Abstract
Eukaryotic DNA is organized into nucleosomes and higher order chromatin structure, which plays an important role in the regulation of many nuclear processes including DNA repair. Non-homologous end-joining, the major pathway for repairing DNA double-strand breaks (DSBs) in mammalian cells, is mediated by a set of proteins including DNA-dependent protein kinase (DNA-PK). DNA-PK is comprised of a large catalytic subunit, DNA-PKcs, and its regulatory subunit, Ku. Current models predict that Ku binds to the ends of broken DNA and DNA-PKcs is recruited to form the active kinase complex. Here we show that DNA-PK can be activated by nucleosomes through the ability of Ku to bind to the ends of nucleosomal DNA, and that the activated DNA-PK is capable of phosphorylating H2AX within the nucleosomes. Histone acetylation has little effect on the steps of Ku binding to nucleosomes and subsequent activation of DNA-PKcs. However, acetylation largely enhances the phosphorylation of H2AX by DNA-PK, and this acetylation effect is observed when H2AX exists in the context of nucleosomes but not in a free form. These results suggest that the phosphorylation of H2AX, known to be important for DSB repair, can be regulated by acetylation and may provide a mechanistic basis on which to understand the recent observations that histone acetylation critically functions in repairing DNA DSBs.
INTRODUCTION
DNA double-strand breaks (DSBs) are generated by exposure to ionizing radiation, and also naturally occur as a result of oxidative metabolism, stalled replication forks and V(D)J recombination. The major pathway for repairing DSBs in mammalian cells is the non-homologous end- joining (NHEJ), which is mediated by a set of cellular proteins that include DNA-PK, DNA ligase IV and XRCC4 (1). DNA-PK is composed of an ∼470 kDa catalytic subunit, DNA-PKcs and a heterodimer of Ku70 and Ku80 subunits. Mammalian cells that lack either component of the DNA-PK complex are defective in repairing DNA DSBs and hence are sensitive to the effects of exposure to ionizing radiation (2).
Ku is an abundant nuclear protein that exists in a heterodimeric form consisting of 70 (Ku70) and 80 kDa (Ku80) subunits. A unique property of Ku is its ability to bind with very high affinity to the ends of double-stranded DNA, independent of the sequence or precise structure of the DNA ends (2). Once bound to the DNA ends, Ku was shown to translocate inward thus allowing for multiple binding of the protein on a single DNA fragment (3). These properties of Ku reflect its characteristic structure with a preformed ring that encircles double helix while binding to the DNA ends (4). Because of its abundance and high affinity for DNA ends, Ku is thought to be the first protein to bind to the broken DNA ends during the process of NHEJ.
DNA-PKcs is a Ser/Thr kinase that belongs to the phosphatidylinositol-3 (PI-3) kinase family (2). The kinase domain of DNA-PKcs is encoded by the C-terminal 380 amino acids of the 4128 amino acids and its kinase activity is required for DSB repair and V(D)J recombination (5). The unusual property of DNA-PKcs is the requirement of DNA for its kinase activity. DNA-PKcs alone is capable of binding to DNA ends but the presence of Ku is required for its full activation (2). Current models predict that Ku binds to the ends of DNA and then recruits DNA-PKcs to form the active kinase complex. The XRCC4/DNA ligase IV complex is then recruited to the site of damage for rejoining of the DSB (1,2). In support of this model, a recent electron microscopic study has revealed that DNA-PKcs is capable of bringing DNA ends together to form DNA synapsis and this synaptic formation is greatly enhanced by Ku (6).
In eukaryotic cells, DNA is organized into nucleosomes and higher order chromatin structures that provide a regulatable impediment to the access of proteins to their target DNA (7–9). The nucleosomes, consisting of the histone octamer (two molecules of each core histone, H2A, H2B, H3 and H4) wrapped around by 146 bp DNA, are subjected to the various mode of structural changes, of which reversible acetylation is the best-known modification that critically functions in the regulation of transcription (10,11). In general, acetylation is known to render chromatin more accessible to proteins whereas deacetylation is associated with repressive chromatin (12,13). Acetylation exerts its effect by both modulating the compactness of higher order chromatin and directly inducing a conformational change at the level of a single nucleosome.
By analogy with its crucial function in the regulation of transcription, the chromatin is presumed to have roles in DNA repair. Indeed, evidence from recent studies strongly suggests that histone acetylation plays an important role in the process of DSB repair (14–18). For example, acetylation of histone H4 has been shown to be required for repairing DSBs by the NHEJ pathway and another pathway termed as replication-coupled repair (17). Another evidence for a direct link between DSB repair and chromatin comes from the finding that H2AX, a variant of histone H2A, is phosphorylated in response to ionizing radiation and other agents that introduce DSBs (19,20). H2AX is phosphorylated at the C-terminal at Ser139. The two amino acids that follow Ser139 are glutamine and glutamate, thus making it a consensus SQE motif for phosphorylation by the PI-3-like family of kinases including DNA-PK, ATM and ATR. This family of PI-3-like kinase is involved in DNA damage response and may play a role in the phosphorylation of H2AX in response to DNA DSBs (20). γ-H2AX, the phosphorylated form of H2AX, has been suggested to spread over a megabase range of chromatin and serve to recruit a number of repair proteins to the site of DNA damage (21,22). Recent gene targeting experiments highlighted the importance of the formation of γ-H2AX in DSB repair and hence radiation resistance of cells; however, its role in DSB repair remains unknown (23–25).
