Abstract
The in vivo association of transcription factors with the metallothionein-I promoter was examined using chromatin immunoprecipitation (ChIP) assays. The results demonstrated that c-fos is rapidly recruited along with the metal response element-binding transcription factor-1 (MTF-1) to this promoter in response to zinc or cadmium, and that this recruitment is reversed in the visceral yolk sac by a zinc-deficient diet in vivo, and in cultured cells after lowering the zinc concentration in the medium or during prolonged zinc exposure. In contrast, the interactions of c-jun, USF-1, USF-2 and Sp1 with this promoter are metal-independent. Studies of knockout cells revealed that the recruitment of c-fos to the MT-I promoter requires MTF-1, but that c-fos is not essential for recruitment of MTF-1 and metal-induction of MT-I gene expression. Studies of Hepa cells stably-transfected with reporter genes driven by the MT-I promoter suggested two in vivo binding sites for USF-1 and -2. In contrast, Sp1 was apparently associated with a single binding site (upstream of –153 bp). In addition, maximal recruitment of c-fos by metals required sequences and/or other proteins that interact upstream of –153 bp. In summary, these studies extend our understanding of the complexity and dynamics of the transcription factor complex that forms at the MT-I promoter in vivo in response to metals.
INTRODUCTION
The heavy metals zinc and cadmium can induce the expression of a myriad of genes, including those coding for metallothioneins (MT-I) (1). In the mouse, MT-I and MT-II participate in zinc homeostasis and protection against heavy metal toxicity and oxidative stress (2). Studies of the molecular mechanisms of regulation of MT-I gene expression by heavy metals have focused primarily on the MRE-binding transcription factor-1 (MTF-1) (for reviews, see 3–5). However, upstream stimulatory factor-1 (USF-1), Sp1, and as yet unidentified coactivators have also been implicated in regulation of the mouse MT-I gene (5–7).
In the mouse, the availability of dietary zinc regulates MT-I expression in the embryonic visceral endoderm as well as in other tissues, including the intestine and pancreas (8,9). While MTF-1 is required for MT-I gene expression in the visceral endoderm, loss of USF-1 attenuates, but does not eliminate gene expression (10). This strongly suggests functional cooperativity between MTF-1 and USF-1 in regulating MT-I gene expression in response to zinc. MTF-1 is also essential for basal and metal-induced MT-I gene activation in cultured cells (11). Within the proximal promoter, the five copies of MREs confer metal responsiveness, and there is ample evidence that MTF-1 binds in vivo to these elements in response to oxidative stress or zinc or cadmium treatment of cultured cells (6,12,13). In contrast, results from in vitro DNA binding assays identified two potential USF-1 binding sites in the proximal 250 bp of the MT-I promoter; one at an E-Box element (centered at –220 bp) and the other at a more complex site (–89 to –101 bp) that also includes an anti-oxidant response element. This USF/ARE contributes to maximal activation of gene expression by cadmium, but not by zinc, in transiently transfected cells (7). However, results from that study showed increased binding activity of the ARE, but not the USF-binding element, in response to cadmium, suggesting that a transcription factor other than USF may bind to this element. Indirect evidence also suggests that Sp1 may constitutively bind to the proximal MT-I promoter. Two GC-boxes, which are potential Sp1-binding sites, have been identified, one centered at –183 bp, the other overlapping the MRE-d (6,14). However, the functional role of these GC-boxes has not been demonstrated.
Other than USF-1, the co-factors that may cooperate with MTF-1 to induce MT-I gene expression in response to metals remain unidentified. Nrf-2 has been shown to regulate promoters that contain ARE sequences (15,16). Notably, induction of the HO-1 gene by cadmium requires Nrf-2 interactions at these sites (17). The ARE in the MT-I promoter also resembles a consensus AP-1 binding site. Cadmium (10 µM) can activate binding of c-jun to a consensus AP-1 sequence measured in vitro (18) and we have previously shown that phorbol ester treatment, but not oxidative stress, can induce c-jun to bind to the MT-I ARE in vitro (6). Many members of the family of AP-1 transcription factors, including the Fos, Jun and ATF subfamily members, can bind to promoter regions that deviate from the optimal AP-1 recognition sequence (19). The sum of the results from these studies suggests that the USF/ARE may contribute to cadmium-mediated induction of gene expression through recruitment of Nrf and/or AP-1 transcription factors to the promoter.
A limitation of many of the previous studies is that they were carried out using in vitro DNA binding assays (EMSA), in vivo footprinting, and/or assays measuring reporter gene activation in transiently transfected cells. Although EMSA pinpoints sequence-specific binding of a transcription factor, proteins that require DNA secondary structure (DNA looping, for example) for binding may not be revealed by this approach (20). While in vivo footprinting enables the study of protein binding at specific promoter regions within living cells, it is generally not amenable to identifying the specific factors that bind to these sites, or factors recruited to the promoter by protein–protein interactions. The transient transfection approach is also limited insofar as the promoter/reporter construct is not packaged by chromatin. Thus, this approach fails to address the contributions of chromatin remodeling and other chromatin-specific processes to the molecular mechanisms that may significantly affect interactions of transcription factor with promoters, as has been demonstrated for the interferon-β gene (21).
Currently, chromatin immunoprecipitation (ChIP) is a state-of-the-art method to examine transcription factor interactions in vivo with chromatin-packaged promoters (for review, see 20). There are very few studies that describe transcription factor binding to the chromatin-packaged MT-I promoter in vivo. Results of one study indicate that histone deacetylases and DNA methyltransferases are associated with the MT-I promoter in lymphosarcoma cells in which the MT-I gene is completely silenced (22). In other studies, we demonstrate that metal-dependent recruitment of MTF-1 to the MT-I promoter chromatin in vivo is a required step for gene activation, requires an intact zinc-finger domain (13), and occurs independent of de novo protein synthesis (23) or phosphorylation of MTF-1 (H.Jiang and G.K.Andrews, submitted). However, in vivo association of other transcription factors with the MT-I promoter chromatin in response to metals has not been formally demonstrated, nor have the dynamics of this association been examined. Herein we utilized ChIP in combination with complementary approaches (promoter transfections, knock-out cells) to study the formation of the transcription factor complex at the MT-I promoter in vivo. We demonstrate that MTF-1, USF-1, USF-2 and Sp1 interact in vivo with the MT-I promoter chromatin and we also report the novel finding that the MTF-1-dependent recruitment of c-fos to the promoter coincides with robust activation of the MT-I gene by metals. In addition, we demonstrate that the metal-dependent recruitment of MTF-1 and c-fos to the MT-I promoter is reversibly modulated by zinc.
MATERIALS AND METHODS
Animals
All experiments involving animals were conducted in accordance with NIH guidelines for the care and use of experimental animals and were approved by our Institutional Animal Care and Use Committee. Experiments involving dietary zinc deficiency were carried out as described previously (10). On day 14 of pregnancy, animals that had been fed a zinc-deficient or zinc-adequate diet were sacrificed and yolk sacs harvested as described previously (10).
Reporter plasmids
The reporter plasmids –153-Luc and –153Δ-Luc used to create the stably-transfected cell lines described herein were constructed as follows. Luciferase reporter plasmids under control of the proximal mouse MT-I promoter fragments –153 to +66 and –153ΔUSF/ARE to +66 (deletion of –100 to –89 bp; numbers relative to the transcription start point in the MT-I gene) were described previously for transient transfection assays (7). The BglII/BamHI fragment from these vectors, which contained the minimal MT-I promoter and the coding sequence of the Luciferase gene, were subcloned into the pcDNA3.1/Hygro(+) vector (Invitrogen, Carlsbad, CA) that had the CMV promoter removed. The new reporter vectors, which were verified by DNA sequencing, carried the Hygromycin-resistant gene that allowed for selection of stably-transfected cells.
Cell culture and stably-transfected cells
Mouse hepatoma (Hepa cells), wild-type mouse embryo fibroblasts (MEFs), and MTF-1 knockout MEFs derived from MTF-1–/– mice (MTF-KO), described previously (13,23), were cultured in Dulbecco’s modified Eagle’s media (DMEM). MTF-KO and MEFs were maintained and treated with metals using DMEM supplemented with 10% FBS and Hepa cells maintained and treated using media with either 2% or 10% FBS, as indicated. Mouse embryonic fibroblasts with targeted deletions of the c-fos gene (fosB+/+ c-fos–/–) or both c-fos and fosB genes (fosB–/– c-fos–/–) were obtained from Dr Michael Greenberg of Harvard Medical School and have been described previously (24). These cells were maintained in DMEM supplemented with 15% FBS; however, the media was adjusted to 10% FBS 48 h prior to metal treatment in order to keep the serum concentration the same as that used for culturing wild-type cells (MEFs). Stable cell lines were generated by transfecting Hepa cells, cultured in a 10 cm dish at 60–70% confluency, with 1 µg of reporter plasmid using Lipofectamine (Invitrogen) according to the manufacturer’s instructions with modifications. Colonies of cells that survived 700 µg/ml of Hygromycin selection for 14 days were isolated and expanded for use in subsequent experiments.
Reporter gene activation
Eight cell lines were established by antibiotic selection from transfections with either –153-Luc or –153Δ-Luc vectors. These cells (2 × 104) were plated in a 24-well plate, allowed to reach 50–60% confluency and, where indicated, treated for the indicated time by direct addition of ZnSO4 (100 µM) or CdCl2 (10 µM) to the culture medium. Cells were lysed and assayed for Luciferase activity, normalized to total protein, using the luminescence assays as described (25).
