from the CP and rebound to some of the chaperones. Two other recent papers report insights into the assembly process without using the chaperones as starting points19, 45. Hendil, et al. used pulse chase in Hela cells to nicely show a rapid formation of an Rpt3-Rpt6 pair45. Furthermore, they argue for the formation of a precursor complex consisting of CP plus Rpn2, Rpn10, Rpn11, Rpn13 and Txnl1 (the last protein has recently been identified as associating with the proteasome) based on pulse chase and immunoprecipitation experiments. This proposed precursor complex has, however, not been biochemically identified. Their suggestion that pairs of Rpt proteins would subsequently join this assembly fits nicely with an important role for CP as a scaffold in RP assembly and a function for chaperones in regulating CP-Rpt interaction.
The second paper by Thompson, et al. identified three RP sub-complexes from red blood cells19. Consistent with the other studies, each sub-complex contains one Rpt pair. Ps-1 contains Rpt3 and Rpt6 together PAAF-1 and almost all RP subunits, except for Rpt1, 2, 4 and 5. Ps-2 is similar to BP-1, containing Rpn1, Rpt1, Rpt2 and S5b. Ps-3 is formed by Rpt4, Rpt5 and Nas2. In vitro these complexes can be reconstituted efficiently to form RP. While the role of these sub-complexes in assembly in vivo remains to be determined, it is very interesting that, apart from gankyrin, the RP chaperones are present in the different complexes. The role of the chaperones in reconstitution unfortunately has not been studied, but this study shows that CP is not required for this in vitro reconstitution.
In summary, it seems plausible that more than one pathway can lead to assembled 26S proteasomes. In demanding conditions, such as the high temperature used in one of the studies23, a single preferred 26S proteasome assembly pathway might be used. Although the proteasome assembly pathways appear largely conserved from yeast to man, there are probably subtle differences in the process between different organisms. Clearly, more detailed mechanistic and structural studies are needed to completely understand the assembly process.
In addition to CP and chaperones the role of other molecular players in assembly need to be characterized. Hsp90 stimulates lid complex and 26S maintenance and assembly, but it is unclear how46. Nob1 has a controversial role associating CP with RP in the nucleus47, 48. Ecm29 could tether CP and RP as it stabilizes their association even in the absence of ATP18, 49. The proteasome activator Blm10/PA200 has been reported to promote CP assembly and activation50. There is no reason to assume this list is complete and there are likely to be more chaperones or modulators of the assembly process.
Structure of the 26S proteasome
To understand proteasome assembly and function in general, structural information about the proteasome is essential. All high resolution structures of proteasome complexes have so far been of the CP or the CP associated with PA26 or Blm10, both proteasomal activators51–53. How the RP is organized is still unclear. EM studies of CP with the RP subunits Rpn1 and Rpn2 show the Rpn1 subunit of the 19S regulator on an interface with the central surface of the 20S core with the Rpn2 subunit on top of Rpn154. The ATPases then have to surround this Rpn1/2 core structure. However, this proposed architecture of the base has been challenged based on detailed EM structures of complete 26S proteasomes44, 55. These reports indicate that there is insufficient space within the ATPase ring to accommodate Rpn1/2. We also know that the C-termini of the Rpt proteins dock into pockets located on the CP, thus the Rpt proteins need to be in close proximity to the CP40–42. As has been observed previously, and described as “wagging”56, the recent EM structural papers also show a non-symmetrical positioning of the ATPase ring on the CP44, 55. This suggests that each Rpt may interact differently with the CP. Such asymmetry could be coordinated with the catalytic state of the ATPases. In summary, it is clear we still lack important understanding of how the six-member ATPase ring interacts with the seven-member α ring of the CP.
Additionally, a previously proposed order of the proteasomal ATPases of Rpts 2-1-3-5-4-6,43, 57 seems in conflict with some of the assembly data described above. It has also been challenged recently using structural information from PAN-AAA-ATPase structures (Figure 2b)44, 58, 59. Extrapolating from this data, the ATPase ring should be regarded as a trimer of dimers in which the different subunits of each dimer need to have a different conformation of their coiled-coil domain. Based on this information the following order of ATPases has been suggested: Rpt1-2-6-3-4-544, 55. Although this order remains to be confirmed experimentally, it shows nice consistency with the Rpt pairs that form during assembly (Figure 3), the evolutionary relationship between the Rpt proteins60, and many of the published interaction data44, 55.
Function: how does the 26S proteasome perform its role?
Conformation changes in assembled proteasomes
The mechanism by which the 26S proteasome engages ubiquitinated proteins and feeds them into the 20S catalytic core is not understood.