As a step towards understanding the role of chromatin structure in DSB repair, we have investigated the effects of nucleosomes on activation of DNA-PK and its ability to phosphorylate H2AX. Here we show that DNA-PK can be activated by nucleosomes through the ability of Ku to bind to the ends of nucleosomal DNA, and that the activated DNA-PK is capable of phosphorylating H2AX within the nucleosomes. Histone acetylation has little effect on these events. Interestingly, however, acetylation largely enhances the phosphorylation of H2AX by DNA-PK in the context of nucleosomes but not in the form of free histone.
MATERIALS AND METHODS
Proteins and antibodies
Ku and DNA-PKcs were purified to near homogeneity from human placenta as previously described (26). Non-acetylated and hyperacetylated core histones were purified from untreated and butyrate-treated HeLa cells, respectively, as previously described (27). Antibodies against γ-H2AX, acetylated H4, phosphorylated H3 (Ser10) and H2A (acidic patch) were purchased from Upstate Biotechnology.
Nucleosome assembly and chromatin purification
Nucleosomes were assembled by salt dilution method using purified histone octamer and a 150-bp DNA containing the 12RSS (12/TP1), and mononucleosomes were purified through a 5–30% glycerol gradient as previously described (27). H1-depleted oligonucleosomes were purified from HeLa cells essentially as described (28). In brief, HeLa chromatin was digested with 5 U/ml of micrococcal nuclease (Sigma) at 37°C for 5 min, and then adjusted to a final concentration of 0.6 M NaCl to dissociate H1 from oligonucleosome cores. Oligonucleosomes on the average length of two to three nucleosomes were purified through a 10–40% glycerol gradient. To prepare hyperacetylated oligonucleosomes, the chromatin of butyrate-treated HeLa cells was digested with 3 U/ml of micrococcal nuclease (Sigma), and hyperacetylated oligonucleosomes were Mg2+-fractionated as previously described (27), and subjected to a glycerol gradient purification. The extent of acetylation of purified oligonucleosomes was determined by acid urea gel analysis and western blot with specific antibodies.
Gel shift and DNase I footprinting
Gel shift asssays were carried out in 20 µl reactions containing 25 mM Hepes (pH 7.5), 60 mM potassium glutamate, 2 mM CaCl2, 2 mM DTT and 0.1 mg/ml BSA. Ku and/or DNA-PKcs were incubated with 0.5 ng of 32P-labeled template DNA in the free form or in the context of mononucleosomes at room temperature for 20 min. Samples were then subjected to a polyacrylamide gel electrophoresis (PAGE) in 0.5× TBE, 5% acrylamide (29:1) and 5% glycerol for 3 h at 200 V at 4°C. DNase I footprinting was done by subjecting the Ku binding reactions described above to DNase I digestion followed by band separation on an 8% sequencing gel as previously described (29).
DNA-PK kinase assays
Kinase assays were essentially the same as previously described (30), except that PHAS-1 protein, free histones, assembled mononucleosomes or purified oligonucleosomes were used as substrates. Ten microliter volume of kinase reaction contains 25 mM Hepes (pH 7.6), 75 mM KCl, 10 mM MgCl2, 1 mM DTT, 1 mM ATP (omitted when [γ-32P]ATP was used), 1 µCi [γ-32P]ATP (where indicated), 10 µg/ml of fragmented calf thymus (CT) DNA (where indicated), 40 ng DNA-PKcs, 40 ng Ku70/80. To this, substrates were added as follows: 100 ng of PHAS-1, 200 ng of free histones, 0.5–1 ng (in 32P-labeled DNA) of assembled mononucleosomes, 25 ng of purified oligonucleosomes. Free core histones prepared in 2.5 M NaCl (27) were dialyzed against buffer containing 25 mM Hepes (pH 7.6) and 100 mM NaCl prior to addition to kinase reactions. Kinase reactions were incubated at 30°C for 10 min, stopped with SDS sample buffer, and subjected to 15% SDS–PAGE. Gels were either dried followed by exposure to X-ray film for autoradiograph or subjected to western blot analysis.
RESULTS
Ku binds to nucleosomal DNA in the manner that does not permit inward translocation
In vitro, Ku binds to the ends of naked DNA with a characteristic manner: upon the initial binding to the DNA ends, Ku translocates inward thus allowing for multiple proteins binding to the same DNA molecule. We wanted to investigate how Ku would bind to the ends of DNA in the context of nucleosomes. Nucleosomes were assembled using a 150-bp DNA fragment by salt dilution method and mononucleosomes were purified through a glycerol gradient. Purified nucleosomes typically contained 2–10% of free DNA as assessed by native gel analysis.
