Abstract
For decades, hundreds of different human tumor type–specific cell lines have been used in experimental cancer research as models for their respective tumors. The veracity of experimental results for a specific tumor type relies on the correct derivation of the cell line. In a worldwide effort, we verified the authenticity of all available esophageal adenocarcinoma (EAC) cell lines. We proved that the frequently used cell lines SEG-1 and BIC-1 and the SK-GT-5 cell line are in fact cell lines from other tumor types. Experimental results based on these contaminated cell lines have led to ongoing clinical trials recruiting EAC patients, to more than 100 scientific publications, and to at least three National Institutes of Health cancer research grants and 11 US patents, which emphasizes the importance of our findings. Widespread use of contaminated cell lines threatens the development of treatment strategies for EAC.
CONTEXT AND CAVEATS
Prior knowledge
Human tumor cell lines are commonly used in basic cancer research as preclinical models of human cancer. Research on esophageal adenocarcinoma relies heavily on these cell lines because of the limited availability of patient samples and animal models.
Study design
In collaboration with the primary investigators who established the cell lines, the authenticity of all currently available esophageal adenocarcinoma cell lines were examined using data from pathology archives and genotyping assays.
Contribution
Three commonly used cell lines were identified as being contaminated and were confirmed as being tumor types other than esophageal adenocarcinoma. Two of these cell lines have been used in 11 US patents and in more than 100 published studies, which have led to clinical trials of esophageal adenocarcinoma patients. The 10 cell lines whose authenticity was verified will be placed in public repositories to promote future research.
Implications
The development of treatments for esophageal adenocarcinoma may be negatively affected by the widespread use of these contaminated cell lines.
Limitations
It was not possible to include in this analysis studies that have not been published that may also be using the contaminated cell lines or that were based on results from studies using the contaminated cell lines.
From the Editors
Cell lines derived from human cancers have been crucial to building our understanding of the molecular pathophysiology of cancer and its treatment. Of equal importance, they form an in vitro model system for rational drug discovery and development because they are easy to maintain and manipulate in vitro and in animal xenograft models. However, it has been estimated that up to one-third of all cell lines have an origin other than that supposed (1). Cross-contamination between cell lines and mislabeling of cultures lead to unrecognized cell line admixtures (1,2). In the past, the scientific community has recognized this problem, but decisive action has not been taken to date. Results based on experiments using contaminated cell lines might be translated to the clinic, forming the basis for clinical trials, and directly affecting the treatment of patients.