It has been suggested that yeast 26S particles may dissociate during the catalytic cycle61, which might suggest a role of some of the chaperons in the catalytic cycle. However, it appears that (at least mammalian) 26S proteasomes can degrade polyubiquitinated proteins without dissociation62. Also for yeast proteasomes it has been shown that engagement of the proteolytic active sites actually stabilizes the 26S complex18. In either case, the six ATPases in the base of the regulatory particle may undergo conformational changes during ATP binding and hydrolysis63. These conformational changes may be used for substrate unfolding and translocation2.
The binding of polyubiquitinated substrates can cause small structural changes in the proteasome. Substrates targeted for degradation are generally modified with at least an ubiquitin chain-length of four ubiquitins. These chains bind to the 26S particle64 and an unstructured region initiates the degradation of the protein65 or sometimes of a binding partner66. Polyubiquitinated model substrates bound to the proteasome enhance their own degradation by facilitating gate opening and allosterically activating the peptidase activities67. Thus, assembled proteasomes are dynamic structures that undergo conformational changes during degradation as well as substrate binding.
The role of ubiquitin binding and chains
Substrates can be delivered to the proteasome via different paths as well as with different types of ubiquitin chains. At least two subunits of the regulatory particle, Rpn13/ADRM1 and Rpn10/S5a, bind polyubiquitinated proteins, but roles for Rpn1 and Rpt 5 have also been suggested2, 3. In yeast, a large amount of Rpn10/S5a is not associated with the 26S proteasome which may imply that this protein has a function in substrate shuttling to the 26S particle68. There are also many extra-proteasomal ubiquitin binding proteins which may shuttle polyubiquitinated proteins to the 26S proteasome for degradation, e.g. Rad23 and Dsk2. Many of these proteins contain ubiquitin-like (Ubl) domains, which may bind to the proteasomal ubiquitin receptors, and an ubiquitin binding domain (UbA), to bind to polyubiquitinated substrates.
Interestingly, when DSK2 is induced (over-expressed) Lys 48-linked ubiquitin conjugates accumulate with impaired proteolysis and cytotoxicity. These data suggest that the excess of Dsk2 might bind and sequester Lys-48-linked chains away from the proteasome. Alternatively, the binding of Dsk2 to the proteasome, e.g. via Rpn10/S5a, may interfere sterically with the binding of substrate-bound Ubl-containing shuttle proteins or Lys 48-linked chains of so modified substrates69. It is not excluded either that Dsk2 might stimulate the degradation of substrates linked via non-canonical chains, i.e. other than Lys 48-based. Ubiquitin has seven lysine residues (and a free amino terminus) all of which can be involved in ubiquitin chain formation. Chain linkages can be recognised by many proteins containing ubiquitin binding domains70 and by deubiquitinating enzymes71. Under the conditions used in the Dsk2 study the majority of conjugated ubiquitin in the cell lysate was linked via Lys 48 and Lys 6369. Lys-63 linked chains are generally considered to be involved in non-proteolytic functions72. Interestingly, the 26S proteasome does bind Lys 63 as well as Lys 48 polyubiquitinated proteins73. Also substrates modified with Lys 63-linked chains by the yeast ubiquitin protein ligase Rsp5 in vitro, are degraded by the 26S proteasome. Furthermore, the membrane anchored transcription factor precursor Mga2, an Rsp5 substrate, contains high levels of Lys63-linked chains in vivo that are sufficient for Mga2 processing by the 26S proteasome73. Thus, Lys 63-linked chains are not necessary excluded from proteasomal degradation.
In contrast to the earlier described paper69, Xu et al found that unconventionally-linked ubiquitin chains are abundantly attached to proteins in yeast under their experimental conditions, with Lys 11 (28%) being almost as abundant as Lys 48 (29%)74. Their data suggests that all the non-Lys 63-based linkages may target proteins for degradation by the 26S proteasome. Specific roles for Lys 11-linked chains have been reported, e.g. the anaphase promoting complex (APC/C) triggers substrate degradation by assembling Lys 11-linked ubiquitin chains75. Another example, in yeast, is that Ubc6 attaches Lys 11-linked ubiquitin chains to proteins extracted from the endoplasmic reticulum (ER) for ER-associated degradation (ERAD)74.
Recent studies corroborate these findings since UBX proteins that deliver ubiquitinated proteins in ERAD to the cdc48/VCP/p97 complex for transfer to the 26S proteasome associate more prominently with Lys 11 linkages than Lys 48 linkages74. The p97 complex can assemble with all of the 13 known mammalian UBX-domain proteins. The UBX proteins that bind ubiquitin conjugates also interact with dozens of E3 ubiquitin ligases. The large number of ubiquitin ligases found associated with UBX proteins suggests that p97 plays a far broader role than previously anticipated in the global regulation of protein turnover76. This presumably explains why mutations in p97 cause a progressive autosomal dominant disorder called inclusion body myopathy (IBM) associated with Paget disease of the bone (PDB) and frontotemporal dementia (FTD)77.