Northern and western blotting
For northern blotting experiments, visceral yolk sacs (10 per group) from zinc-deficient or zinc-adequate pregnant mice were rapidly thawed in TRIzol reagent (Invitrogen, Carlsbad, CA). Alternatively, Hepa cells or fibroblasts were treated with metals as appropriate, washed in PBS and lysed in Triazol. Total RNA (3 µg) was analyzed by northern blotting as described previously (23). For western blotting experiments, whole cell extracts were prepared as follows: Hepa cell proteins were solubilized by snap freezing and thawing of the cell pellet in nuclear extraction buffer (6). The lysates were cleared by ultracentrifugation (45 000 g for 5 min at 4°C) and the extract was adjusted to 140 mM KCl and protein concentration determined by a Bio-Rad Protein Assay reagent (Bio-Rad, Hercules, CA) according to the manufacturer’s directions. Proteins (10 µg) were resolved by SDS–PAGE and immunoblotting was carried out essentially as described previously (26) using a polyclonal antibody to c-fos (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:1000, and a goat anti-rabbit peroxidase-conjugated antibody (Santa Cruz Biotechnology) diluted 1:10 000. Specific protein complexes were visualized with the enhanced chemiluminescence (ECL) system (Amersham Pharmacia Biotech, Piscataway, NJ). Membranes were stained with Ponceau’s solution to ensure equal loading and transfer of proteins.
Chromatin immunoprecipitation (ChIP)
ChIP assays were carried out as described previously (13,27,28) with modifications. Cells (5 × 106), untreated or treated with zinc or cadmium as indicated were fixed and homogenized as described previously (6) and the nuclei isolated and chromatin sonicated as described previously (13). For ChIP analysis using mouse tissues, chromatin was isolated as described previously (20) with modifications. Visceral yolk sacs (50 mg wet weight per antibody) were fixed in 1% formalin for 10 min and chromatin cross-linked as described above. Tissues were homogenized in PBS using a Dounce homogenizer, washed by centrifugation at 200 g, followed by a second homogenization step in homogenization buffer, and chromatin sheared by sonication as described previously (13). Following clarification by centrifugation (12 000 g for 10 min at 4°C), chromatin was either snap frozen in liquid nitrogen for future ChIP assays, or diluted 10-fold in ChIP dilution buffer (29) and pre-cleared with 15 µl of packed Protein-A-Sepharose beads (Sigma) plus 5 µg of sonicated salmon sperm DNA for 1 h at 4°C. Immunoprecipitation was carried out overnight at 4°C using 1–2 µg of polyclonal antibodies against MTF-1 (26), fra-1, nrf-2, c-fos, c-jun, USF-1 or USF-2 (Santa Cruz Biotech), or with beads alone. Polyclonal antibodies against c-maf or v-maf were kindly provided by Dr Masaharu Sakai, Hokkaido University School of Medicine, Sapporo, Japan. The following morning, Protein A-Sepharose (15 µl of packed beads plus 12 µg salmon sperm DNA) was added and the incubation continued for 1 h at 4°C. The beads were washed, the immune complexes eluted and the cross-links reversed, as described previously (27). The input (5% of the total chromatin) or immunoprecipitated DNA was phenol–chloroform extracted, ethanol precipitated, reconstituted in TE buffer, and analyzed by PCR. For PCR analysis of immunoprecipitated or input DNA, primers were synthesized (Integrated Technologies, Coralville, IA) that amplified the endogenous MT-I promoter, spanning –230 to +123 bp (relative to the transcription start point) or the –153-Luc or –153Δ-Luc vector (spanning from –153 bp of the MT-I promoter to +104 bp of the coding sequence for the Luciferase gene). PCR products were separated by agarose gel electrophoresis, stained with SYBR Green (Sigma Chemicals, St Louis, MO) and specific PCR products visualized and quantified, relative to input PCR products, using a Chemiimager gel documentation system (Alpha Innotech Corp., San Leandro, CA). Samples were subjected to PCR for different numbers of cycles to ensure that amplification was in the linear range.
RESULTS
The in vivo interaction of USF-1 and -2 with the MT-I promoter does not require MTF-1
Although there are several other potential binding sites for transcription factors in the proximal –250 bp of the MT-I promoter (illustrated in Fig. 1A), no studies other than those of MTF-1 (13) have directly examined the in vivo interactions of transcription factors with this chromatin-packaged, metal-inducible promoter. Here we utilized ChIP assays to begin examining metal-dependent and metal-independent transcription factor complexes at the MT-I promoter. Furthermore, to explore potential cell-specificity of the transcription factor complex, we compared Hepa cells and mouse embryonic fibroblasts. Zinc or cadmium strongly induce expression of the MT-I gene in both types of cells (23). Dose response experiments in MEF cells demonstrated maximal recruitment of MTF-1 to the MT-I promoter in vivo by 100 µM zinc or 10 µM cadmium (data not shown). These are the concentrations of metals used in the remainder of the experiments described herein. ChIP assays were carried out using cross-linked chromatin isolated from cells treated with zinc or cadmium. Sheared chromatin was immunoprecipitated with antibodies against MTF-1, USF-1, USF-2, c-fos or c-jun (and other antisera as indicated) and analyzed by PCR, using primers specific for the proximal 250 bp of the MT-I gene (Fig. 1).
Figure 1.
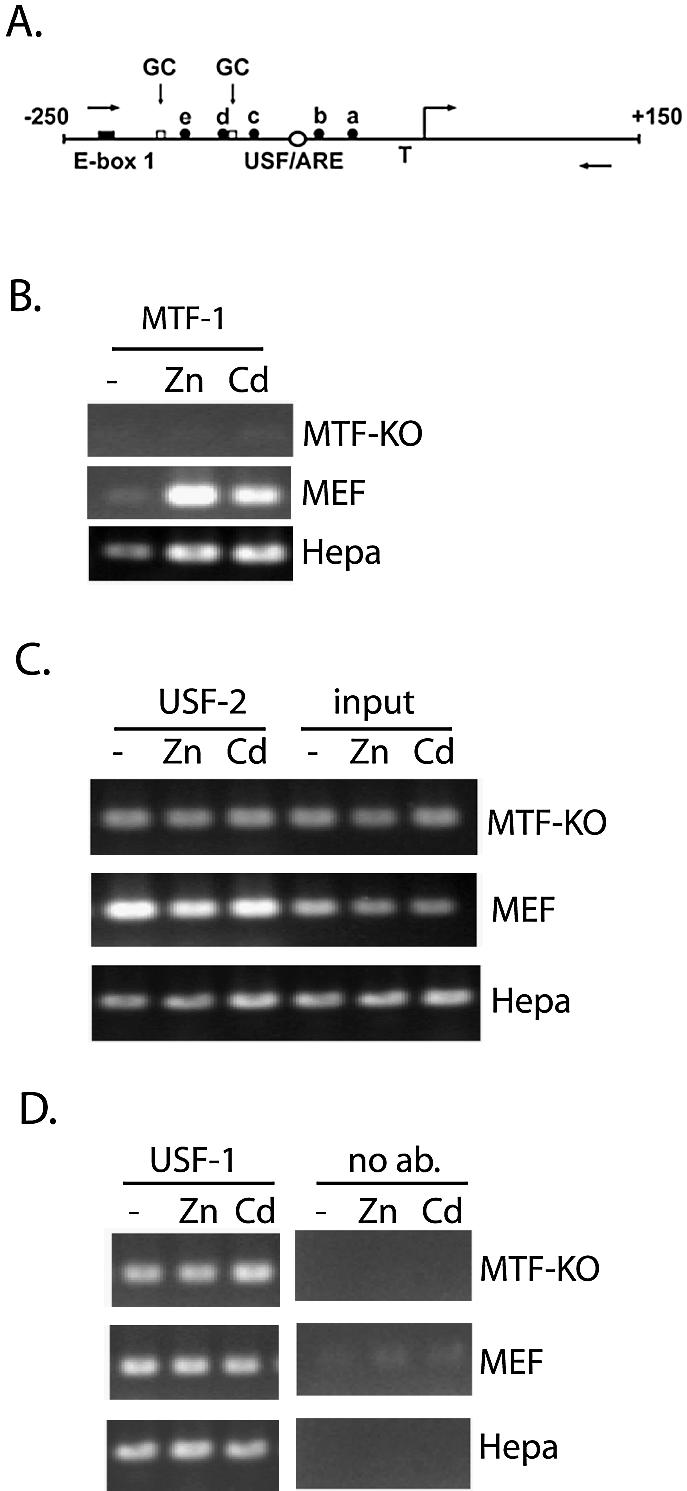
Interactions of USF-1 and -2 in vivo with the MT-I promoter are metal-independent and MTF-1-independent. (A) Schematic of the proximal promoter of the MT-I gene. The nucleotide sequence (bp) is relative to the transcriptional start site (right angle arrow). The relative locations of the USF/ARE (centered at –95 bp), which contains a sequence homologous to an AP-1 consensus binding site, E-box 1 (centered at –220 bp), the multiple MREs (a–e), and the TATA box (T) are illustrated. Two GC-boxes (one centered at –183 bp; the other immediately downstream of MRE-d) represent putative binding sites for Sp transcription factors. The locations of PCR primers used in the ChIP assay are indicated using forward (sense) and reverse (antisense) arrows. (B–D) Chromatin immunoprecipitation (ChIP) assay of the proximal MT-I promoter. Chromatin was isolated from Hepa cells, wild-type mouse embryonic fibroblasts (MEF) or fibroblasts lacking expression of MTF-1 (MTF-KO) that remained untreated (–), or were treated with 100 µM zinc (Zn) or 10 µM cadmium (Cd) for 3 h. ChIP was carried out using antibodies against MTF-1 (B), USF-2 (C), or USF-1 (D), as described in the text. Shown in (C), a portion of the chromatin (0.5% of that used for all of these immunoprecipitations) was purified and analyzed by PCR prior to the immunoprecipitation step (input). Fold-induction for metal-induced binding of transcription factors to the chromatin, enumerated in the text, was calculated from triplicate PCRs after normalizing antibody-specific PCR products to input PCR products.