Model research on esophageal adenocarcinoma (EAC), which is the cancer type showing the steepest rise in incidence in the Western world over recent years (3), relies entirely on a relatively small set of established tumor cell lines. Appropriate animal models and familial cases for EAC are lacking (4). Cell lines are very useful to investigate molecular pathways that are involved in EAC tumorigenesis and to test experimental drugs on EAC cells in vitro and in vivo. Despite intensive efforts to culture EAC cells in vitro, only 14 permanent cell lines have been established: SEG-1, BIC-1, and FLO-1 (5); SK-GT-4, SK-GT-5, and BE-3 (6); KYAE-1 (7); OE19 and OE33 (8); JH-EsoAd1 (9); OACP4C and OACM5.1 (10); and two newly established cell lines ESO26 and ESO51 (by Grupo de Estudos de Esófago de Barrett do IPOLFG, Lisbon, Portugal). In collaboration with the primary investigators who established the cell lines, the original EAC tissues for 13 of the 14 cell lines were traced in pathology archives and made available for study (anonymously): The original tissue for cell line BE-3 (6) was not found. The availability of the primary tissues made it possible to authenticate these EAC cell lines by comparing the genotype of the cell line with the genotypes of patient’s normal and tumor tissue (see Supplementary Materials and Methods, available online, for detailed methods). Genotyping was performed by short tandem repeat profiling using the polymerase chain reaction–based Powerplex 16 System (Promega, Madison, WI) (1). To further verify the authenticity of the cell lines, TP53 mutation analysis was performed (11). All exons and intron–exon boundaries of the TP53 gene were sequenced in all the EAC cell lines (Asper Biotech Ltd, Tartu, Estonia). The TP53 (GenBank accession number AF307851.1) mutations identified in the cell lines were then investigated in the original tumor tissues from which the cell lines had been derived. Ten of the 13 cell lines unambiguously had the same genotype and harbored the same TP53 mutation(s) as the original tissues, proving their correct derivation (Table 1 and Supplementary Table 1, available online). The most frequently used EAC cell lines SEG-1 and BIC-1 and the SK-GT-5 cell line had genotypes different from the original tissue, of which the cell line was stated. Comparison of the genotypes of SEG-1, BIC-1, and SK-GT-5 with genotypes available from databases (http://www.lgcstandards-atcc.org/ATCCCulturesandProducts/CellBiology/STRProfileDatabase/tabid/986/Default.aspx) revealed that SEG-1 is lung carcinoma (large cell lung cancer) cell line H460 (ATCC_HTB-177) and BIC-1 is colorectal adenocarcinoma cell line SW620 (ATCC_CCL-227). The genotype and TP53 mutation of the SK-GT-5 cell line matched with the tissue from which the cell line SK-GT-2 was derived, indicating that cell line SK-GT-5 is actually the gastric fundus carcinoma cell line SK-GT-2 (Supplementary Table 2, available online). In independent experiments, the researcher who established cell lines SEG-1 and BIC-1 (D. G. Beer) confirmed our results using the earliest passages of these cell lines. Clearly, contamination occurred early during establishment of the cell lines, and all of the cultures that were distributed subsequently to different laboratories were contaminated. We obtained the earliest available passage of cell line SK-GT-5 (1993) from D. S. Schrump (on referral by N. K. Altorki). This sample matched tissue from which cell line SK-GT-2 was derived, indicating that cross-contamination had occurred at the site of origin or during the early interinstitutional exchange of cell line SK-GT-5.
Table 1.
Short tandem repeat profiles (number of repeats at each locus is indicated) of 14 established esophageal adenocarcinoma cell lines*
| Cell line | Short tandem repeat locus |
|||||||||||||||
| Penta E | D18S51 | D21S11 | TH01 | D3S1358 | FGA | TPOX | D8S1179 | vWA | Penta D | CSF1PO | D16S539 | D7S820 | D13S317 | D5S818 | Amg | |