Finally, there appear to be differences in the linkage-repertoire of ubiquitin chains in proteins in yeast and mammalian cells. Exemplifying this, when the yeast 26S proteasome is inhibited chemically or genetically there is no change in Lys 63 linkage levels74. In contrast, in cultured mammalian cells accumulation of Lys 63-linked chains occurs, although at a slower rate than Lys 48 or Lys 11 chains, when proteasomes are inhibited by MG13278. In Huntington's disease, Lys 63- and Lys 11-linked polyubiquitin chains also accumulate in human brain (with an early accumulation of Lys 48 chains) as well as in brains of the R6/2 mouse model of the disease79. Lys 63 specific antibodies show the accumulation of Lys 63-linked ubiquitinated proteins in inclusions in Huntington's disease and in neurofibrillary tangles in Alzheimer's disease but rarely in Lewy bodies in Parkinson's disease. However, neuronal inclusions in brain, resembling “pale bodies”, which are considered precursors of Lewy bodies, contain Lys 63-linked chains in a mouse model of neurodegeneration80 when the 26S proteasome is genetically obliterated81, 82. These contrasting findings may indicate that Lys 63 linked ubiquitin chains are utilised more frequently for proteasomal degradation in mammalian cells than in yeast74.
Promiscuous 26S proteasomal subunits
To understand the full range of proteasomal functions it is necessary to discover all the proteins in the cell that interact with the 26S proteasome as well as those proteins that may exist freely or in other complexes in the cell besides interacting with the 26S proteasome (in addition to proteins that may bring ubiquitinated proteins to the proteasome for degradation, e.g Rpn10 and RAD23). In yeast, quantitative analyses of tandem affinity-purified cross-linked protein complexes has shown that at least 471 proteins, including weak and transient interactors, associate with the proteasome83. As might be expected, there are also an increasing number of proteins that are found in other complexes in the cell as well as in the 26S proteasome. Many of these proteins are known to have roles in human disease (Table 2).
Table 2.
26S proteasomal interactors and Human Disease
| Name | Disease |
|---|---|
| DSS1/SEM1 | Developmental abnormality (Deleted in split hand/split foot) |
| Gankyrin/Nas6p | overexpresed oncoprotein (liver and colon cancer) |
| P97/cdc48 | Paget's disease of bone, dementia, inclusion body myositis |
See text for functions of the proteins
One such example is the human DSS1 protein (deleted in split hand/split foot-1) known in yeast as suppressor of exocytosis mutations (SEM1). The Sem1 protein is found in at least three protein complexes in the cell: with the COP9/signalosome-associated protein Csn12 in an mRNA splicing complex, in a Thp1/Sac3 mRNA export/transcription complex, and in the proteasome84, 85. Some protein subunits of these complexes are structurally related, e.g. the different subunits of the proteasomal lid and those of the COP9/signalosome are structurally highly homologous. In the proteasome Sem1/DSS1 binds to the Rpn3 subunit of the lid sub-complex and maintains the stability of the 26S proteasome86–89.
Chromatin immunoprecipitation in yeast shows that Sem1 is recruited along with 19S and 20S proteasomes and double-strand break (DSB) repair proteins to DSBs in vivo suggesting a direct role for proteasomes in DSB repair90. Interestingly, DSS1 interacts with the breast cancer protein BRCA2 (breast cancer associated gene 2)91, 92 and both DSS1 and BRCA2 participate in DSB repair92, 93. BRCA2 is part of a large BRCC (BRCA1- and BRCA2-containing complex) complex that has ubiquitin protein ligase activity94. Thus, DSS1 may serve to anchor an ubiquitin ligase involved in DNA repair to the 26S proteasome. The very acidic region in Sem1/Dss1 might control access of ssDNA to BRCA2 during double strand break repair or more generally control access of nucleic acids to Sem1/DSS1 containing complexes84, 92. Adding to the multiplicity of its role is the recent finding that Sem1 and the proteasome-associated deubiquitinating enzyme Ubp6 are involved in telomeric silencing by modulating histone ubiquitination and acetylation and therefore recruitment of silencing factors95. On top of this, it appears that 26S proteasomes containing DSS1 bind and target p53 for ubiquitin-mediated degradation via the gankyrin-MDM2 pathway88.
The oncoprotein gankyrin, which is over-expressed in hepatocellular carcinoma, also seems to have multiple distinct functions. It is one of the RP chaperones described earlier, but it also binds to the retinoblastoma protein (pRb), cyclin-dependent kinase 4 (CDK4), the p53-ubiquitylating enzyme MDM2 and the RelA subunit of NFκB32, 96–98.