The results demonstrated that MTF-1 was significantly recruited by zinc or cadmium to the proximal MT-I promoter in MEFs or Hepa cells, but not in cells that do not express MTF-1 (MTF-KO; see Fig. 1B). These results verify the specificity of the antisera against MTF-1. Our previous study also demonstrated that the interaction of USF-1 with the MT-I promoter in vivo was metal-independent in cells that overexpress MTF-1 (13). Herein, we also determined if USF-2 interacts at this promoter in vivo and if the interaction of USF transcription factors at this promoter was MTF-1-dependent. ChIP assays using antibodies specific for either USF-1 or -2 demonstrated a metal-independent interaction of both USF factors with this promoter in vivo in Hepa cells, MEFs and MTF-KO cells (Fig. 1C and D), establishing that USF transcription factors do not require MTF-1 to interact with the promoter.
Metals induce recruitment of c-fos to the MT-I promoter in an MTF-1-dependent manner, whereas recruitment of MTF-1 does not require c-fos
Results from in vivo footprinting and in vitro DNA binding assays suggest an increased recruitment of a factor(s) at the ARE in the MT-I promoter in response to metals (6,7). Although the identity of this factor(s) is unknown, several candidate transcription factors have been shown to interact with ARE-like sequences (15,17). Therefore, ChIP assays were carried out utilizing antibodies against several proteins, including c-fos, c-jun, fra-1, nrf-2, c-maf or v-maf, that are known to bind in vitro to ARE and AP-1 consensus sequences. The results demonstrated that among these proteins, only c-fos was recruited to the MT-I promoter in vivo by cadmium (Fig. 2A and B) or zinc (Fig. 2B) in both Hepa cells and MEFs. However, the magnitude of recruitment was more robust in Hepa cells (5-fold) than in MEFs (3-fold) and this difference was reproducible in three separate experiments, although the reason for this difference between these types of cells is unclear. Differences in c-fos protein levels in the cell are unlikely to be an explanation, as the expression of c-fos was essentially the same in MEFs and in Hepa cells in western blotting assays (data not shown). In contrast, c-jun interacted in a metal-independent manner in these cells (Fig. 2B). In addition, PCR products from c-fos immunoprecipitates were absent in c-fos knockout cells, which verified the specificity of the antisera against c-fos that was used in these ChIP assays (Fig. 2A).
Figure 2.
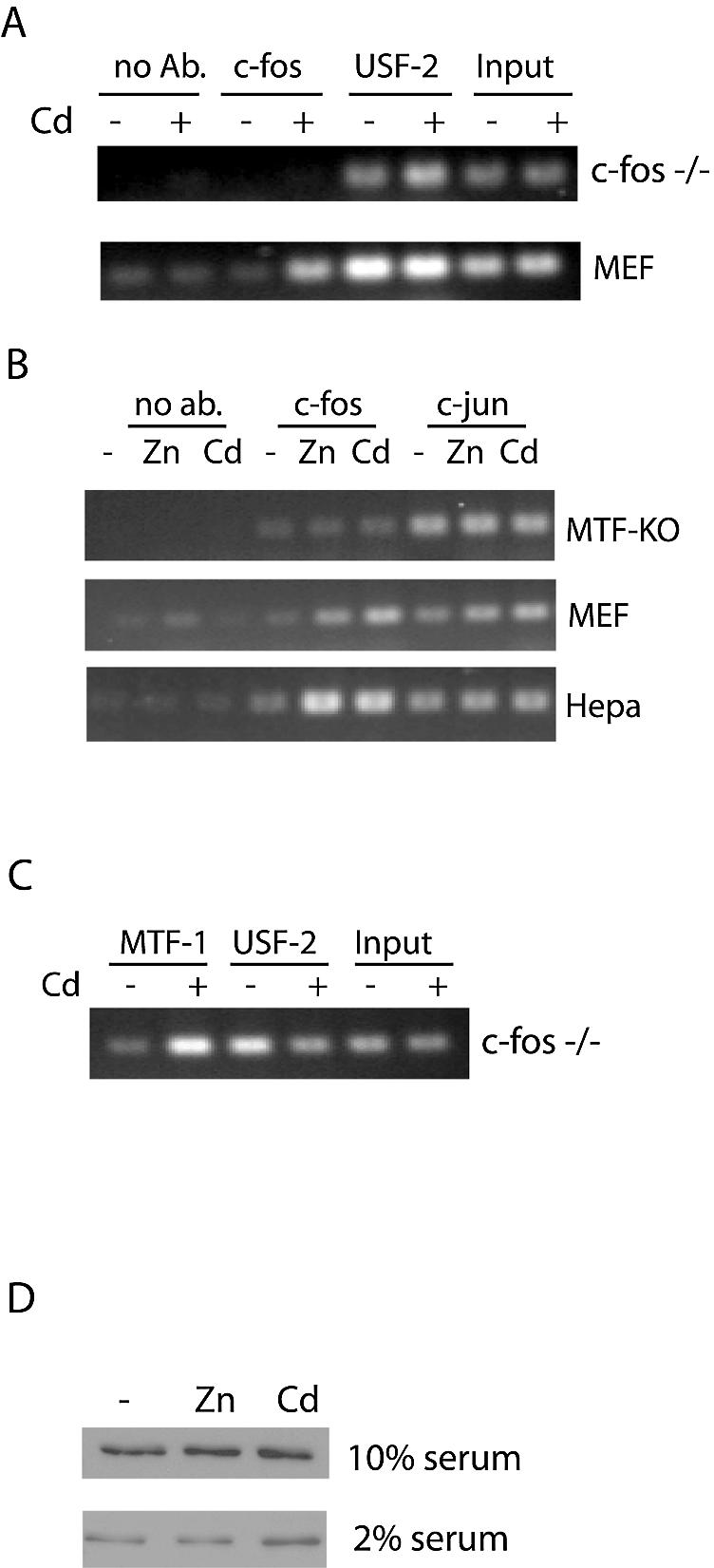
Metal-dependent recruitment of c-fos to the MT-I promoter requires MTF-1, whereas the metal-dependent recruitment of MTF-1 does not require c-fos. (A–C) Mouse embryonic fibroblasts (c-fos knockout (c-fos–/–), MTF-KO, or wild-type MEFs) or Hepa cells remained untreated (–) or were treated for 3 h with 100 µM zinc (Zn) or 10 µM cadmium (Cd) prior to chromatin isolation. Chromatin immunoprecipitation was carried out using antibodies against the indicated transcription factors (c-fos, c-jun, USF-2) or beads alone (no ab). Products from triplicate PCRs were quantified relative to input DNA as described in the legend to Figure 1. Similar results were obtained using zinc or cadmium [shown only for cadmium in (A)]. (C) c-fos–/– mouse embryonic fibroblasts remained untreated or were treated with cadmium (10 µM) for 3 h and ChIP was carried out using the antibodies against MTF-1 or USF-2. (D) Western blot analysis of the cellular levels of c-fos. Hepa cells cultured in DMEM containing 10% or 2% FBS as indicated, were untreated or treated for 3 h with zinc or cadmium and whole cell extracts (10 µg) were prepared. After western blotting, the membrane was stained with Ponceau’s solution to ensure equal loading and transfer of protein. ECL was carried out using the same antibody against c-fos that was used in the ChIP experiments.
To determine whether MTF-1 and c-fos required each other in vivo to interact with the MT-I promoter, we carried out ChIP assays in MTF-KO and c-fos–/– fibroblasts. PCR products were absent in c-fos immunoprecipitates of chromatin from metal-treated MTF-KO cells (Fig. 2B). In contrast, recruitment of MTF-1 to the MT-I promoter by metals was unaffected by the absence of c-fos (Fig. 2C). These results suggest that MTF-1 and c-fos may physically interact, but we have been unable to demonstrate this using a coimmunoprecipitation approach (data not shown). The sum of these results suggests that MTF-1 does not require c-fos in order to interact with the MT-I promoter. In contrast, recruitment of c-fos, but not c-jun, to this promoter requires MTF-1, but may not depend on direct protein contact between these factors.
Increased c-fos in the cell does not govern its recruitment to the MT-I promoter
Cadmium can activate expression of the c-fos gene (30); therefore, cadmium treatment may result in higher levels of this transcription factor in the cell, which could contribute to its increased recruitment to the MT-I promoter. However, western blotting revealed no detectable increase in c-fos protein levels in response to metal treatment when the cells were cultured in media containing 10% serum (Fig. 2D). Because the concentration of serum can also affect the expression of c-fos (31), these experiments were repeated using extracts from cells that were cultured in media containing 2% serum. In this case, there was a slight increase in the amounts of c-fos protein after cadmium treatment, but not after zinc treatment (Fig. 2D). That zinc treatment of cells resulted in enhanced recruitment of c-fos to the MT-I promoter without increasing the amount of c-fos protein in the cell indicates that the increased amount of cellular c-fos protein is not a key, determining factor for its increased recruitment to the MT-I promoter in response to metals.
c-Fos and MTF-1 are rapidly co-recruited to the MT-I promoter in response to metals
Northern blotting experiments demonstrate that zinc and cadmium treatment increases the steady-state levels of MT-I mRNA in Hepa cells and MEFs within 3 h (23), while nuclear run-on assays indicate that zinc increases the relative rate of transcription of the MT-I gene in Hepa cells within 1 h (32). Given this rapid induction of MT-I transcription by zinc, we hypothesized that a pre-initiation complex that contains c-fos and MTF-1 may be rapidly formed after metal treatment. To test this hypothesis, we analyzed the time course of recruitment of c-fos and MTF-1 to the chromatin in response to metals. Preliminary experiments indicated that culturing the cells in media containing 2%, rather than 10% serum, hastened and enhanced the magnitude of recruitment of MTF-1 and c-fos to the MT-I promoter in response to metals. In contrast, serum concentrations in the medium had no effect on the metal-independent binding of c-jun, USF-1 or USF-2 to the promoter (time-course data shown only for 2% serum conditions). ChIP results revealed that both c-fos and MTF-1 were rapidly (within 20 min) recruited to the promoter in response to zinc or cadmium (Fig. 3A). However, MTF-1 recruitment by metals was at or near maximal by 20 min, whereas c-fos continued to accumulate for one to two more hours (see Fig. 3C for fold-recruitment). These results indicate that MTF-1 and c-fos are co-recruited to the proximal MT-I promoter within 20 min of zinc or cadmium treatment. In contrast, USF-2 (Fig. 3B), and USF-1 (not shown) were constitutively bound to the promoter and the magnitude of binding was unchanged in response to metals.