| SEG-1 | 5 | 13, 15 | 30 | 9.3 | 15, 18 | 21, 23 | 8 | 12 | 17 | 11, 13 | 11, 12 | 9 | 9, 12 | 13 | 9, 10 | X, Y |
| BIC-1 | 10 | 13 | 30, 30.2 | 8 | 15, 16 | 24 | 11 | 13 | 16 | 9, 15 | 13, 14 | 9, 13 | 8, 9 | 12 | 13 | X |
| FLO-1 | 5, 17 | 14, 16 | 30, 32.2 | 6 | 15 | 21 | 9, 11 | 13 | 16 | 11, 12 | 11 | 12, 13 | 8 | 11 | 12, 14 | X |
| KYAE-1 | 5, 8 | 14, 15 | 29, 31 | 6 | 15, 16 | 18 | 8 | 13, 14 | 14 | 9, 10 | 11 | 11 | 11, 12 | 11, 12 | 10, 13 | X |
| OE19 | 5, 8 | 12, 13 | 30 | 8, 9 | 15, 18 | 23, 26 | 8 | 13, 15 | 16, 17, 18 | 9 | 11, 13 | 12, 13 | 8 | 9, 11 | 11, 14 | X |
| OE33 | 12, 18 | 12 | 29, 31.2 | 7, 8 | 18 | 23 | 8, 11 | 10, 11, 12 | 17 | 9, 11 | 10, 11 | 12 | 9, 10 | 14 | 11 | X |
| OACM5.1 | 7, 14 | 16 | 28, 31 | 6, 9.3 | 16, 17 | 22 | 8 | 13, 14 | 19, 20 | 10 | 10, 13 | 10, 11 | 8 | 11, 12 | 12 | X |
| OACP4C | 20 | 12, 13 | 30 | 9 | 18 | 20 | 11 | 13 | 16 | 12 | 11 | 12 | 9, 11 | 12 | 9 | X |
| SK-GT-4 | 7 | 14 | 31.2 | 6, 9.3 | 17 | 22, 23 | 8, 10 | 13 | 17, 19 | 9 | 11, 15 | 11, 12, 13 | 7, 11 | 9, 10 | 12 | X |
| SK-GT-5 | 15, 17 | 14, 15 | 32.2 | 8, 9 | 15 | 26 | 9, 12 | 15 | 15, 18 | 9 | 10 | 11, 12 | 9, 10 | 12, 13 | 10 | X |
| BE-3 | 10, 16 | 17 | 30.2 | 9 | 16, 17 | 22 | 8, 10 | 12, 14 | 18 | 13 | 11 | 12 | 8, 11 | 11, 12 | 11, 13 | X |
| JH-EsoAd1 | 11 | 12 | 30 | 6, 7 | 16 | 24 | 8, 9 | 10 | 18, 19 | 14 | 10 | 10, 12 | 10, 12 | 11 | 11 | X |
| ESO26 | 12, 14 | 17 | 30 | 9 | 15 | 21 | 7, 9 | 13, 14 | 14, 18 | 10 | 9, 10 | 9 | 11 | 9, 11 | 12 | X, Y |
| ESO51 | 17 | 17, 20 | 30, 33.2 | 9 | 15, 16 | 21, 22 | 8 | 10, 11, 12 | 14, 15 | 9, 13 | 10 | 13 | 8 | 11 | 11 | X |
The profile of cell line SEG-1 is identical to large cell lung carcinoma cell line H460. The profile of cell line BIC-1 is identical to colon adenocarcinoma cell line SW620. The profile of cell line SK-GT-5 is identical to gastric adenocarcinoma cell line SK-GT-2. Amg = Amelogenin.
After hundreds, perhaps thousands, of culture passages of the 10 verified EAC cell lines, it could be questioned how representative these cell lines still are as models for their original EAC tumors. In the 10 verified EAC cell lines, 11 TP53 mutations were identified, and all were found to be present in the respective original tumor tissues (Supplementary Table 3, available online), indicating consistency of the TP53 mutations during decades of in vitro propagation of the cell lines. Furthermore, xenografts obtained from nine of the 10 verified EAC cell lines all showed a phenotype identical to the original EAC tumors, demonstrating that the cellular features and architecture of the tumor are preserved even after long term in vitro culture (Figure 1).
Figure 1.
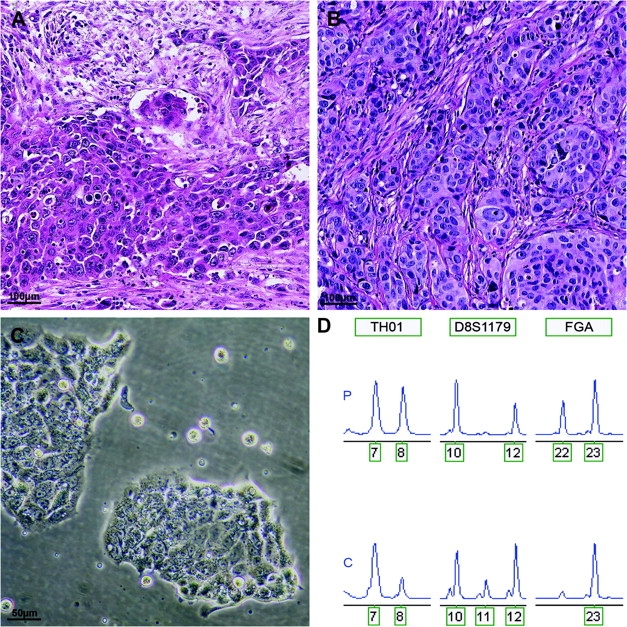
Authentication of human esophageal adenocarcinoma cell line OE33. A and B) Hematoxylin- and eosin-stained sections of the original tissue and xenograft of OE33 showing a poorly differentiated adenocarcinoma. C) In vitro growth pattern of cell line OE33. D) Short tandem repeat profile of the primary normal tissue (P) and cell line OE33 (C) indicating correct derivation of the cell line. Short-term repeat loci are indicated in boxes above electropherogram; the number of repeat units is indicated below the peaks. Note: loss of heterozygosity in the cell line at loci TH01 and FGA. The additional allele (11 repeat units) of D8S1179 observed in cell line OE33 is a known phenomenon and is probably due to somatic mutation or localized chromosomal rearrangements at this heterozygous locus.