It is currently unclear how gankyrin, a chaperone involved in 19S regulator biogenesis, yet not present in 26S proteasomes, can connect 26S proteasomes to the stability and function of pRb, CDK4, MDM2 and RelA. However, when the C-terminus of Rpt3 does not engage in its complementary pocket in the CP, gankyrin/Nas6 remains bound to the 26S proteasome21, 25 providing a potential mechanism for gankyrin association with the 26S particle. Furthermore pRb, CDK4, MDM2 and RelA are not found in yeast, leaving open the possibility of pleiotypic functions of gankyrin and the 26S proteasome in higher eukaryotic cells and their relationship to cell physiology and disease. How these different functions of gankyrin are regulated and how they relate to the oncogenicity of gankyrin remain important questions to address.
A third protein with apparent functions in different complexes is the 26S proteasomal lid subunit Rpn11 (human POH1). This deubiquitylating enzyme has a function through its C-terminal residues in controlling mitochondrial structure and function independent of the N-terminal MPN+/JAMM catalytic motif99. A pool of non-proteasomal Rpn11 that associates with mitochondria may exert this function100. Clearly, given the importance of mitochondrial dynamics in normal cell biology and disease processes, e.g. optic atrophy101, further studies to corroborate the non-proteasomal activities of Rpn11/POH1 and mitochondria are needed in yeast and in human cells.
20S proteasomes and protein degradation in the cell
Studies in reticulocyte lysates have indicated that added purified 20S proteasomes can degrade more than 20% of extracted proteins102. More specifically, 20S proteasomes may degrade oxidized or otherwise damaged proteins103, 104 as found in several diseases such as inflammation, ischemia, and neurodegeneration. Interestingly, in yeast an aberrant form of free 20S proteasomes has been observed in mutants of the 20S chaperone PAC3-PAC4. Mutants also show an increased resistance to high levels of cadmium14. Further studies will have to address if the aberrant 20S proteasome has a role in the acquired resistance and whether wild type cells can regulate the formation of this 20S form. Studying neurons in mouse brains, the conditional regional ablation of a proteasomal ATPase gene causes depletion of 26S but not 20S proteasomes and recapitulates the neuropathology and neurochemical deficits of Parkinson's disease and dementia with Lewy bodies. There might be many reasons why reduced 26S proteasome levels in these neurons leads to cell death because of the extensive different activities of 26S proteasomes in the cell. However, remaining 20S proteasomes are not sufficient for neuronal viability81, 105. Besides important cellular roles for 26S proteasomes, free 20S proteasomes, and free RP has several reported roles within the cell, for example in transcriptional elongation106. The RP-chaperones described earlier might play a role in regulating cellular free RP, however this aspect has only briefly been touched upon up to now25. It will be interesting to see how functions of free RP are impacted in yeast strains with the strongly reduced RP levels in chaperone mutants. With the presence of cellular functions for free RP and 20S proteasomes, in addition to all the important roles of the 26S proteasome, it is clear that it will be essential to understand what regulates the levels of 20S, RP, and 26S in cells. These levels will depend on the assembly pathways (with important roles for the chaperones), but will also be affected by the equilibrium between 20S proteasomes and other activators such as BLM10/PA200 or PA28/REG/11S. These dynamics and interactions are still poorly understood.
Concluding remarks
In the last 21 years it has become clear that the cellular “multicatalytic proteinase”, conserved from archaebacteria to eukaryotes107, has central roles in all aspects of cell physiology. The assembly, structure and function of the protein degradative 26S proteasome is now seen to be as sophisticated as its cellular protein synthesising counterpart the ribosome. The discovery of chaperones involved in the biogenesis of both the CP and RP serves to illustrate the complexity of the assembly process but still leaves open questions concerning alternative routes to the building of the mega structure. There is still a need to determine the complete structure of the 26S proteasome, but given the natural dynamic fluctuation of the complete particle56, this may prove difficult. In higher eukaryotic cells with multiple compartments the outstanding questions include the elucidation of the interactions of 26S proteasomes with cellular organelles and movement between compartments (Box 2). Nowhere can this complexity be better seen than in the nervous system where the movement of 26S proteasomes into dendritic spines108 to facilitate synaptic plasticity and memory is controlled by the interaction of the particles with an abundant regulatory enzyme of the nervous system called calcium calmodulin dependent kinase IIα that regulates memory deposition109. The world of proteasomal interactors and dynamics deserves much more investigation.
Text Box 1 The ubiquitin proteasome system.