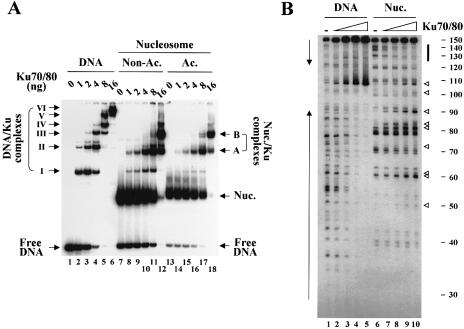
First, we examined the ability of Ku to bind to nucleosomes by gel shift assays. Incubation of free template DNA with moderate concentrations of Ku generated two shifted bands (complexes I and II) and these bands were further shifted by incubation with higher concentrations of Ku (complexes III–VI) (Fig. 1A, lanes 1–6). This is the typical pattern of Ku binding to free DNA, reflecting the inward translocation and multiple binding of Ku proteins (3). When incubated with nucleosomes, Ku was able to bind to the nucleosomes (Fig. 1A, lanes 7–12). In contrast to that on free DNA, however, Ku binding to nucleosomes resulted in the generation of only two shifted bands (complexes A and B); the minor bands are due to Ku binding to the free DNA present in our nucleosome preparations (lanes 7–12). These two bands did not further shift even at the highest concentration of Ku tested (lane 12), indicating that multiple binding of Ku is not permitted on a nucleosome. Incubation of nucleosomes with very high concentrations of Ku (from 64 to 256 ng) resulted in most of the radioactivity being left unresolved in the well (data not shown).
Figure 1.
Ku binds to nucleosomes. (A) Gel mobility shifts assays. Indicated amounts of Ku were incubated with in vitro assembled mononucleosomes in either a non-acetylated (lanes 7–12) or an acetylated state (13–18), or with free template DNA as a control (lanes 1–6) and the reactions were subjected to 5% native gel analysis. The bands representing the complexes of Ku/DNA (complexes I–VI) and of Ku/nucleosomes (complexes A and B) are indicated. (B) DNase I footprinting assays. The binding reactions in which increasing concentrations of Ku were incubated with free DNA (lanes 1–5) or non-acetylated nucleosomes (6–10) were subjected to DNase I digestion followed by band separation on an 8% denaturing gel. DNase I protection on free template DNA is indicated by vertical arrows on the left of the gel. The typical 10-bp digestion pattern of rotationally positioned nucleosomes is noted on the far right side of the gel. The vertical solid bar indicates the region of nucleosomal DNA protected by Ku. The open triangles indicate the bands that were enhanced by Ku binding to nucleosomes.
We next analyzed Ku binding to nucleosomes by DNase I footprinting assays. Free template DNA as a control showed the expected pattern of DNase I digestion, such that, as Ku increased, DNase I protection extended from the termini to the center of the DNA molecule (Fig. 1B, lanes 1–5). DNase I digestion of nucleosomes without Ku generated the pattern of 10-bp ladder through the length of DNA, indicative of rotationally positioned nucleosomes (lane 6). Incubation of nucleosomes with Ku followed by DNase I digestion resulted in the protection of the sequence covering 20–30 bp at the DNA ends (lanes 7–10). These data demonstrated that Ku binds throughout free DNA but only to the ends of nucleosomal DNA, in keeping with the results from the gel shift experiments. Interestingly, Ku binding caused enhancement of DNase I digestion at many sites out of the protected region (lanes 7–10; noted by open triangles), suggestive of induction of structural changes on nucleosomes (possibly distortions of DNA double helix) upon Ku binding. Nonetheless, the overall structure of the nucleosomes appeared to remain unchanged after Ku binding as diagnosed by the retained 10-bp DNase I digestion.
Acetylation does not significantly affect Ku binding to nucleosomes
While acetylation is thought to have a major effect on the interaction between individual nucleosomes, its direct effect on a single nucleosome has also been documented. For example, acetylation has been shown to enhance the access of RAG proteins and thereby V(D)J cleavage on DNA organized into a nucleosome (29). We therefore asked whether acetylation could affect Ku binding to nucleosomes. Acetylated nucleosomes were prepared in the same way as non-acetylated nucleosomes by using hyperacetylated core histones purified from butyrate-treated HeLa cells. Gel shift analysis showed that Ku bound to acetylated nucleosomes in much the same way it did to non-acetylated nucleosomes; formation of the complexes A and B only (Fig. 1A, lanes 13–18). The pattern of DNase I footprinting for Ku binding to acetylated nucleosomes was almost identical to that for non-acetylated nucleosomes (data not shown). These results indicate that inward translocation of Ku is not permitted even on acetylated nucleosomes. In addition, the results also indicate that the levels of Ku binding to nucleosomes are not much affected by acetylation. Consistent with these results, the subsequent recruitment and activation of DNA-PKcs following Ku binding on nucleosomes was not affected by acetylation (see Fig. 5). The discussion of the results below with regard to this issue thus focuses on those experiments with non-acetylated nucleosomes.
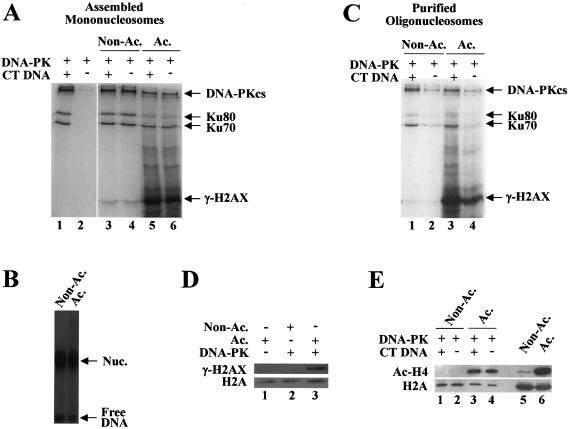
Figure 5.