The impact of our findings is illustrated by two clinical trials that are currently recruiting EAC patients based on experimental results using the contaminated cell lines SEG-1 and BIC-1. The first trial (http://clinicaltrials.gov/ct2/show/NCT00619242) investigates the effect of sorafenib (BAY 43-9006), a potent competitive small-molecule multikinase inhibitor of the Raf/MAPK/ERK pathway, on Barrett-related EAC. Several studies suggested that exposure of cell line SEG-1 to acid increased proliferation and decreased apoptosis by activating the Raf/MAPK/ERK pathway (12–14). In addition, treatment of SEG-1 cells with Raf/MAPK/ERK inhibitors resulted in pronounced antiproliferative effects (15–18). In this study, we have proved that cell line SEG-1 does not represent EAC but large cell lung carcinoma. This means that there is scant scientific evidence for activation of the Raf/MAPK/ERK pathway by acid or bile exposure in Barrett-related adenocarcinoma. The use of sorafenib in patients with Barrett-related EAC and the recruitment of patients for this clinical trial should therefore be reconsidered.
Another potential target for therapy in EAC, which is based mainly on research with contaminated cell lines SEG-1 and BIC-1, is telomerase (19,20). In a recent study, investigators demonstrated that treatment of SEG-1 xenografts with a specific telomerase inhibitor, GRN163L, led to loss of telomerase activity, reduction of telomere length, and inhibition of cell growth through induction of both senescence and apoptosis (21). Currently, EAC patients (among patients with other cancer types) are being recruited within a phase I clinical trial to study the effects of this telomerase inhibitor (http://clinicaltrials.gov/ct2/show/NCT00310895?term=GRN163L&rank=2). Our findings suggest that there is little scientific evidence for treatment of EAC patients with telomerase inhibitor GRN163L.
We have identified more than 100 scientific publications in which the contaminated cell lines SEG-1, BIC-1, or SK-GT-5 were used (Supplementary Tables 4 and 5, available online). Almost half of these reports were based solely on the use of cell lines not representative for EAC and should therefore be reevaluated because these cell lines in reality represent large cell lung carcinoma (SEG-1), colon adenocarcinoma (BIC-1), or gastric fundus adenocarcinoma (SK-GT-5). In addition, at least three National Institutes of Health grants have been assigned to esophageal cancer research projects (http://crisp.cit.nih.gov/), and 11 US patents (http://www.uspto.gov/) have been granted based on the use of cell lines SEG-1 and BIC-1. Following the lead of Walter Nelson-Rees, the first person who urged scientists to stop using contaminated cell lines (22,23), this report is a call for all scientists to authenticate their cell lines. Recent advances in DNA-profiling techniques make it possible to genotype cell lines simply and cheaply. The use of verified cell lines is a shared responsibility of scientists, editorial boards of scientific journals, and clinical and basic cancer research funding agencies.