The ubiquitin proteasome system is the major cellular system for the regulated degradation of proteins in the cell. Proteins are ubiquitinated as a signal for degradation by the 26S proteasome. Protein ubiquitination has many other roles in the cell including the regulation of chromatin structure, nucleic acid metabolism, autophagy, and receptor mediated endocytosis111.
The ubiquitination of proteins is carried out by the sequential activities of specific enzymes: ubiquitin activating enzymes (E1), ubiquitin conjugating enzymes (E2), and ubiquitin protein ligases (E3). There are also many deubiquitinating enzymes that remove ubiquitin from proteins. The E3 ligases are the ultimate arbiters of protein stability in the cell. There are similar numbers of E3 ligases in the cell (~600 hundred) as there are protein kinases. Protein ubiquitination/deubiquitination and protein phosphorylation/dephosphorylation are intimately linked to control all aspects of cell physiology, as are other functional group and functional protein post-translational protein modifications.
The Proteasome
The proteasome is the major cellular protease. The proteolytically active sites reside within the 20S core particle (CP), a cylindrically shaped structure formed by four stacked rings, ordered α1–7, β1–7, β1–7, α1–7. Subunits β2, β5 and β1 have proteolytic activity, displaying trypsin-like, chymotrypsin-like and caspase-like peptidase activity respectively. Importantly, these active sites are sequestered to the inside of the CP to protect the cell from non-specific degradation. The N-termini of the αsubunits close the ends of the cylinder, often referred to as the gate. While some proteins can be degraded by the CP, most degradation depends on ATP. The only ATP hydrolyzing proteins in the proteasome are the six ATPases present in the regulatory particle (RP). The 19 subunits of the RP are divided between two sub-structures, the base and lid complexes. The lid contains the Rpn11 subunit that has deubiquitinating activity. The base consists of a ring of AAA-ATPases. The C-termini of these ATPases dock into pockets present between the α subunits of the CP. This results in a translocation of the α tails and an opening of the gate. An interesting architectural problem is that this six-membered ATPase ring sits on a seven-membered α–ring. Although sequence alignments suggest that one pocket remains empty, it's not clear how these rings interdigitate. The CP can associate with one or two RPs, but there are other activators that may also bind to the 20S CP particle. These include the Blm10/PA200 protein that is a large heat-repeat containing protein and the 11S/PA28 complex, itself a heteroheptamer, either of which may dock directly with the CP.
Box 2 Outstanding Questions.
Outstanding questions in proteasomal cell biology are: (i) how many protein assemblies, including 19S, 20S and 26S proteasomal complexes, control key events in the cell like DNA synthesis, DNA repair and transcription?; (ii) how are the presence and dynamics of these different interdependent assemblies regulated?; and (iii) when are 26S proteasomes (or sub-complexes) involved in these processes in a non-proteolytic or proteolytic manner?
Acknowledgements
We would like to thank Daniel Finley and Stella Y. Lee for critically reading the manuscript. J.R. is supported in part by grant number P20 RR016475 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). L.B. and R.J.M. thank the Parkinson's Disease Society and the Alzheimer's Research Trust for support of some of the studies.
Glossary Box
- Proteostasis(protein homeostasis)
a process regulating proteins within the cell in order to maintain the health of both proteome and the organism itself and maintained by a highly inter-connected network comprising pathways that control protein synthesis, folding, trafficking, aggregation, disaggregation, and degradation.
- Ubiquitin
a small covalent protein modifier that is used to modify (ubiquitinate) proteins in the cell by a process similar to that of protein phosphorylation.
- Ubiquitin chain
an ubiquitin attached to a lysine within another ubiquitin creates an ubiquitin chain. Often chains are referred to based on the sequence number of the lysine in ubiquitin to which a subsequent ubiquitin is attached. Chains can be found with single and mixed linkages (e.g. K11 and K48 linkage together in a chain). Besides linear chains, also forked chains exist, in which one ubiquitin has two or more ubiquitins directly attached to different lysines within a single ubiquitin. Ubiquitin chains can be attached to proteins or found free in the cell110.
- 26S proteasome
a large protein complex that recognizes and degrades polyubiquitinated proteins.
- 20S proteasome
the catalytic core of the 26S proteasome consisting of a α7β7β7α7 cylindrical complex. Commonly referred to as core particle (CP).
- 19S regulator
a multiprotein complex that can be attached to one or both ends of the 20S CP for the recognition of ubiquitinated proteins, their deubiquitination and their subsequent degradation in the 20S core. Commonly referred to as regulatory particle (RP).
- Polyubiquitination
modification of a protein by a chain of polyubiquitin anchored to a single lysine residue on a target protein.
- 19S regulator structure
this consists of a base complex and lid complex. The base contains a putative ring of six ATPases and two non-ATPases. The lid contains many proteins, including those with deubiquitinating activity, involved in the recognition of ubiquitinated proteins and their processing for eventual degradation.