Histone acetylation stimulates DNA-PK phosphorylation of H2AX in the context of nucleosomes. (A) Kinase reactions were performed under the standard conditions using the assembled mononucleosomes whose radioactivity had been decayed by storing at 4°C for about 3 months. The gel shown here is from a 4 h exposure. (B) Native gel analysis of the nucleosomes used in (A) with a prolonged exposure (7 days) shows that they are still intact. (C) Oligonucleosomes (two to three nucleosomes on the average) purified from untreated (non-acetylated) or butyrate-treated (acetylated) HeLa cells were used as substrates in the kinase reactions under the standard conditions. (D) A 5-fold scale-up of the same reactions as lanes 2 and 4 of (C) except substituting cold ATP for [γ-32P]ATP was subjected to SDS–PAGE followed by western blot with γ-H2AX antibody (lanes 2 and 3). The control reaction in which non-acetylated oligonucleosomes were incubated without DNA-PK was also included (lane 1). (E) The same reactions as in (C) except substituting cold ATP for [γ-32P]ATP were subjected to SDS–PAGE followed by western blot with γ-H2AX antibody (lanes 1–4). The extent of histone acetylation of the purified oligonucleosomes was determined by western blot using acetylated H4 antibody (lanes 5 and 6).
DNA-PKcs can bind to Ku-bound nucleosomes
Although DNA-PKcs alone is able to bind to DNA ends to some extent, a prior binding of Ku is required for the maximal binding and activation of DNA-PKcs (31). In addition, the property of inward translocation of Ku has been suggested to facilitate subsequent recruitment of DNA-PKcs (32). Given the results that inward translocation of Ku did not occur on nucleosomal DNA, it was interesting to see whether DNA-PKcs still could bind to the nucleosomes following Ku binding.
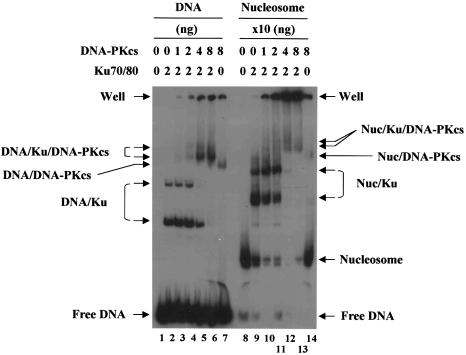
To address this issue, we performed gel shift assays using highly purified human DNA-PKcs. As previously shown (33), DNA-PKcs readily bound to free DNA when the DNA was preoccupied by Ku, and it also bound to DNA in the absence of Ku to some extent (Fig. 2, lanes 1–7). Incubation of nucleosomes with DNA-PKcs in the presence of Ku consistently resulted in the generation of smeared bands migrating slower than the nucleosome/Ku complexes (lanes 8–13). We believe that these bands are likely to represent the complexes of DNA-PKcs/nucleosome/Ku although they could not be maximally detected in our experimental conditions, partly due to the poorly resolved bands left in the well. More experiments will be necessary for better defining these bands together with the band detected from the incubation of nucleosomes with DNA-PKcs in the absence of Ku (lane 14).
Figure 2.
Binding of DNA-PKcs to Ku-bound nucleosomes. DNA-PKcs were incubated with free DNA or nucleosomes (non-acetylated) in the presence or absence of Ku as indicated, and the reactions were subjected to 5% native gel analysis. Note that 10 times more Ku and DNA-PKcs were used for the reactions with nucleosomes as compared to free DNA.
DNA-PK can be activated by nucleosomes
The observations that Ku and DNA-PKcs bound to nucleosomes prompted us to ask if nucleosomes could stimulate the activity of DNA-PK. To address this question we performed in vitro kinase assays using PHAS-1, a protein that has been previously shown to be a potent substrate of DNA-PK (34). As expected, DNA, fragmented CT DNA in this case, was required for the activation of DNA-PK as assessed by measuring the autophosphorylation of DNA-PKcs/Ku (lanes 4–5 in Fig. 3C) or the phosphorylation of PHAS-1 (data not shown). Addition of nucleosomes to the kinase reactions lacking CT DNA resulted in robust stimulation of the phosphorylation of PHAS-1, with the magnitude of stimulation being proportional to the concentrations of added nucleosomes (Fig. 3B, lanes 1–6).
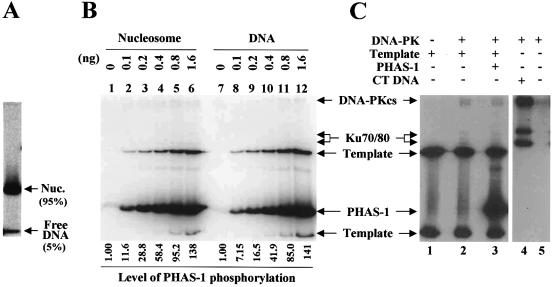
Figure 3.