Figure 3.
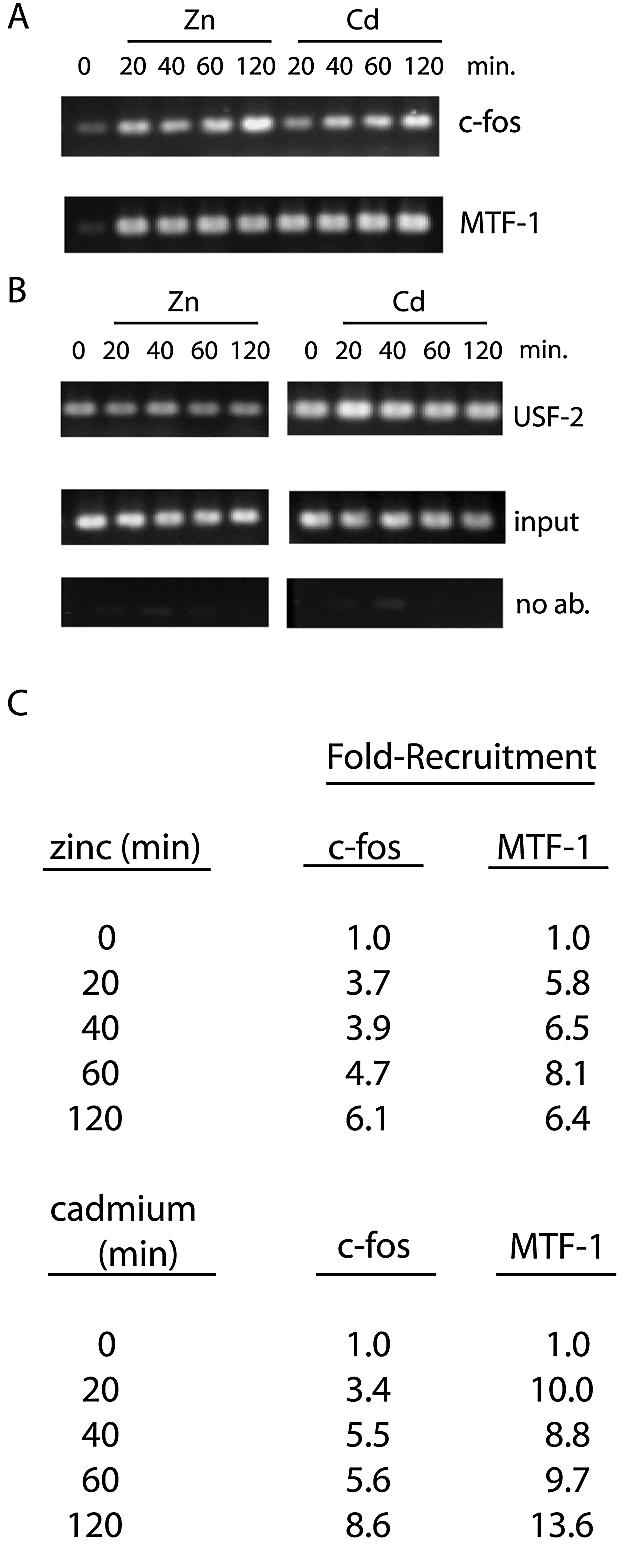
Metal-dependent recruitment of c-fos and MTF-1 to the MT-I promoter is rapid and sustained. (A and B) ChIP was carried out as described in the text and in the legend for Figure 1. Hepa cells, cultured in DMEM medium containing 2% serum, remained untreated or were treated for the indicated times with zinc (100 µM) or cadmium (10 µM), and chromatin was isolated and immunoprecipitated using antibodies against the indicated transcription factors. (C) Fold-recruitment was calculated from triplicate PCRs and normalized to input samples.
Zinc-mediated recruitment of MTF-1 and c-fos to the MT-I promoter is reversible
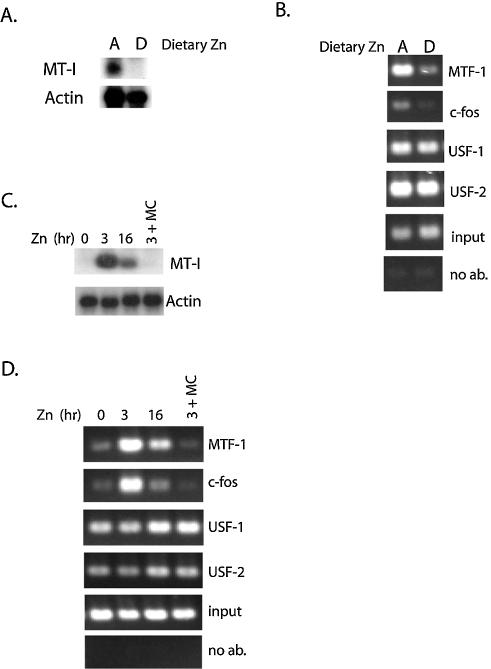
Little is known about the dynamics (formation or dissociation) of protein interactions with the MT-I promoter. We addressed this issue both in tissues from mice and in cultured cells. The availability of dietary zinc has a significant impact on MT-I gene expression in the visceral yolk sac of the embryonic mouse and this expression is MTF-1- and zinc-dependent (10,23). To determine whether dietary zinc affects the interaction of transcription factors with the MT-I promoter in vivo, we isolated chromatin from yolk sacs obtained from embryos in pregnant mice that had been fed a zinc-deficient diet, and ChIP assays were then performed using primers to the endogenous MT-I promoter. The results demonstrate that a zinc-deficient diet significantly decreases the interaction of both c-fos and MTF-1 with the MT-I promoter (Fig. 4B), concomitant with the loss of detectable MT-I mRNA (Fig. 4A). In contrast, dietary zinc had no effect on the constitutive interaction of USF-1 and USF-2 with the chromatin (Fig. 4B). These data strongly suggest that metal-dependent recruitment of MTF-1 and c-fos to the MT-I promoter is a mechanism of gene activation that is present in multiple cell-types and tissues in the mouse.
Figure 4.
Zinc-dependent recruitment of MTF-1 and c-fos to the MT-I promoter is reversible in the visceral yolk sac and in cultured cells. (A and B) Visceral yolk sacs were obtained on day 14 of pregnancy from mice that had been fed with a zinc-adequate (Dietary zinc A) or zinc-deficient (Dietary zinc D) diet for the previous 6 days. (A) Total RNA was extracted from yolk sacs and MT-I and actin mRNA levels were assayed by northern blotting. (B) Chromatin was isolated from yolk sacs, immunoprecipitated using antibodies against the indicated transcription factors or Sepharose beads only (no ab) and subjected to PCR using primers to the MT-I gene (Fig. 1). (C and D) Hepa cells remained untreated (0 h) or were treated with zinc (50 µM) for the indicated times. Alternatively, medium from cells that had been pretreated with zinc (50 µM) for 3 h was replaced with fresh medium containing 0.75 µM zinc (DMEM with 2% FBS), and cells were cultured for an additional 16 h (3 + MC). (C) RNA was extracted from Hepa cells and MT-I and actin mRNAs assayed by northern blotting. (D) ChIP carried out using antibodies against the indicated transcription factors.
Our previous studies suggested that a transient induction of MT-I mRNA occurs in Hepa cells exposed to zinc (23). Therefore, ChIP was used to monitor MTF-1 and c-fos interaction with the MT-I promoter during extended zinc (50 µM) treatment. Northern blotting revealed that MT-I mRNA levels were induced 11-fold by 3 h of zinc treatment but declined to 4-fold above control levels by 16 h of exposure (Fig. 4C). ChIP assays demonstrated that the recruitment of MTF-1 to the MT-I promoter by zinc was increased by 6.8-fold at 3 h and by 4.1-fold at 16 h of extended zinc treatment. Remarkably, the recruitment of c-fos returned to basal (untreated) levels at 16 h of zinc treatment (Fig. 4D). These data indicate that during the adaptation of cells to prolonged zinc treatment, recruitment of MTF-1 to the promoter is reduced, concomitant with a similar decrease in gene expression and loss of c-fos binding.
Another approach was to determine whether zinc-induced MT-I gene expression and transcription factor binding to the promoter chromatin were reversible in Hepa cells after removal of zinc from the culture medium. After a 3-h incubation in medium containing 50 µM zinc, the concentration of zinc was lowered to 0.75 µM. The incubation was then continued for another 16 h before the cells were analyzed using northern blotting and ChIP. The results revealed that MT-I gene expression and recruitment of MTF-1 and c-fos had returned to basal levels under these conditions (Fig. 4C and D; see far right lanes in each panel). Thus, the recruitment of MTF-1 and c-fos to the promoter chromatin is completely reversible, concomitant with the loss of MT-I gene expression.
c-Fos is not required for induction of MT-I gene expression by zinc
Previous studies demonstrated that the loss of function of c-fos does not inhibit MT-I induction in response to 50 µM cadmium (33). However, this concentration of cadmium is much higher than what is required to induce MT-I gene expression. In order to determine whether c-fos has a functional role in the induction of MT-I gene expression by zinc, we treated wild-type embryonic fibroblasts or c-fos deficient (fosB+/+ c-fos–/–) fibroblasts with zinc (100 µM) for 3 h. Following RNA isolation, northern blot analysis revealed that induction of MT-I mRNA by zinc was similar to that seen in wild-type cells (Fig. 5). In addition, fosB can compensate for a lack of c-fos expression in cell cycle entry and expression of cell cycle regulatory proteins (24). However, zinc still induced MT-I gene expression in cells lacking both c-fos and fosB (Fig. 5). Therefore, the precise functional role, if any, of the interaction of c-fos with the MT-I promoter remains unclear. It is worth noting that these experiments do not necessarily address the potential role of c-fos in regulating MT-I gene expression in Hepa cells, where the recruitment of c-fos by metals was much more robust than in fibroblasts (Fig. 2A).