- ATPase
an enzyme that hydrolyses ATP for a biochemical process.
- Proteasomal ATPases
proteasome subunits that are members of the AAA (ATPases associated with various cellular activities) superfamily of ATPases that control numerous processes in the cell.
- Chaperone
a protein that assists in the (un)folding or (dis)assembly of other proteins, but that is not part of the final biological complex.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Balch WE, et al. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murata S, et al. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 4.Kusmierczyk AR, Hochstrasser M. Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol Chem. 2008;389:1143–1151. doi: 10.1515/BC.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marques AJ, et al. Catalytic mechanism and assembly of the proteasome. Chem Rev. 2009;109:1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 6.Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Apcher GS, et al. The alpha4 and alpha7 subunits and assembly of the 20S proteasome. FEBS Lett. 2004;569:211–216. doi: 10.1016/j.febslet.2004.05.067. [DOI] [PubMed] [Google Scholar]
- 8.Gerards WL, et al. The human alpha-type proteasomal subunit HsC8 forms a double ringlike structure, but does not assemble into proteasome-like particles with the beta-type subunits HsDelta or HsBPROS26. J Biol Chem. 1997;272:10080–10086. doi: 10.1074/jbc.272.15.10080. [DOI] [PubMed] [Google Scholar]
- 9.Yao Y, et al. alpha5 subunit in Trypanosoma brucei proteasome can self-assemble to form a cylinder of four stacked heptamer rings. Biochem J. 1999;344(Pt 2):349–358. doi: 10.1042/0264-6021:3440349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velichutina I, et al. Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. Embo J. 2004;23:500–510. doi: 10.1038/sj.emboj.7600059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirano Y, et al. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- 12.Hirano Y, et al. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol Cell. 2006;24:977–984. doi: 10.1016/j.molcel.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Hirano Y, et al. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. Embo J. 2008;27:2204–2213. doi: 10.1038/emboj.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kusmierczyk AR, et al. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat Struct Mol Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 15.Le Tallec B, et al. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol Cell. 2007;27:660–674. doi: 10.1016/j.molcel.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 16.Yashiroda H, et al. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat Struct Mol Biol. 2008;15:228–236. doi: 10.1038/nsmb.1386. [DOI] [PubMed] [Google Scholar]
- 17.Effantin G, et al. Electron microscopic evidence in support of alpha-solenoid models of proteasomal subunits, Rpn1 and Rpn2. J Mol Biol. 2009;386:1204–1211. doi: 10.1016/j.jmb.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleijnen MF, et al. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat Struct Mol Biol. 2007;14:1180–1188. doi: 10.1038/nsmb1335. [DOI] [PubMed] [Google Scholar]
- 19.Thompson D, et al. Subcomplexes of PA700, the 19 S regulator of the 26 S proteasome, reveal relative roles of AAA subunits in 26 S proteasome assembly and activation and ATPase activity. J Biol Chem. 2009;284:24891–24903. doi: 10.1074/jbc.M109.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Tallec B, et al. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Park S, et al. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Funakoshi M, et al. Multiple Assembly Chaperones Govern Biogenesis of the Proteasome Regulatory Particle Base. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeki Y, et al. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko T, et al. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 25.Roelofs J, et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isono E, et al. Rpn7 Is required for the structural integrity of the 26 S proteasome of Saccharomyces cerevisiae. J Biol Chem. 2004;279:27168–27176. doi: 10.1074/jbc.M314231200. [DOI] [PubMed] [Google Scholar]
- 27.DeMartino GN, et al. Identification, purification, and characterization of a PA700-dependent activator of the proteasome. J Biol Chem. 1996;271:3112–3118. doi: 10.1074/jbc.271.6.3112. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe TK, et al. cDNA cloning and characterization of a human proteasomal modulator subunit, p27 (PSMD9) Genomics. 1998;50:241–250. doi: 10.1006/geno.1998.5301. [DOI] [PubMed] [Google Scholar]
- 29.Gorbea C, et al. Mapping subunit contacts in the regulatory complex of the 26 S proteasome. S2 and S5b form a tetramer with ATPase subunits S4 and S7. J Biol Chem. 2000;275:875–882. doi: 10.1074/jbc.275.2.875. [DOI] [PubMed] [Google Scholar]
- 30.Hori T, et al. cDNA cloning and functional analysis of p28 (Nas6p) and p40.5 (Nas7p), two novel regulatory subunits of the 26S proteasome. Gene. 1998;216:113–122. doi: 10.1016/s0378-1119(98)00309-6. [DOI] [PubMed] [Google Scholar]
- 31.Verma R, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawson S, et al. Gankyrin is an ankyrin-repeat oncoprotein that interacts with CDK4 kinase and the S6 ATPase of the 26 S proteasome. J Biol Chem. 2002;277:10893–10902. doi: 10.1074/jbc.M107313200. [DOI] [PubMed] [Google Scholar]