DNA-PK is activated by nucleosomes. (A) Native gel analysis shows that ∼5% of free DNA is present in the nucleosome preparation (non-acetylated) used in the kinase assays described below. (B) In vitro kinase assays were performed under the conditions described in Materials and Methods. Indicated amounts of nucleosomes (non-acetylated) and free template DNA were incubated with DNA-PKcs/Ku in the presence of PHAS-1 and [γ-32P]ATP. Reactions were subjected to SDS–PAGE followed by autoradiograph. The amounts of nucleosomes are in DNA. The relative levels of the PHAS-1 phosphorylation, shown at the bottom of the gel, were drawn from the quantitation of PhosphoImage of the gel. (C) Control experiments identified the bands of phosphorylated PHAS-1 on the gel by distinguishing them from those of template DNA. The phosphorylated PHAS-1 band was detected only when PHAS-1 protein was added to the reactions (lane 3).
Since the nucleosome preparation used in the assays contained ∼5% of free DNA as assessed by a native gel (Fig. 3A), it was necessary to determine how much the free DNA would contribute to the levels of the observed stimulation. We performed the titration experiments in which a range of the concentrations of free template DNA was tested for its effect on the phosphorylation of PHAS-1 (Fig. 3B, lanes 7–12). The results indicated that the robust stimulation of the PHAS-1 phosphorylation by nucleosomes was attributed to nucleosomes per se but not to the contaminating free DNA. For instance, the activity supported by 1.6 ng of nucleosomes (in DNA) was about 20 times higher than that supported by 0.1 ng of free DNA (compare lane 6 and lane 8 in Fig. 3B); the amount of free DNA present in 1.6 ng of nucleosomes is expected to be 0.08 ng. We therefore concluded that DNA-PK can be activated by nucleosomes. Histone acetylation did not affect the magnitude with which nucleosomes stimulated the activity of DNA-PK as assessed by either the PHAS-1 phosphorylation (data not shown) or the phosphorylations of DNA-PKcs and Ku (see Fig. 5A and C).
DNA-PK phosphorylates H2AX both in a free form and in the context of nucleosomes
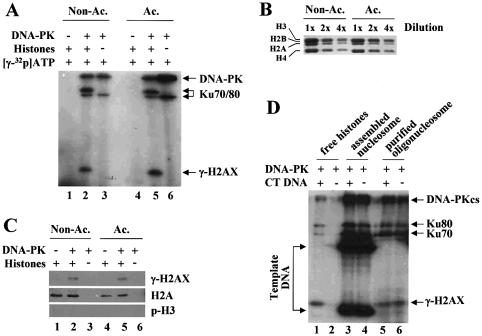
Having found that DNA-PK could be activated by nucleosomes, we asked whether the activated DNA-PK would phosphorylate H2AX in the context of nucleosomes. Prior to addressing this question, we wanted to confirm the previous observation that H2AX is a substrate of DNA-PK, and carried out kinase assays with purified HeLa core histones, where ∼2.4% of the total H2A protein level is H2AX (19). DNA-PK phosphorylated a histone migrating at the position of H3 (Fig. 4A, lanes 1–3), and this phosphorylated histone was identified as γ-H2AX by western blot analysis using γ-H2AX antibody (Fig. 4C, lanes 1–3). The phosphorylation of the free form of H2AX by DNA-PK required the presence of DNA as expected (Fig. 4D, lanes 1 and 2).
Figure 4.
DNA-PK phosphorylates H2AX both in a free form and in the context of nucleosomes. (A) Kinase assays were performed using as substrates the core histones purified from untreated (non-acetylated) or butyrate-treated (acetylated) HeLa cells. γ-H2AX, which was verified by western blot (see below), and phosphorylated DNA-PKcs and Ku are indicated. (B) Coomassie-stained SDS–PAGE for serially diluted core histones shows that equal amounts of the two types of histones were used in the reactions described in (A). (C) Reactions for kinase assays were performed under the standard conditions except substituting cold ATP for [γ-32P]ATP, and were subjected to SDS–PAGE followed by western blot analysis with the antibodies specific for γ-H2AX, H2A or phosphorylated H3 (Ser10). (D) Kinase assays were performed with the indicated substrates (non-acetylated form) in the presence or absence of fragmented CT DNA. The phosphorylation of H2AX by DNA-PK was clearly detected when H2AX was in a free form (lanes 1 and 2) or in the context of oligonucleosomes (two to three nucleosomes on the average, lanes 5 and 6). The bands of γ-H2AX from the reactions containing freshly assembled mononucleosomes (lanes 3 and 4) are not apparent due to the interfering bands of 32P-labeled template DNA.