In summary, cell lines SEG-1, BIC-1, and SK-GT-5 are not EAC cell lines but large cell lung cancer cell line H460, colorectal adenocarcinoma cell line SW620, and gastric fundus carcinoma cell line SK-GT-2, respectively. Cell lines FLO-1, KYAE-1, SK-GT-4, OE19, OE33, JH-EsoAd1, OACP4C, OACM5.1, ESO26, and ESO51 are derived from human EACs. All of these 10 verified EAC cell lines, together with their genotyping information, will be deposited in publicly available cell line repositories in the United States (http://www.lgcpromochem-atcc.com), Europe (www.ecacc.org.uk), and Japan (www.brc.riken.jp) to promote and facilitate future solid research on EAC.
Funding
Revolving Fund (FC0991) of the Erasmus MC Rotterdam and SK foundation.
Supplementary Material
Footnotes
The sponsors had no role in the data collection and analysis, interpretation of the findings, the preparation of the manuscript, or the decision to submit the manuscript for publication. None of the authors has a conflict of interest.
We thank W. M. van Weerden (Erasmus MC), B. A. Grotenhuis (Erasmus MC), A. W. van der Velden (Erasmus MC), and R. Yantiss (Weill Medical College of Cornell University) for contributions and H. Clevers (Hubrecht Institute), J. H. J. Hoeijmakers (Erasmus MC), and P. C. Ewing-Graham (Erasmus MC) for critical reading.
References
- 1.Masters JR, Thomson JA, Daly-Burns B, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc Natl Acad Sci U S A. 2001;98(14):8012–8017. doi: 10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masters JR. Human cancer cell lines: fact and fantasy. Nat Rev Mol Cell Biol. 2000;1(3):233–236. doi: 10.1038/35043102. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 4.Paulson TG, Reid BJ. Focus on Barrett's esophagus and esophageal adenocarcinoma. Cancer Cell. 2004;6(1):11–16. doi: 10.1016/j.ccr.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 5.Trauth BC, Klas C, Peters AM, et al. Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science. 1989;245(4915):301–305. doi: 10.1126/science.2787530. [DOI] [PubMed] [Google Scholar]
- 6.Altorki N, Schwartz GK, Blundell M, Davis BM, Kelsen DP, Albino AP. Characterization of cell lines established from human gastric-esophageal adenocarcinomas. Biologic phenotype and invasion potential. Cancer. 1993;72(3):649–657. doi: 10.1002/1097-0142(19930801)72:3<649::aid-cncr2820720305>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 7.Kan T, Shimada Y, Sato F, et al. Gene expression profiling in human esophageal cancers using cDNA microarray. Biochem Biophys Res Commun. 2004;286(4):792–801. doi: 10.1006/bbrc.2001.5400. [DOI] [PubMed] [Google Scholar]
- 8.Rockett JC, Larkin K, Darnton SJ, Morris AG, Matthews HR. Five newly established oesophageal carcinoma cell lines: phenotypic and immunological characterization. Br J Cancer. 1997;75(2):258–263. doi: 10.1038/bjc.1997.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez H, Koorstra JB, Hong SM, et al. Establishment and characterization of a bona fide Barrett esophagus-associated adenocarcinoma cell line. Cancer Biol Ther. 2008;7(11):1753–1755. doi: 10.4161/cbt.7.11.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Both NJ, Wijnhoven BP, Sleddens HF, Tilanus HW, Dinjens WNM. Establishment of cell lines from adenocarcinomas of the esophagus and gastric cardia growing in vivo and in vitro. Virchows Arch. 2001;438(5):451–456. doi: 10.1007/s004280000358. [DOI] [PubMed] [Google Scholar]