- 33.Park Y, et al. Proteasomal ATPase-associated factor 1 negatively regulates proteasome activity by interacting with proteasomal ATPases. Mol Cell Biol. 2005;25:3842–3853. doi: 10.1128/MCB.25.9.3842-3853.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lassot I, et al. The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol Cell. 2007;25:369–383. doi: 10.1016/j.molcel.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 35.Dawson S, et al. Gankyrin: a new oncoprotein and regulator of pRb and p53. Trends Cell Biol. 2006;16:229–233. doi: 10.1016/j.tcb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Schultz J, et al. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letunic I, et al. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ammelburg M, et al. Classification of AAA+ proteins. J Struct Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Davey MJ, et al. Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J Biol Chem. 2003;278:4491–4499. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- 40.Gillette TG, et al. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabl J, et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith DM, et al. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartmann-Petersen R, et al. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch Biochem Biophys. 2001;386:89–94. doi: 10.1006/abbi.2000.2178. [DOI] [PubMed] [Google Scholar]
- 44.Forster F, et al. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem Biophys Res Commun. 2009;388:228–233. doi: 10.1016/j.bbrc.2009.07.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hendil KB, et al. The 20S proteasome as an assembly platform for the 19S regulatory complex. J Mol Biol. 2009;394:320–328. doi: 10.1016/j.jmb.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 46.Imai J, et al. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. Embo J. 2003;22:3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tone Y, Toh EA. Nob1p is required for biogenesis of the 26S proteasome and degraded upon its maturation in Saccharomyces cerevisiae. Genes Dev. 2002;16:3142–3157. doi: 10.1101/gad.1025602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fatica A, et al. Nob1p is required for cleavage of the 3' end of 18S rRNA. Mol Cell Biol. 2003;23:1798–1807. doi: 10.1128/MCB.23.5.1798-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leggett DS, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 50.Marques AJ, et al. The C-terminal extension of the beta7 subunit and activator complexes stabilize nascent 20 S proteasomes and promote their maturation. J Biol Chem. 2007;282:34869–34876. doi: 10.1074/jbc.M705836200. [DOI] [PubMed] [Google Scholar]
- 51.Groll M, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 52.Whitby FG, et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 53.Sadre-Bazzaz K, et al. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol Cell. 37:728–735. doi: 10.1016/j.molcel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenzweig R, et al. The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nickell S, et al. Insights into the molecular architecture of the 26S proteasome. Proc Natl Acad Sci U S A. 2009;106:11943–11947. doi: 10.1073/pnas.0905081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walz J, et al. 26S proteasome structure revealed by three-dimensional electron microscopy. J Struct Biol. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- 57.Ferrell K, et al. Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem Sci. 2000;25:83–88. doi: 10.1016/s0968-0004(99)01529-7. [DOI] [PubMed] [Google Scholar]
- 58.Djuranovic S, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 59.Zhang F, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wollenberg K, Swaffield JC. Evolution of proteasomal ATPases. Mol Biol Evol. 2001;18:962–974. doi: 10.1093/oxfordjournals.molbev.a003897. [DOI] [PubMed] [Google Scholar]
- 61.Babbitt SE, et al. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. doi: 10.1016/j.cell.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 62.Kriegenburg F, et al. Mammalian 26S proteasomes remain intact during protein degradation. Cell. 2008;135:355–365. doi: 10.1016/j.cell.2008.08.032. [DOI] [PubMed] [Google Scholar]
- 63.Horwitz AA, et al. ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J Biol Chem. 2007;282:22921–22929. doi: 10.1074/jbc.M702846200. [DOI] [PubMed] [Google Scholar]
- 64.Pickart CM. Ubiquitin in chains. Trends Biochem Sci. 2000;25:544–548. doi: 10.1016/s0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- 65.Prakash S, et al. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 66.Prakash S, et al. Substrate selection by the proteasome during degradation of protein complexes. Nat Chem Biol. 2009;5:29–36. doi: 10.1038/nchembio.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bech-Otschir D, et al. Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome. Nat Struct Mol Biol. 2009;16:219–225. doi: 10.1038/nsmb.1547. [DOI] [PubMed] [Google Scholar]
- 68.Fu H, et al. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J Biol Chem. 1998;273:1970–1981. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- 69.Matiuhin Y, et al. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell. 2008;32:415–425. doi: 10.1016/j.molcel.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dikic I, et al. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Komander D, et al. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 72.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 73.Saeki Y, et al. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. Embo J. 2009;28:359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin L, et al. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alexandru G, et al. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kimonis VE, et al. VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim Biophys Acta. 2008;1782:744–748. doi: 10.1016/j.bbadis.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 78.Meierhofer D, et al. Quantitative analysis of global ubiquitination in HeLa cells by mass spectrometry. J Proteome Res. 2008;7:4566–4576. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bennett EJ, et al. Global changes to the ubiquitin system in Huntington's disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 80.Paine S, et al. Immunoreactivity to Lys63-linked polyubiquitin is a feature of neurodegeneration. Neuroscience Letters. 2009 doi: 10.1016/j.neulet.2009.05.074. [DOI] [PubMed] [Google Scholar]