We then examined whether DNA-PK could phosphorylate H2AX in the context of the nucleosomal structure. Two types of nucleosomes were used as substrates; oligonucleosomes (two to three nucleosomes on the average) purified from HeLa cells and in vitro assembled mononucleosomes. As shown in Figure 4D (lanes 5–6), DNA-PK was able to phosphorylate H2AX within the oligonucleosomes, which was also verified by western blot analysis (data not shown). The phosphorylation of H2AX in the context of oligonucleosomes did not require the presence of DNA, again demonstrating that nucleosomes are sufficient to activate DNA-PK (Fig. 4D, lanes 5 and 6, and also see Fig. 5). Demonstrating the phosphorylation of H2AX by DNA-PK using assembled mononucleosomes was problematic because 32P-labeled DNA template migrates at the same molecular weight as H2AX in SDS–PAGE gels (see Fig. 4D, lanes 3 and 4). We overcame this problem either by subjecting nucleosomes to an intensive DNase I digestion prior to gel analysis (data not shown) or by using the nucleosomes whose radioactivity had been decayed (see below).
Acetylation stimulates the phosphorylation of H2AX by DNA-PK in the context of nucleosomes but not in a free form
We next asked whether acetylation could affect the phosphorylation of H2AX by DNA-PK. First, the effect of acetylation on the phosphorylation of free H2AX was examined by subjecting the equal amounts of non-acetylated and acetylated core histones to kinase assays (Fig. 4B). As shown in Figure 4A and C, acetylation did not have an effect on the phosphorylation of H2AX when it existed as a free histone.
Again, the two types of nucleosomes were examined with respect to the effect of acetylation on the phosphorylation of H2AX. As described above, the use of the nucleosomes assembled with freshly prepared 32P-labeled DNA resulted in the template DNA interfering with detecting the phosphorylation of H2AX (Fig. 4D, lanes 3 and 4). To avoid this problem, the radioactivity of assembled nucleosomes had been decayed by storing them at 4°C for a few months prior to being subjected to kinase assays. Native gel analysis with a prolonged exposure showed that the nucleosomes were still intact although the percentage of free DNA increased up to ∼20% (Fig. 5B). We found, using these nucleosomes, that DNA-PK could phosphorylate H2AX within the assembled nucleosomes, and that the phosphorylation of H2AX was greatly enhanced by acetylation (Fig. 5A, lanes 3–6). The magnitude with which DNA-PK was activated by nucleosomes, as assessed by the phosphorylations of DNA-PKcs and Ku in the reactions, was not increased by acetylation (Fig. 5A and also see Fig. 5C).
To confirm the acetylation effect on the H2AX phosphorylation, highly acetylated oligonucleosomes (two to three nucleosomes on the average) were purified from butyrate-treated HeLa cells. The extent of acetylation was determined by acid urea gel (data not shown) and western blot analyses (Fig. 5E, lanes 5 and 6). The phosphorylation of H2AX was barely detectable on the non-acetylated oligonucleosomes (Fig. 5C, lanes 1 and 2). However, a robust increase of the phosphorylation of H2AX was observed for the acetylated oligonucleosomes (Fig. 5C, lanes 3 and 4). This acetylation effect was also demonstrated by western blot analysis of the same reactions as the above except that cold ATP was substituted for [γ-32P]ATP (Fig. 5D). Similar experiments showed that the extent of acetylation remained unchanged during the kinase reactions (Fig. 5E, lanes 1–4). These results together demonstrate that histone acetylation stimulates the phosphorylation of H2AX by DNA-PK when H2AX is in the context of nucleosomes but not in a free form.
DISCUSSION
We have studied the effects of chromatin structure on the activation of DNA-PK and phosphorylation of its substrate, H2AX, with a highly purified system employing in vitro assembled mononucleosomes and cell-purified oligonucleosomes. Here we show that DNA-PK can be activated by nucleosomes through the ability of Ku to bind to the ends of nucleosomal DNA, and that the activated DNA-PK is capable of phosphorylating H2AX within the nucleosomes. Interestingly, histone acetylation has little effect on the steps of Ku binding to nucleosomes and subsequent activation of DNA-PKcs. However, acetylation largely stimulates the phosphorylation of H2AX by DNA-PK, and this acetylation effect is seen when H2AX exists in the context of nucleosomes but not in a free form. These results suggest that the phosphorylation of H2AX, thought to be important for DSB repair, can be regulated by histone acetylation.
The observation that Ku can readily bind to the termini of nucleosomal DNA may not be unexpected since the DNA at the entry/exit point of nucleosome has been suggested to be in a dynamic state whereby it continues to dissociate and reassociate with the histone octamer (35). Ku could easily come to catch the ends of nucleosomal DNA when they are in the off state. Unlike on free DNA, however, Ku binding is limited in the termini of nucleosomal DNA with the protein being unable to translocate beyond the distal 20–30 bp region. This is likely due to the inability of Ku to overcome the energy barrier that exists 25 bp from the terminus of nucleosomal DNA as has been previously suggested (36). Nonetheless, the Ku binding on nucleosomes supported the activation of DNA-PKcs, indicating that the distal 20–30 bp DNA of nucleosome is sufficient for the formation of the active kinase complex. In support of this argument, photo-crosslinking studies indicated that, prior to DNA-PKcs binding, Ku resides at 10-bp region of the DNA terminus, and that Ku shifts inward another 10 bp upon DNA-PKcs binding (32).