Figure 5.
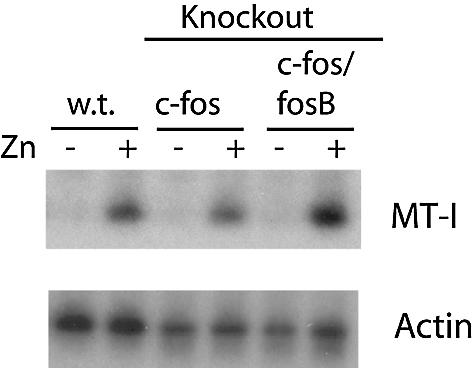
c-Fos is not required for induction of the MT-I gene by zinc in mouse embryo fibroblasts. Northern blot detection analysis of MT-I and actin mRNAs isolated from mouse embryonic fibroblasts treated with or without zinc (100 µM) for 3 h. The fibroblasts used were MEFs (wt) or fibroblasts with targeted deletions of the c-fos gene only, or both c-fos and fosB genes (c-fos/fosB), as indicated.
In vivo mapping of protein interactions with the MT-I promoter
Using ChIP, one cannot identify the specific location on an endogenous promoter where a given transcription factor interacts. In order to address this issue, at least with regard to the MT-I proximal promoter (–153 bp), Hepa cells were transfected with Luciferase reporter genes (illustrated in Fig. 6A) in which expression was driven by the proximal 153 bp of the MT-I promoter [plus or minus the USF/ARE (–153ΔLuc)] and stably-transfected cell lines were selected. For each transfected vector, eight cell lines that survived antibiotic selection were screened to find those in which the integrated reporter gene retained metal-responsiveness (data not shown). Two cell lines for each vector were selected for further studies. Time course experiments revealed that the activation of reporter genes by metals was markedly increased (10- and 16-fold by 6 h) in two –153Luc cell lines (4 and 6). In contrast, more modest (3- to 4-fold) inductions were noted for two –153ΔLuc cell lines (1 and 7; Fig. 6B and C). In all four of these cell lines, cadmium treatment resulted in essentially the same induction of reporter gene activity as that seen for zinc (Fig. 6B and C). In contrast, there were no apparent differences in basal reporter gene activity among these four cell lines (Fig. 6B and C). That the inductions of gene expression by metals in both of the –153Luc cell lines were much greater than those of the –153ΔLuc lines (note difference in y-axes in Fig. 6B), particularly at 6 h of metal treatment suggests that the USF/ARE contributes to more robust levels of MT-I gene activation by zinc or cadmium within the context of chromatin-packaging.
Figure 6.
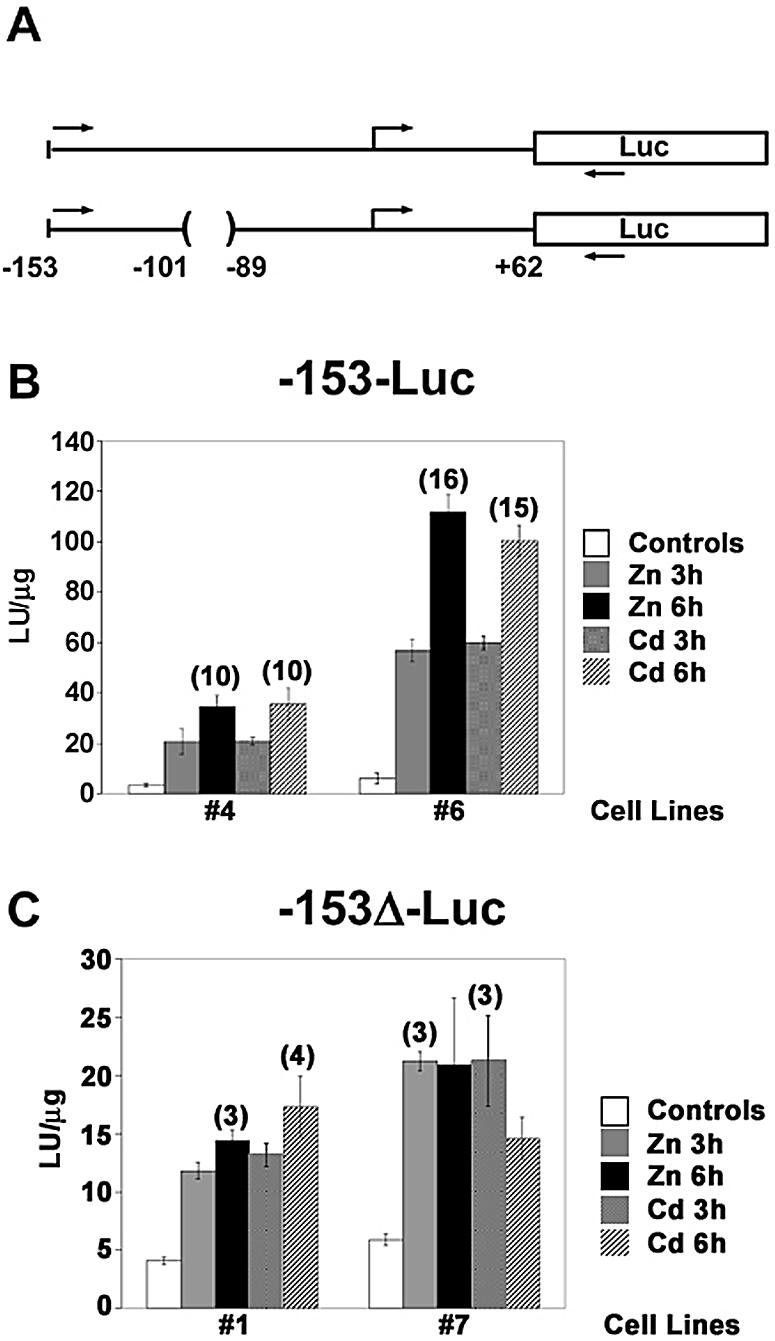
Characterization of stably-transfected cells used for in vivo mapping studies. (A) Diagram of the MT-I promoter-Luciferase fusion vectors. Structures of the Luciferase (Luc) reporter genes under the control of a minimal (–153 to +62 bp) MT-I promoter with (top) or without (bottom) the USF/ARE (–100 to –90 bp) are illustrated and are referred to as (–153-Luc) or (–153Δ-Luc), respectively. The relative location of the PCR primers used in the ChIP assays described in Figure 7 are indicated by the forward (sense) and reverse (antisense) arrows. (B and C) Hepa cells were stably-transfected with the –153-Luc or the –153Δ-Luc vectors. Two stably-transfected cell lines for each reporter gene were cultured in medium containing 2% serum and remained untreated (controls) or were treated with zinc (100 µM) or cadmium (10 µM) for 3 or 6 h as indicated. Whole cell extracts from triplicate cell cultures were analyzed for Luciferase reporter gene activity, which was normalized to protein content (LU/µg protein). For each cell line and metal treatment, the maximal fold-inductions of reporter gene activity, rounded to the nearest whole number, are indicated above the error bar. Please note the different y-axis values for the (–153-Luc) cell lines (B), and (–153Δ-Luc) cell lines (C).
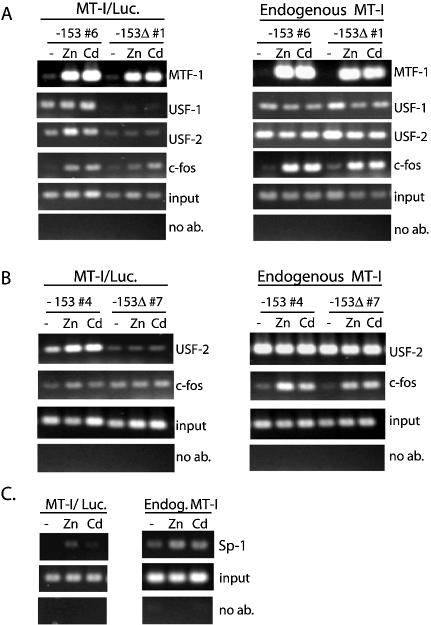
That the USF/ARE is not required for modest increases in MT-I gene expression by metals suggests that MTF-1 is recruited independent of the presence of this element. To address this hypothesis, ChIP was carried out using PCR primers that span the minimal MT-I promoter and the 5′ end of the Luciferase reporter gene (illustrated in Fig. 6A), as well as primers that allow amplification of the endogenous MT-I promoter as described above. The results revealed that recruitment of MTF-1 to the minimal promoter was essentially the same in the –153Luc and –153ΔLuc cell lines (Fig. 7A, left top panel). Recruitment to these transfected promoters was essentially as robust as that to the endogenous promoter (Fig. 7A; compare left and right panels). Thus, neither the USF/ARE nor the MRE-e (–166 bp to –174 bp) is required for MTF-1 recruitment.
Figure 7.
Mapping interactions of transcription factors (MTF-1, USF, c-fos and Sp1) with MT-I promoter in vivo. ChIP analyses, using the stably- transfected Hepa cell lines –153-Luc-6 and –153Δ-Luc-1 (A), or –153-Luc-4 and –153Δ-Luc-7 (B), were carried out using the indicated antibodies and primers specific for the endogenous MT-I gene (right panels) or the integrated reporter genes (left panels). Cells remained untreated (–) or were treated for 2 h with zinc (100 µM) or cadmium (10 µM). For the number 4 and number 7 cell lines, ChIP results from USF-1 and MTF-1 immunoprecipitations (not shown) were similar to those shown for the number 6 and number 1 cell lines, respectively (A). In (C), chromatin was isolated from the –153-Luc-4 cells that were treated with zinc or cadmium for 2 h, ChIP carried out using an antibody against Sp1, and PCRs performed using primers to the transfected –153-Luciferase construct (MT-I/Luc) or the endogenous MT-I gene (Endog. MT-I).
