Abstract
A substantial number of studies have reported the development of gene expression–based prognostic signatures for lung cancer. The ultimate aim of such studies should be the development of well-validated clinically useful prognostic signatures that improve therapeutic decision making beyond current practice standards. We critically reviewed published studies reporting the development of gene expression–based prognostic signatures for non–small cell lung cancer to assess the progress made toward this objective. Studies published between January 1, 2002, and February 28, 2009, were identified through a PubMed search. Following hand-screening of abstracts of the identified articles, 16 were selected as relevant. Those publications were evaluated in detail for appropriateness of the study design, statistical validation of the prognostic signature on independent datasets, presentation of results in an unbiased manner, and demonstration of medical utility for the new signature beyond that obtained using existing treatment guidelines. Based on this review, we found little evidence that any of the reported gene expression signatures are ready for clinical application. We also found serious problems in the design and analysis of many of the studies. We suggest a set of guidelines to aid the design, analysis, and evaluation of prognostic signature studies. These guidelines emphasize the importance of focused study planning to address specific medically important questions and the use of unbiased analysis methods to evaluate whether the resulting signatures provide evidence of medical utility beyond standard of care–based prognostic factors.
Lung cancer is one of the most frequent human cancers and the leading cause of cancer-related deaths in the United States (1). The 5-year relative survival rate among patients diagnosed with lung cancer is only 15%. The vast majority of lung cancer cases (approximately 80%) are non–small cell lung cancers (NSCLC), and the remaining fraction is small cell lung cancers.
The TNM staging system is currently used to guide treatment decisions and predict prognosis for patients with NSCLC (2). Surgical resection, if possible, is the first line of treatment. Although adjuvant chemotherapy after surgical resection has been shown to improve survival in patients with stage II or IIIA disease, its benefit in stage I patients remains controversial (3). National Comprehensive Cancer Network (NCCN) guidelines stipulate additional factors that should be considered when making adjuvant treatment decisions in NSCLC (4).
According to NCCN guidelines, the most important risk factor besides stage for considering adjuvant chemotherapy is the extent of residual tumor after resection. In the case of completely resected stage IA NSCLC, risk factors that signal the need for adjuvant chemotherapy include poor differentiation, vascular invasion, wedge resection, and minimal margins. However, the fact that disease relapse rates are as high as 30%, even among stage IA patients, has led to an interest in identifying additional prognostic factors for NSCLC. In the case of completely resected stage IB tumors as well, disagreement prevails over the possible benefit of adjuvant chemotherapy (3).
For completely resected stage II tumors, although inadequate mediastinal lymph node dissection, extracapsular spread, multiple positive hilar nodes, and close margins have been suggested as factors to be considered for deciding on the extent of adjuvant therapy (4), it is possible that additional molecular factors might help identify patients with good prognosis who could be spared chemotherapy. Thus, improving the existing decision criteria for selecting patients for adjuvant treatment in NSCLC is an important unmet medical need.
Some of the early microarray studies in NSCLC noted an association between patient survival and gene expression profiles (5–7). The primary objectives of some of these early studies were molecular classification and subclassification of lung tumors (5,6). Subsequently, however, several groups designed gene expression studies whose primary aim was to identify prognostic signatures using global gene expression profiling of surgically excised tumor samples. Although a review of these studies was recently published (8), we believe that a critical evaluation of the statistical design, analysis, and usefulness of their results in improving clinical decisions is important, not only to provide a thorough assessment of the current state of the art but also to identify methodological problems and set the trend for future research.
Current guidelines for making adjuvant treatment decisions for patients with completely resected NSCLC are based on factors that can be easily measured after surgery, such as tumor stage. Thus, for a new prognostic signature to be accepted and widely used by the medical community, it should provide therapeutically relevant information for each tumor stage. Moreover, a new prognostic signature for NSCLC can be considered clinically useful if it is either 1) more effective than standard prognostic factors (ie, tumor size, differentiation, vascular invasion, and margin status [negative margins vs positive or close margins]) in identifying high-risk completely resected stage I patients who might benefit from adjuvant chemotherapy or 2) identifies stage II patients who have a low risk of recurrence in the absence of chemotherapy. Also, although the real test of the clinical usefulness of a new prognostic signature is its validation in a prospective clinical trial, in its initial stages of development, the signature must demonstrate utility for a specific intended use when tested retrospectively on large clinical datasets to warrant a prospective trial.
In this review, we critically evaluate studies that reported prognostic gene expression signatures in NSCLC. Our evaluation was made on the basis of defined criteria that provide some of the key parameters for determining whether the study was planned and conducted in a manner that provides evidence of clinical utility for the signature beyond that obtained by existing treatment guidelines.