Our results showed that the magnitude with which Ku bound to nucleosomes was not significantly affected by acetylation. Consistent with these results, the activation of DNA-PKcs following Ku binding to nucleosomes was not enhanced by acetylation (see Fig. 5A and C). These results could be somewhat unexpected in that acetylation, in general, is known to have direct effect on nucleosomes in a way to render the nucleosomal DNA more accessible to proteins. However, acetylation may not be crucial for proteins such as Ku that bind to nucleosomes by gaining access through the ends of nucleosomal DNA. This interpretation is supported by the fact that histone acetylation occurs mainly on the N-terminal tails of H3 and H4, and these tails make contact with primarily the central 120-bp but not the distal region of the nucleosomal DNA (37). The access of Ku to the ends of nucleosomal DNA and subsequent activation of DNA-PKcs thus may not be dependent on histone acetylation.
When DNA DSB occurs on chromatin, it could exit in many different ways with respect to its relevant positions within a single nucleosome and/or the higher order chromatin structure. The DNA ends positioning at the entry/exit points of the nucleosome, as appeared in the mononucleosomes reconstituted on a 150-bp DNA fragment used in this study, may represent one way of DSB existing on the chromatin in the cell. In this particular case, the results from this work suggest that neither histone modification such as acetylation nor disruption of nucleosome such as removal of the histone octamer is required for DNA-PK to be activated in response to DSBs. It remains to be determined, although technically challenging, whether this would be the case for DSBs positioned away from the entry/exit points, for example, on the dyad.
While the kinase activity of DNA-PK is required for NHEJ, its physiological targets have remained elusive. DNA-PKcs appears to serve as its own target such that autophosphorylation of DNA-PKcs is required for repairing DSBs by NHEJ (38). H2AX is thought to be another target of DNA-PK, which implicates for a direct link between chromatin modification and DSB repair. Our data show that H2AX is an in vitro substrate of DNA-PK both in free form as previously reported (19) and in the context of nucleosome.
Interestingly, we find that acetylation largely stimulates phosphorylation of H2AX by DNA-PK when H2AX is in the context of nucleosomes. Since the acetylation effect is not seen for the free form of H2AX, the structural constraints imposed by the nucleosomal organization are likely to render DNA-PK targeting of H2AX to be sensitive to acetylation rather than H2AX molecule per se becomes more susceptible to the phosphorylation by DNA-PK when it is acetylated. Although the lack of the structural information on H2AX-containing nucleosome makes it difficult to interpret these results, a few plausible explanations are possible. A simple explanation would be that acetylation induces a conformational change of nucleosome in a way to make the SQE motif on the C-terminal tail of H2AX more accessible to DNA-PK. Alternatively, but not mutually exclusively, the overall structure of acetylated nucleosome might better fit DNA-PK for the enzymatic function than its non-acetylated counterpart.
Recent genetic and biochemical studies have provided implications for direct involvement of histone acetylation in DSB repair (reviewed in 18). For example, acetylation of histones H3/H4 and the histone acetyltransferases responsible for these modifications have been demonstrated to be required for DSB repair (16,17). Our results, showing that the phosphorylation of H2AX on nucleosomes is dependent on acetylation, may provide a mechanistic framework within which to understand those observations. Acetylation could serve as a signal for the process of expanding γ-H2AX over a megabase range of chromatin at the site of DSB as previously suggested (21). It is also possible that acetylation functions as a simple modifier of charge on chromatin to induce its structural changes in a way to facilitate the access of repair proteins to the site of DNA damage. Alternatively, as postulated by the histone code hypothesis (39,40), acetylation and γ-H2AX, individually or in combination, could serve as binding sites for repair factors or proteins responsible for chromatin remodeling. In this regard, it will be of interest to examine whether acetylation of histones at the site of DNA damage is dependent on H2AX phosphorylation. Future experiments to test these possibilities and others will provide insights into the better understanding of the roles of chromatin and its modifications in DSB repair.
Acknowledgments
ACKNOWLEDGEMENTS
D.W.C was supported by postdoctoral fellowships from the Canadian Institutes of Health Research and the Alberta Heritage Foundation for Medical Research. M.A.O. was supported by the Leukemia and Lymphoma Scholars Program. This work was supported by the Brain Korea 21 Project (J.K.).