- 81.Bedford L, et al. Is malfunction of the ubiquitin proteasome system the primary cause of alpha-synucleinopathies and other chronic human neurodegenerative disease? Biochim Biophys Acta. 2008;1782:683–690. doi: 10.1016/j.bbadis.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 82.Bedford L, et al. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci. 2008;28:8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guerrero C, et al. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci U S A. 2008;105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilmes GM, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Faza MB, et al. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J Cell Biol. 2009;184:833–846. doi: 10.1083/jcb.200810059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sone T, et al. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J Biol Chem. 2004;279:28807–28816. doi: 10.1074/jbc.M403165200. [DOI] [PubMed] [Google Scholar]
- 87.Funakoshi M, et al. Sem1, the yeast ortholog of a human BRCA2-binding protein, is a component of the proteasome regulatory particle that enhances proteasome stability. J Cell Sci. 2004;117:6447–6454. doi: 10.1242/jcs.01575. [DOI] [PubMed] [Google Scholar]
- 88.Wei SJ, et al. Identification of a specific motif of the DSS1 protein required for proteasome interaction and p53 protein degradation. J Mol Biol. 2008;383:693–712. doi: 10.1016/j.jmb.2008.08.044. [DOI] [PubMed] [Google Scholar]
- 89.Sharon M, et al. Structural organization of the 19S proteasome lid: insights from MS of intact complexes. PLoS Biol. 2006;4:e267. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krogan NJ, et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 91.Marston NJ, et al. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol. 1999;19:4633–4642. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang H, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 93.Kojic M, et al. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2003;12:1043–1049. doi: 10.1016/s1097-2765(03)00367-8. [DOI] [PubMed] [Google Scholar]
- 94.Dong Y, et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12:1087–1099. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 95.Qin S, et al. Sem1p and Ubp6p orchestrate telomeric silencing by modulating histone H2B ubiquitination and H3 acetylation. Nucleic Acids Res. 2009;37:1843–1853. doi: 10.1093/nar/gkn1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Higashitsuji H, et al. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat Med. 2000;6:96–99. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- 97.Higashitsuji H, et al. The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell. 2005;8:75–87. doi: 10.1016/j.ccr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 98.Higashitsuji H, et al. The oncoprotein gankyrin interacts with RelA and suppresses NF-kappaB activity. Biochem Biophys Res Commun. 2007;363:879–884. doi: 10.1016/j.bbrc.2007.09.072. [DOI] [PubMed] [Google Scholar]
- 99.Rinaldi T, et al. Dissection of the carboxyl-terminal domain of the proteasomal subunit Rpn11 in maintenance of mitochondrial structure and function. Mol Biol Cell. 2008;19:1022–1031. doi: 10.1091/mbc.E07-07-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rinaldi T, et al. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem J. 2004;381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Knott AB, et al. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baugh JM, et al. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J Mol Biol. 2009;386:814–827. doi: 10.1016/j.jmb.2008.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Strosova M, et al. Limited degradation of oxidized calmodulin by proteasome: formation of peptides. Arch Biochem Biophys. 2008;475:50–54. doi: 10.1016/j.abb.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 104.Jung T, Grune T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life. 2008;60:743–752. doi: 10.1002/iub.114. [DOI] [PubMed] [Google Scholar]
- 105.Bedford L, et al. The UPS and autophagy in chronic neurodegenerative disease: six of one and half a dozen of the other--or not? Autophagy. 2009;5:224–227. doi: 10.4161/auto.5.2.7389. [DOI] [PubMed] [Google Scholar]
- 106.Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 107.Dahlmann B, et al. The multicatalytic proteinase (prosome) is ubiquitous from eukaryotes to archaebacteria. FEBS Lett. 1989;251:125–131. doi: 10.1016/0014-5793(89)81441-3. [DOI] [PubMed] [Google Scholar]
- 108.Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–1148. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- 109.Bingol B, et al. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 140:567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 110.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Welchman RL, et al. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nature Reviews in Molecular Cell Biology. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]