USF-1 binds in vitro to the USF/ARE sequence as well as to E-box1 in the MT-I promoter (6,7,10). To determine whether USF-1 and/or USF-2 interact in vivo with the chromatin at the USF/ARE, ChIP assays were performed for the transfected promoters using antibodies against USF-1 and USF-2. The results demonstrated that USF-1 and USF-2 interact with the intact promoter but not with the promoter in which the USF/ARE (–153ΔLuc) was deleted (Fig. 7A). Thus, both USF-1 and USF-2 bind in vivo to the USF/ARE. Metal induction of USF-2 binding to the USF/ARE in vivo was minimal (2- to 3-fold; Fig. 7A and B) but no increase in USF-1 recruitment was noted (Fig. 7A). In parallel PCRs from the same immunoprecipitated chromatin samples, but using primers to the endogenous MT-I gene, USF-1 (Fig. 7A, right panel) and USF-2 (Fig. 7A and B, right panels) were constitutively bound to the endogenous MT-I promoter in all cell lines, demonstrating that chromatin immunoprecipitations using the anti-USF antibodies were equally efficient in both cell lines. Furthermore, the amount of the endogenous MT-I promoter immunoprecipitated by the antibody against USF-2 was always greater than that of the transfected promoter, which suggests a second USF binding site upstream of –153 bp (Fig. 7B; compare right and left panels). These results and those from our recent studies (7,10), taken together, suggest that USF-1 and USF-2 form a heterodimer at both the USF/ARE and the E Box-1 regions of the MT-I promoter. The fact that USF-1 or USF-2 do not bind to the minimal MT-I promoter without the USF/ARE establishes that MTF-1 binds to the MT-I promoter independent of USF-1 or USF-2.
We hypothesized that c-fos may be recruited to the USF/ARE by metals. To test this hypothesis, ChIP assays were performed on the transfected promoter as described above. Surprisingly, the results revealed PCR products specific for the transfected DNA in both the –153Luc-6 and the –153ΔLuc-1 cell lines (Fig. 7A; left panels). Thus, c-fos can interact at the proximal (–153 bp) promoter independent of the USF/ARE. Recruitment of c-fos to the transfected promoter by zinc or cadmium was 5- to 6-fold in the –153Luc-6 cells. In contrast, in the –153ΔLuc-1 cells, recruitment of c-fos by zinc or cadmium was 2-fold and 4-fold. Although the interaction of c-fos at the transfected proximal promoter was also detected in the –153Luc-4 and the –153ΔLuc-7 cell lines, the metal induction of recruitment was weak (2- to 3-fold) in the number 4 cell line and absent in the number 7 cell line (Fig. 7B; left panels). In contrast, for all four lines, the metal-induced recruitment of c-fos to the endogenous MT-I gene was robust (Fig. 7A and B; right panels). The finding that the recruitment of c-fos was enhanced in the presence of the USF/ARE (Fig. 7A, left panel) indicates that c-fos may interact weakly with the USF/ARE. However, we have been unable to show interaction of c-fos with this element in vitro using supershift EMSA (data not shown). Nevertheless, the collective ChIP results from these stably-transfected cell lines demonstrate that c-fos binds to the proximal (–153 bp) promoter at a site other than, or in addition to, the USF/ARE, and may bind upstream of the proximal promoter as well. In contrast, interaction of c-jun with either the transfected or endogenous MT-I promoter was constitutive in all four lines (data not shown), indicating that this factor binds to the proximal promoter independent of the USF/ARE or metal treatment.
Previous in vitro DNA-binding assays provide evidence that Sp1 or possibly other members of the Sp family of transcription factors constitutively interacts at GC-rich sequences in the MT-I promoter (6,14). One GC-rich site is centered at –183 bp in the MT-I promoter and the other overlaps the MRE-d. In the context of the MT-I promoter, the best studied member of the Sp family is Sp1. Therefore, to determine whether Sp1 interacts within the proximal (–153 bp) part of the MT-I promoter, we examined the recruitment of Sp1 to the transfected minimal promoter and to the endogenous MT-I promoter. Sp1 interacted with the endogenous MT-I promoter in control cells and slightly more (2-fold increase) in metal treated cells (Fig. 7C; right panel). However, no significant binding occurred to the transfected minimal promoter (Fig. 7C; left panel). These results are consistent with a major binding site for Sp1 upstream of the –153 bp, but not at the MRE-d. Consistent with this observation, an in vitro DNA-binding assay indicated that recombinant Sp1 binds poorly to MRE-d (unpublished data).
DISCUSSION
There have been numerous studies of the mouse MT-I gene that were designed to identify members of the transcription factor complex at this promoter in response to metals, and significant progress has occurred particularly in our understanding of the roles of MTF-1 in this complex. Many studies of the MT-I promoter relied on in vitro binding assays and in vivo footprinting assays to deduce the identity of binding factors and to map sites of protein–DNA interactions within this promoter, respectively (6,7,12,34). Herein we have extended those studies to include ChIP assays, and the results have allowed us to more clearly define the in vivo dynamics of protein interactions with this interesting promoter. Taken together with previous studies, our results confirm that multiple transcription factors bind to or interact with the proximal 230 bp of the mouse MT-I promoter. A model of these interactions is presented in Figure 8 and is discussed below.
Figure 8.
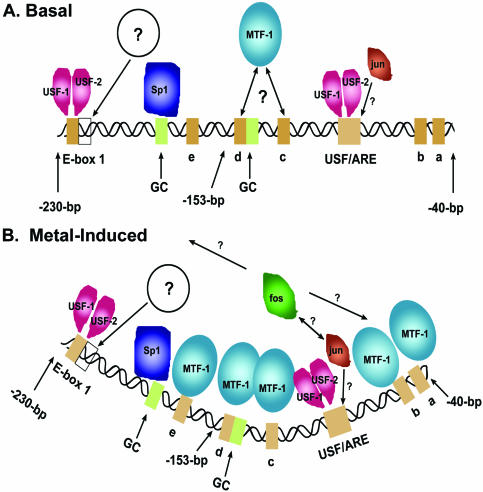
A model depicting the dynamic transcription factor complexes found at the proximal region of the mouse MT-I promoter. (A) ‘Basal’ or uninduced complex. (B) Metal-induced complex. The approximate positions of base pairs (–230 to –40 bp) relative to the transcription start site are indicated. The multiple MREs are indicated (a–e), and the other promoter elements shown here are described in the text and in the legend to Figure 1. An open box indicates the site of a constitutive in vivo footprint downstream of E-box 1, and an open circle with a question mark inside indicates an unidentified transcription factor(s) that binds to this sequence. The apparently weaker or transient association of MTF-1 with multiple MREs under basal conditions is indicated using a double-headed arrow in (A). In contrast, all the MREs are occupied under metal-induced conditions (B). USF-1, USF-2 and Sp1 binding sites are indicated. Protein–protein and protein–DNA interactions for fos and jun are not well-defined and are indicated by arrows with question marks. The looping of the promoter DNA is hypothetical.
Approximately 10 sites of protein–DNA interactions have been identified within the proximal region (–230 to –40 bp) of the mouse MT-I promoter (Fig. 8A). Our results demonstrate that USF-1 and USF-2, Sp1 and MTF-1 each bind to this region of the promoter in vivo. In addition, ChIP assays suggest that c-jun and c-fos also interact within this promoter, perhaps in a sequence-independent manner, although binding of c-jun to the USF/ARE cannot be excluded (6). In addition, in vivo footprinting assays suggest that an as yet unidentified factor(s) interacts just downstream of E-Box 1 (7). When all of these 10 binding sites are occupied, as appears to be the case in cells exposed to excess metal (6), then a vast majority of the promoter DNA is actually bound by proteins in metal-treated cells (Fig. 8B). The fact that protein–DNA interactions occur throughout this 190 bp region of the MT-I promoter suggests that it is not packaged into a nucleosome.
Many of these transcription factors interact with the MT-I promoter in uninduced cells (basal; Fig. 8A), as well as in metal-treated cells (induced; Fig. 8B). It is the degree of interaction, specifically of MTF-1 and c-fos, which changes most dramatically in response to zinc or cadmium. It is important to note that cells cultured in 10% serum-containing medium are growing in the presence of 3–4 µM zinc. This concentration is sufficient to drive expression of the MT-I gene and this ‘basal’ level of activity is known to be dependent on MTF-1 (11). ChIP assays confirm the interaction of MTF-1 with this promoter in uninduced cells. However, previous in vivo footprinting studies do not reveal a significant footprint over any of the MREs in these cells (6). This suggests that MTF-1 does not preferentially associate tightly with a subset of the MREs; rather it may associate weakly with most of these MREs in uninduced cells (Fig. 8A). In contrast, USF-1 and -2 interact strongly with this promoter regardless of exposure to excess zinc or the presence of MTF-1. The extensive, constitutive binding of transcription factors to the MT-I promoter suggests that it is maintained in an open chromatin configuration in cells in which this gene is metal-responsive (22). In contrast, in cells in which this gene is silenced and unresponsive to metals, the MT-I gene is methylated and packaged into a condensed heterochromatin state (35).