- 11.Boonstra JJ, van der Velden AW, Beerens EC, et al. Mistaken identity of widely used esophageal adenocarcinoma cell line TE-7. Cancer Res. 2007;67(17):7996–8001. doi: 10.1158/0008-5472.CAN-07-2064. [DOI] [PubMed] [Google Scholar]
- 12.Souza RF, Shewmake K, Terada LS, Spechler SJ. Acid exposure activates the mitogen-activated protein kinase pathways in Barrett's esophagus. Gastroenterology. 2002;122(2):299–307. doi: 10.1053/gast.2002.30993. [DOI] [PubMed] [Google Scholar]
- 13.Souza RF, Shewmake K, Pearson S, et al. Acid increases proliferation via ERK and p38 MAPK-mediated increases in cyclooxygenase-2 in Barrett's adenocarcinoma cells. Am J Physiol Gastrointest Liver Physiol. 2004;287(4):G743–G748. doi: 10.1152/ajpgi.00144.2004. [DOI] [PubMed] [Google Scholar]
- 14.Jaiswal K, Tello V, Lopez-Guzman C, Nwariaku F, Anthony T, Sarosi GA., Jr Bile salt exposure causes phosphatidyl-inositol-3-kinase-mediated proliferation in a Barrett's adenocarcinoma cell line. Surgery. 2004;136(2):160–168. doi: 10.1016/j.surg.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Morgan C, Alazawi W, Sirieix P, Freeman T, Coleman N, Fitzgerald R. In vitro acid exposure has a differential effect on apoptotic and proliferative pathways in a Barrett's adenocarcinoma cell line. Am J Gastroenterol. 2004;99(2):218–224. doi: 10.1111/j.1572-0241.2004.04054.x. [DOI] [PubMed] [Google Scholar]
- 16.Vona-Davis L, Frankenberry K, Cunningham C, et al. MAPK and PI3K inhibition reduces proliferation of Barrett's adenocarcinoma in vitro. J Surg Res. 2005;127(1):53–58. doi: 10.1016/j.jss.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Delgado JS, Mustafi R, Yee J, et al. Sorafenib triggers antiproliferative and pro-apoptotic signals in human esophageal adenocarcinoma cells. Dig Dis Sci. 2008;53(12):3055–3064. doi: 10.1007/s10620-008-0294-y. [DOI] [PubMed] [Google Scholar]
- 18.Keswani RN, Chumsangsri A, Mustafi R, Delgado J, Cohen EE, Bissonnette M. Sorafenib inhibits MAPK-mediated proliferation in a Barrett's esophageal adenocarcinoma cell line. Dis Esophagus. 2008;21(6):514–521. doi: 10.1111/j.1442-2050.2007.00799.x. [DOI] [PubMed] [Google Scholar]
- 19.Shammas MA, Koley H, Beer DG, Li C, Goyal RK, Munshi NC. Growth arrest, apoptosis, and telomere shortening of Barrett's-associated adenocarcinoma cells by a telomerase inhibitor. Gastroenterology. 2004;126(5):1337–1346. doi: 10.1053/j.gastro.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 20.Shammas M, Koley H, Batchu R, Bertheau R, Protopopov A, Munshi N, Goyal R. Telomerase inhibition by siRNA causes senescence and apoptosis in Barrett's adenocarcinoma cells: Mechanism and therapeutic potential. Mol Cancer. 2005;4 doi: 10.1186/1476-4598-4-24. doi: 10.1186/1476-4598-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shammas MA, Qazi A, Batchu RB, et al. Telomere maintenance in laser capture microdissection-purified Barrett's adenocarcinoma cells and effect of telomerase inhibition in vivo. Clin Cancer Res. 2008;14(15):4971–4980. doi: 10.1158/1078-0432.CCR-08-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson-Rees WA, Flandermeyer RR, Daniels DW. T-1 cells are HeLa and not of normal human kidney origin. Science. 1980;209(4457):719–720. doi: 10.1126/science.7394535. [DOI] [PubMed] [Google Scholar]
- 23.Nelson-Rees WA, Daniels DW, Flandermeyer RR. Cross-contamination of cells in culture. Science. 1981;212(4493):446–452. doi: 10.1126/science.6451928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