REFERENCES
- 1.Jackson S.P. (2002) Sensing and repairing DNA double-strand breaks. Carcinogenesis, 23, 687–696. [DOI] [PubMed] [Google Scholar]
- 2.Smith G.C.M. and Jackson,S.P. (1999) The DNA-dependent protein kinase. Genes Dev., 13, 916–934. [DOI] [PubMed] [Google Scholar]
- 3.Dynan W.S. and Yoo,S. (1998) Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res., 26, 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker J.R., Corpina,R.A. and Goldberg,J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature, 412, 607–614. [DOI] [PubMed] [Google Scholar]
- 5.Kurimasa A., Kumano,S., Boubnov,N.V., Story,M.D., Tung,C.S., Peterson,S.R. and Chen,D.J. (1999) Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol. Cell. Biol., 19, 3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeFazio L.G., Stansel,R.M., Griffith,J.D. and Chu,G. (2002) Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J., 21, 3192–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Workman J.L. and Kingstone,R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- 8.Kornberg R.D. and Lorch,Y. (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell, 98, 285–294. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad K. and Henikoff,S. (2002) Epigenetic consequences of nucleosome dynamics. Cell, 111, 281–284. [DOI] [PubMed] [Google Scholar]
- 10.Wolffe A.P. and Hayes,J.J. (1999) Chromatin disruption and modification. Nucleic Acids Res., 27, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- 12.Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- 13.Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- 14.Barlev N.A., Poltoratsky,V., Owen-Hughes,T., Ying,C., Liu,L., Workman,J.L. and Berger,S.L. (1998) Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol. Cell. Biol., 18, 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikura T., Ogryzko,V.V., Grigoriev,M., Groisman,R., Wang,J., Horikoshi,M., Scully,R., Qin,J. and Nakatani,Y. (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell, 102, 463–473. [DOI] [PubMed] [Google Scholar]
- 16.Qin S. and Parthun,M.R. (2002) Histone H3 and histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol., 22, 8353–8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bird A.W., Yu,D.Y., Pray-Grant,M.G., Qiu,Q., Harmon,K.E., Megee,P.C., Grant,P.A., Smith,M.M. and Christman,M.F. (2002) Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature, 419, 411–415. [DOI] [PubMed] [Google Scholar]
- 18.Hasan S. and Hottiger,M.O. (2002) Histone acetyltransferases: a role in DNA repair and DNA replication. J. Mol. Med., 80, 463–474. [DOI] [PubMed] [Google Scholar]
- 19.Rogakou E.P., Pilch,D.R., Orr,A.H., Ivanova,V.S. and Bonner,W.M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem., 273, 5858–5868. [DOI] [PubMed] [Google Scholar]
- 20.Redon C., Pilch,D., Rogakou,E., Sedelnikova,O., Newrock,K. and Bonner,W.M. (2002) Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev., 12, 162–169. [DOI] [PubMed] [Google Scholar]
- 21.Rogakou E.P., Boon,C., Redon,C. and Bonner,W.M. (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol., 146, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paull T.T., Rogakou,E.P., Yamazaki,V., Kirchgessner,C.U., Gellert,M. and Bonner,W.M. (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol., 10, 886–895. [DOI] [PubMed] [Google Scholar]
- 23.Downs J.A., Lowndes,N.F. and Jackson,S.P. (2000) A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature, 408, 1001–1004. [DOI] [PubMed] [Google Scholar]
- 24.Celeste A. et al. (2002) Genomic instability in mice lacking histone H2AX. Science, 296, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bassing C.H., Chua,K.F., Sekiguchi,J., Suh,H., Whitlow,S.R., Fleming,J.C., Monroe,B.C., Ciccone,D.N., Yan,C., Vlasakova,K., Livingston,D.M., Ferguson,D.O., Scully,R. and Alt,F.W. (2002) Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl Acad. Sci. USA, 99, 8173–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan D.W., Mody,C.H., Ting,N.S. and Lees-Miller,S.P. (1996) Purification and characterization of the double-stranded DNA-activated protein kinase, DNA-PK, from human placenta. Biochem. Cell Biol., 74, 67–73. [DOI] [PubMed] [Google Scholar]
- 27.Kwon J., Morshead,K.B., Guyon,J.R., Kingston,R.E. and Oettinger,M.A. (2000) Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell, 6, 1037–1048. [DOI] [PubMed] [Google Scholar]
- 28.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1989) Current Protocols in Molecular Biology. Greene & Wiley Interscience, New York, NY. [Google Scholar]
- 29.Kwon J., Imbalzano,A.N., Matthews,A. and Oettinger,M,A. (1998) Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol. Cell, 2, 829–839. [DOI] [PubMed] [Google Scholar]
- 30.Chan D.W. and Lees-Miller,S.P. (1996) The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem., 271, 8936–8941. [DOI] [PubMed] [Google Scholar]
- 31.Yaneva M., Kowalewski.T. and Lieber,M.R. (1997) Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J., 16, 5098–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo S. and Dynan,W.S. (1999) Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res., 27, 4679–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dvir A., Peterson,S.R., Knuth,M.W., Lu,H. and Dynan,W.S. (1993) Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl Acad. Sci. USA, 89, 11920–11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan D.W., Son,S.C., Block,W., Ye,R., Khanna,K.K., Wold,M.S., Douglas,P., Goodarzi,A.A., Pelley,J., Taya,Y., Lavin,M.F. and Lees-Miller,S.P. (2000) Purification and characterization of ATM from human placenta. A manganese-dependent, wortmannin-sensitive serine/threonine protein kinase. J. Biol. Chem., 275, 7803–7810. [DOI] [PubMed] [Google Scholar]
- 35.Widom J. (1998) Structure, dynamics and function of chromatin in vitro. Annu. Rev. Biophys Biomol. Struct., 27, 285–327. [DOI] [PubMed] [Google Scholar]
- 36.Studitsky V.M., Clark,D.J. and Felsenfeld,G. (1995) Overcoming a nucleosomal barrier to transcription. Cell, 83, 19–27. [DOI] [PubMed] [Google Scholar]
- 37.Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 38.Chan D.W., Chen,B.P., Prithivirajsingh,S., Kurimasa,A., Story,M.D., Qin,J. and Chen,D.J. (2002) Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev., 16, 2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 40.Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]