The most dynamic changes in protein interactions with the MT-I promoter were noted in cells exposed to metals (zinc or cadmium) and during withdrawal from exposure to excess zinc. In cells exposed to an increase in exogenous zinc or cadmium, c-fos and MTF-1 were rapidly recruited to this promoter consistent with a rapid increase in the relative rate of transcription of the MT-I gene (32). In contrast, during withdrawal from exposure to zinc, the association of these proteins with the MT-I promoter was markedly reduced concurrent with a decrease in MT-I gene expression. These results are consistent with current models of the mechanisms of action of MTF-1 that suggest that the reversible binding of zinc with its zinc-finger domain modulates its stable association with the MT-I promoter (36). Although it has been assumed that MTF-1 disassociates from the MT-I promoter when zinc availability is reduced, this aspect of MTF-1 action had not been investigated previously. The fate of MTF-1 released from the promoter during zinc withdrawal remains to be determined.
The novel finding that c-fos is recruited in an MTF-1-dependent manner to the MT-I promoter suggests that this bZIP transcription factor plays a role in metal responsiveness of this gene. The coordinate binding and disassociation of MTF-1 and c-fos with the MT-I promoter suggest that these proteins may also physically interact. c-Fos can potentially interact with dozens of other transcription factors, including c-jun which was found to constitutively be associated with the MT-I promoter. However, its interaction with MTF-1 was not confirmed in preliminary co-immunoprecipitation studies (P.J.Daniels and G.K.Andrews, unpublished results). Remarkably, the recruitment of c-fos was not dependent on the USF/ARE, but was enhanced in the intact promoter relative to the –153 bp promoter. This suggests that a DNA sequence(s) or proteins binding in that upstream region of the promoter aid in the recruitment of c-fos in response to metals. Analysis of the MT-I promoter revealed another potential AP-1 site at –544 bp. Given that chromatin was sheared to an average of 1000 bp during the ChIP assay, c-fos bound to the –544 bp site would have been included in the immunoprecipitated chromatin. Whether AP-1 can bind to this site remains to be determined. However, our results suggest that DNA-looping (Fig. 8B) may present a mechanism to facilitate stable interactions of c-fos with the more proximal MT-I promoter where MTF-1 binds. Interestingly, the binding of MTF-1 has been shown to introduce a conformational change in the MRE-d (37). The simultaneous occupancy of all of the MREs in the MT-I promoter could significantly alter the structure of that region of the promoter. This altered structure may facilitate c-fos recruitment. That recruitment appears to be a highly ordered process with maximal MTF-1 binding preceding that of c-fos.
A role for c-fos in metal regulation was not confirmed since the MT-I gene remains metal-responsive in c-fos knockout cells, consistent with a previous study of the mouse MT-I gene (33). Despite this result, numerous studies suggest a role for members of the AP-1 family of transcription factors in the regulation of the human MT-IIa gene. A c-fos/c-jun heterodimer has been shown to bind in vitro to the hMT-IIa promoter (38) and ribozyme-mediated cleavage of the c-fos protein decreases the responsiveness of this gene to dexamethasone (39). In the mouse MT-I promoter the USF/ARE has been shown to provide a binding site for c-jun, induced by TPA treatment of NIH 3T3 cells. Furthermore, changes in the in vivo footprint over that USF/ARE have been noted after metal or oxidative stress treatment of cells (6). However, AP-1 binding activity to the USF/ARE in vivo has not been conclusively demonstrated by the studies reported herein. It should also be noted that other AP-1 factors (e.g. fra-1) may compensate for the lack of c-fos expression (40,41). Thus, a member of the AP-1 family other that c-jun or fos-B may compensate for the lack of c-fos in regulating MT-I gene expression in c-fos–/– cells. The possibility that c-fos may also function in a cell-specific manner was not addressed in these studies. However, the recruitment of c-fos to the MT-I promoter was much more robust in Hepa cells compared with MEF cells. In addition, the activity of c-fos/c-jun heterodimers depends on the cell-type and the stimulus the cell receives (reviewed in 42).
USF-1 and USF-2 are bHLH proteins that have been suggested to play a role in MT-I gene expression. DNA-binding assays demonstrated that USF-1 and -2 can bind in vitro to E-Box1 and to the USF/ARE, and in vivo footprinting assays revealed the constitutive occupancy of these promoter sequences in tissues (43) and in cultured cells (7). However, many members of the bHLH protein family can bind to E-box sequences, and USF proteins can positively or negatively regulate gene expression, depending on the gene, cell-type and the particular USF homodimer or heterodimer that is formed (44–46). The ChIP assay data reported herein confirm that USF-1 and -2 proteins are, in fact, constitutively bound in vivo to this promoter. Furthermore, promoter-mapping studies demonstrated that USF-1 and -2 are bound to the USF/ARE, as well as to sequences upstream of –153 bp in the MT-I promoter. Within the upstream promoter region that would have been detected in our ChIP assays, a high affinity USF binding site is found only at E-box1. Taken together with the studies discussed above, our results support the hypothesis that these proteins are important components of the MT-I promoter complex. In that regard, the USF/ARE has been shown to enhance basal expression of the MT-I promoter, as well as to participate in cadmium induction of the MT-I gene (7). Furthermore, studies using transgenic mice suggest that E-box1 cooperates with the proximal MT-I promoter to drive high-level expression of the MT-I gene in the embryonic visceral yolk sac (10). This expression is dependent on MTF-1 and zinc. Thus, we speculate that USF-1 and -2 heterodimers facilitate the functions of MTF-1, perhaps by maintaining an open promoter configuration that facilitates the interaction of MTF-1 with the proximal promoter.
Results of ChIP assays indicated that metal treatment does not significantly increase USF-1 binding to the promoter, but a modest (2-fold) increase in the recruitment of USF-2 in vivo to the USF/ARE was suggested. This may indicate that USF-1 homodimers are replaced with USF-1/USF-2 heterodimers at the USF/ARE in response to metal induction. That we were unable to detect an increased recruitment of USF-2 to the endogenous MT-I promoter chromatin after metal treatment may reflect the stronger, constitutive binding of USF-1 and USF-2 to E-Box 1. This could mask recruitment of USF-2 to the USF/ARE. Binding competition assays suggest that USF binds about 10-times more avidly in vitro to E-box1 than it does to the USF/ARE. Furthermore, zinc-treatment does not change the in vivo footprint over the E Box-1 region of the MT-I promoter, whereas increased occupancy of the USF/ARE was noted (7).
A novel finding in these studies was the mapping of an apparent single site of interaction of the zinc-finger protein Sp1 with the MT-I promoter. ChIP assays revealed that Sp1 binds upstream of –153 bp. Within the upstream promoter region that would have been detected in our ChIP assay, a high affinity Sp1 binding site is found only at –183 bp. Mobility shift assays and in vivo footprinting assays previously demonstrated constitutive protein interactions with this GC-rich sequence (6). The results of the ChIP assay are consistent with the concept that Sp1 constitutively binds to this site. It has also been widely assumed that Sp1 binds to another GC-box that overlaps MRE-d (Fig. 8). Oligonucleotide competition in mobility shift assays suggested that Sp1 binds in vitro to MRE-d independent of metal treatment of cells, whereas MTF-1 also binds to MRE-d after metal treatment (14). However, the ChIP assay data reported herein do not support the notion that Sp1 interacts with this site. Consistent with this conclusion, in vivo footprinting studies of the MT-I promoter do not indicate a strong, constitutive footprint specifically over the GC-box at MRE-d (6). Furthermore, recent studies of the binding interactions of the purified zinc-finger domain of MTF-1 with MRE-d revealed that fingers 5 and 6 interact with bases in this GC-box sequence in a zinc-dependent manner (37). Thus, MTF-1 binding might be expected to compete for binding of another protein to this GC-box in MRE-d. Finally, it was recently demonstrated that Sp1 does not affect the in vitro transcription of an MRE-d-driven reporter gene or contribute to its metal response (47). Taken together with the ChIP assays, these findings suggest that Sp1 may not bind to MRE-d in vivo. However, our results cannot exclude the possibility that another member of the Sp-like family of transcription factors binds to this site. Over 15 members of this family have now been identified that share highly homologous DNA binding motifs that bind to similar sequences (48).
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Gary Lin and Jim Geiser for excellent technical assistance. This work was supported, in part, by a grant from the NIH to G.K.A. (ES05704). An NRSA fellowship provided support for P.J.D. (F32 HD40705).
REFERENCES
- 1.Palmiter R.D. (1987) Molecular biology of metallothionein gene expression. EXS, 52, 63–80. [DOI] [PubMed] [Google Scholar]
- 2.Andrews G.K. (2000) Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem. Pharmacol., 59, 95–104. [DOI] [PubMed] [Google Scholar]
- 3.Giedroc D.P., Chen,X. and Apuy,J.L. (2001) Metal response element (MRE)-binding transcription factor-1 (MTF-1): structure, function, and regulation. Antioxid. Redox Signal., 3, 577–596. [DOI] [PubMed] [Google Scholar]
- 4.Lichtlen P. and Schaffner,W. (2001) Putting its fingers on stressful situations: the heavy metal-regulatory transcription factor MTF-1. Bioessays, 23, 1010–1017. [DOI] [PubMed] [Google Scholar]
- 5.Andrews G.K. (2001) Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals, 14, 223–237. [DOI] [PubMed] [Google Scholar]
- 6.Dalton T.P., Li,Q.W., Bittel,D., Liang,L.C. and Andrews,G.K. (1996) Oxidative stress activates metal-responsive transcription factor-1 binding activity—Occupancy in vivo of metal response elements in the metallothionein-I gene promoter. J. Biol. Chem., 271, 26233–26241. [DOI] [PubMed] [Google Scholar]
- 7.Li Q.W., Hu,N.M., Daggett,M.A.F., Chu,W.A., Bittel,D., Johnson,J.A. and Andrews,G.K. (1998) Participation of upstream stimulatory factor (USF) in cadmium-induction of the mouse metallothionein-I gene. Nucleic Acids Res., 26, 5182–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hempe J.M., Carlson,J.M. and Cousins,R.J. (1991) Intestinal metallothionein gene expression and zinc absorption in rats are zinc-responsive but refractory to dexamethasone and interleukin 1α. J. Nutr., 121, 1389–1396. [DOI] [PubMed] [Google Scholar]
- 9.Dalton T.P., Fu,K., Enders,G.C., Palmiter,R.D. and Andrews,G.K. (1996) Analysis of the effects of over-expression of metallothionein-I in transgenic mice on the reproductive toxicology of cadmium. Environ. Health Perspect., 104, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews G.K., Lee,D.K., Ravindra,R., Lichtlen,P., Sirito,M., Sawadogo,M. and Schaffner,W. (2001) The transcription factors MTF-1 and USF1 cooperate to regulate mouse metallothionein-I expression in response to the essential metal zinc in visceral endoderm cells during early development. EMBO J., 20, 1114–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heuchel R., Radtke,F., Georgiev,O., Stark,G., Aguet,M. and Schaffner,W. (1994) The transcription factor MTF-I is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J., 13, 2870–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller P.R., Salser,S.J. and Wold,B. (1988) Constitutive and metal-inducible protein:DNA interactions at the mouse metallothionein I promoter examined by in vivo and in vitro footprinting. Genes Dev., 2, 412–427. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H., Daniels,P.J. and Andrews,G.K. (2003) Putative zinc-sensing zinc fingers of metal-response element-binding transcription factor-1 stabilize a metal-dependent chromatin complex on the endogenous metallothionein-I promoter. J. Biol. Chem., 278, 30394–30402. [DOI] [PubMed] [Google Scholar]
- 14.Westin G. and Schaffner,W. (1988) A zinc-responsive factor interacts with a metal-regulated enhancer element (MRE) of the mouse metallothionein-I gene. EMBO J., 7, 3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venugopal R. and Jaiswal,A.K. (1996) Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl Acad. Sci. USA, 93, 14960–14965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venugopal R. and Jaiswal,A.K. (1998) Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene, 17, 3145–3156. [DOI] [PubMed] [Google Scholar]
- 17.Alam J., Stewart,D., Touchard,C., Boinapally,S., Choi,A.M.K. and Cook,J.L. (1999) Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem., 274, 26071–26078. [DOI] [PubMed] [Google Scholar]
- 18.Shukla G.S., Chiu,J.F. and Hart,B.A. (2000) Cadmium-induced elevations in the gene expression of the regulatory subunit of gamma-glutamylcysteine synthetase in rat lung and alveolar epithelial cells. Toxicology, 151, 45–54. [DOI] [PubMed] [Google Scholar]
- 19.McBride K. and Nemer,M. (1998) The C-terminal domain of c-fos is required for activation of an AP-1 site specific for jun-fos heterodimers. Mol. Cell. Biol., 18, 5073–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wells J. and Farnham,P.J. (2002) Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods, 26, 48–56. [DOI] [PubMed] [Google Scholar]
- 21.Agalioti T., Lomvardas,S., Parekh,B., Yie,J., Maniatis,T. and Thanos,D. (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell, 103, 667–678. [DOI] [PubMed] [Google Scholar]
- 22.Ghoshal K., Datta,J., Majumder,S., Bai,S.M., Dong,X.C., Parthun,M. and Jacob,S.T. (2002) Inhibitors of histone deacetylase and DNA methyltransferase synergistically activate the methylated metallothionein I promoter by activating the transcription factor MTF-1 and forming an open chromatin structure. Mol. Cell. Biol., 22, 8302–8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmade S.J., Ravindra,R., Daniels,P.J. and Andrews,G.K. (2000) The transcription factor MTF-1 mediates metal regulation of the mouse ZnT1 gene. J. Biol. Chem., 275, 34803–34809. [DOI] [PubMed] [Google Scholar]
- 24.Brown J.R., Nigh,E., Lee,R.J., Ye,H., Thompson,M.A., Saudou,F., Pestell,R.G. and Greenberg,M.E. (1998) Fos family members induce cell cycle entry by activating cyclin D1. Mol. Cell. Biol., 18, 5609–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson J.A. and Nathanson,N.M. (1994) Differential requirements for p21ras and protein kinase C in the regulation of neuronal gene expression by nerve growth factor and neurokines. J. Biol. Chem., 269, 18856–18863. [PubMed] [Google Scholar]
- 26.Smirnova I.V., Bittel,D.C., Ravindra,R., Jiang,H. and Andrews,G.K. (2000) Zinc and cadmium can promote the rapid nuclear translocation of MTF-1. J. Biol. Chem., 275, 9377–9384. [DOI] [PubMed] [Google Scholar]
- 27.Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. [DOI] [PubMed] [Google Scholar]
- 28.Boyd K.E., Wells,J., Gutman,J., Bartley,S.M. and Farnham,P.J. (1998) c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl Acad. Sci. USA, 95, 13887–13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H., Lin,R.J., Xie,W., Wilpitz,D. and Evans,R.M. (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- 30.Joseph P., Muchnok,T.K., Klishis,M.L., Roberts,J.R., Antonini,J.M., Whong,W.Z. and Ong,T. (2001) Cadmium-induced cell transformation and tumorigenesis are associated with transcriptional activation of c-fos, c-jun, and c-myc proto-oncogenes: role of cellular calcium and reactive oxygen species. Toxicol. Sci., 61, 295–303. [DOI] [PubMed] [Google Scholar]
- 31.Angel P., Imagawa,M., Chiu,R., Stein,B., Imbra,R.J., Rahmsdorf,H.J., Jonat,C., Herrlich,P. and Karin,M. (1987) Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell, 49, 729–739. [DOI] [PubMed] [Google Scholar]
- 32.Dalton T.P., Palmiter,R.D. and Andrews,G.K. (1994) Transcriptional induction of the mouse metallothionein-I gene in hydrogen peroxide-treated Hepa cells involves a composite major late transcription factor/antioxidant response element and metal response promoter elements. Nucleic Acids Res., 22, 5016–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu E., Mueller,E., Oliviero,S., Papaioannou,V.E., Johnson,R. and Spiegelman,B.M. (1994) Targeted disruption of the c-fos gene demonstrates c-fos-dependent and -independent pathways for gene expression stimulated by growth factors or oncogenes. EMBO J., 13, 3094–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan M., Heuchel,R., Radtke,F., Hunziker,P.E. and Kägi,J.H.R. (1995) Regulation of metallothionein gene expression in Cd- or Zn-adapted RK-13 cells. Experientia, 51, 606–611. [DOI] [PubMed] [Google Scholar]
- 35.Ghoshal K., Majumder,S. and Jacob,S.T. (2002) Analysis of promoter methylation and its role in silencing metallothionein I gene expression in tumor cells. Methods Enzymol., 353, 476–486. [DOI] [PubMed] [Google Scholar]
- 36.Dalton T.D., Bittel,D. and Andrews,G.K. (1997) Reversible activation of the mouse metal response element-binding transcription factor-1 DNA binding involves zinc interactions with the zinc-finger domain. Mol. Cell. Biol., 17, 2781–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X.H., Chu,M.H. and Giedroc,D.P. (1999) MRE-binding transcription factor-1: Weak zinc-binding finger domains 5 and 6 modulate the structure, affinity, and specificity of the metal-response element complex. Biochemistry, 38, 12915–12925. [DOI] [PubMed] [Google Scholar]
- 38.Abate C., Luk,D., Gentz,R., Rauscher,F.J.,III and Curran,T. (1990) Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc. Natl Acad. Sci. USA, 87, 1032–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scanlon K.J., Jiao,L., Funato,T., Wang,W., Tone,T., Rossi,J.J. and Kashani-Sabet,M. (1991) Ribozyme-mediated cleavage of c-fos mRNA reduces gene expression of DNA synthesis enzymes and metallothionein. Proc. Natl Acad. Sci. USA, 88, 10591–10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleischmann A., Hafezi,F., Elliott,C., Reme,C.E., Ruther,U. and Wagner,E.F. (2000) Fra-1 replaces c-Fos-dependent functions in mice. Genes Dev., 14, 2695–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuo K., Owens,J.M., Tonko,M., Elliott,C., Chambers,T.J. and Wagner,E.F. (2000) Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nature Genet., 24, 184–187. [DOI] [PubMed] [Google Scholar]
- 42.van Dam H. and Castellazzi,M. (2001) Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene, 20, 2453–2464. [DOI] [PubMed] [Google Scholar]
- 43.Lee D.K., Carrasco,J., Hidalgo,J. and Andrews,G.K. (1999) Identification of a signal transducer and activator of transcription (STAT) binding site in the mouse metallothionein-I promoter involved in interleukin-6-induced gene expression. Biochem. J., 337, 59–65. [PMC free article] [PubMed] [Google Scholar]
- 44.Viollet B., Lefrancois Martinez,A.M., Henrion,A., Kahn,A., Raymondjean,M. and Martinez,A. (1996) Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J. Biol. Chem., 271, 1405–1415. [DOI] [PubMed] [Google Scholar]
- 45.Reisman D. and Rotter,V. (1993) The helix–loop–helix containing transcription factor USF binds to and transactivates the promoter of the p53 tumor suppressor gene. Nucleic Acids Res., 21, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lun Y., Sawadogo,M. and Perry,M. (1997) Autoactivation of Xenopus MyoD transcription and its inhibition by USF. Cell Growth Differ., 8, 275–282. [PubMed] [Google Scholar]
- 47.Zhang B., Georgiev,O., Hagmann,M., Günes,C., Cramer,M., Faller,P., Vasák,M. and Schaffner,W. (2003) Activity of metal responsive transcription factor-1 (MTF-1) by toxic heavy metals and H2O2in vitro is modulated by metallothionein. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cook T., Gebelein,B. and Urrutia,R. (1999) Sp1 and its likes: Biochemical and functional predictions for a growing family of zinc finger transcription factors. Ann. N. Y. Acad. Sci., 880, 94–102. [DOI] [PubMed] [Google Scholar]