Methods
Selection of Studies
We conducted a search of the PubMed medical literature database (http://www.ncbi.nlm.nih.gov/pubmed/) using the search term “prognostic gene expression signature lung cancer” to identify articles that involved the analysis of gene expression data for developing prognostic signatures in NSCLC. We then screened the resulting abstracts for relevance. Articles that were published in English between January 1, 2002, and February 28, 2009, on gene expression profiling of NSCLC patients were considered for this review. Studies that were based on real-time quantitative polymerase chain reaction (RT-qPCR) assays as well as microarray platforms were included in this review. Studies that did not address patient outcome were excluded, as were studies that combined data for multiple different primary tumor sites or that contained fewer than 50 NSCLC patients. We also checked the reference lists of the relevant articles to identify any additional publications that might have been missed during the initial search. As a result of our search, a total of 16 studies (9–24) were selected for this review.
Scoring of Studies
Studies were scored on three major criteria: 1) the appropriateness of the study protocol, 2) the statistical validation of the prognostic models and presentation of results, and 3) whether there was a demonstration of medical utility for the prognostic signature. Because at present there seems to be no consensus in the statistical and machine learning communities about what types of models are best for modeling gene expression data (25), this aspect was not considered for the assessment.
Appropriateness of the study protocol was scored on four subcriteria: 1) whether the sample size was planned (ie, statistical power calculations were included); 2) whether appropriate patient selection criteria were used; 3) whether a description of patient characteristics was included; and 4) whether there were adequate protocols for tissue handling. Because the goal of developing a new prognostic signature is to improve adjuvant treatment decisions for patients with completely resected tumors, we gave a study a score of 1 for appropriate patient selection only if it adhered to consecutive enrollment of patients with completely resected tumors who did not receive any adjuvant chemotherapy; otherwise, the score for appropriate patient selection was 0. We gave a study a score of 1 if its description of patient characteristics included, at the minimum, age, sex, tumor stage, and follow-up time; otherwise, the score for description of patient characteristics was 0. A major source of variability in gene expression data is inappropriate tissue processing (26). According to the Tumor Analysis Best Practices Working Group (26), all tissue samples should be flash frozen within minutes of surgery and stored at a maximum temperature of −80°C. A study was given a score of 1 for adequate tissue handling only if it reported that samples were handled in this way; otherwise, the score for adequate tissue handling was 0.
Statistical validation of models and presentation of results were scored on three subcriteria: 1) whether the study had avoided presenting biased “resubstitution” statistics for the training set that was used to develop the models; 2) whether model validation was conducted on an independent dataset; and 3) whether there was complete specification of the prognostic model for future evaluation. Each of these criteria was given a score of 1 if the study was in compliance and a score of 0 if it was not.
In addition to providing risk stratifications for stage I and stage II patients, a new prognostic signature should show increased predictive accuracy compared with using a combination of age and other risk factors that are already part of current treatment guidelines (4). Hence, a demonstration of medical utility for the prognostic signature was scored on three subcriteria: 1) whether there was statistically significant risk separation on validation for stage IA and stage IB samples; 2) whether there was statistically significant risk separation on validation for stage II samples; and 3) whether the signature demonstrated improved predictive value over and above a combination of age and other known NCCN-defined risk factors. Again, each of these criteria was given a score of 1 if it was demonstrated in the study; otherwise, a score of 0 was given. If validation results were presented only for stage I overall, a score of 1 was given for validation on stage IA and stage IB samples only if the study also showed the predictions to be statistically significantly better than what could be obtained by tumor size information alone (ie, using information on whether the tumor stage was IA or IB).
To address the question of whether the gene expression signature improves upon the predictions obtained using standard risk factors, hazard ratios or regression coefficients (univariate or multivariate) or tests of statistical significance of these measures are inadequate (25,27). Hazard ratios and regression coefficients are measures of association, not of predictive power. Several techniques that have been suggested for comparing two prognostic factors: an analysis of the change in concordance index (28), an analysis of the change in the area under the time-dependent receiver operating characteristic curve (29), or an analysis comparing the positive predictive values and negative predictive values for predicting failure time outcome [PPV(t) and NPV(t), respectively] (30). The concordance index and the area under the receiver operating characteristic curve are quite similar measures: They both represent the probability that given two randomly selected patients, the patient with the worse outcome is, in fact, predicted to have a worse outcome. PPV(t) represents the probability that the outcome (say, recurrence or death) occurs by time t, given a high predicted risk, and NPV(t) represents the probability that the outcome does not occur by time t, given a low predicted risk. Testing for differences in the PPV(t) and NPV(t) of two prognostic factors has also been outlined previously (30). In this review, for demonstrating improved predictive value for the signature, a score of 1 was given only if the study demonstrated statistical significance of the gene expression signature over a combination of standard risk factors using any of these measures.
Results
The scores obtained by each study are presented in Table 1. The immediate striking finding from this table is that none of the studies succeeded in showing improvement in predictive power for the gene expression signatures over and above known risk factors. In fact, the majority of the risk factors outlined by the NCCN were not even considered by most of the studies. For example, according to NCCN guidelines (4), completeness of resection is the most important decision variable after stage; it has also been shown to statistically significantly influence survival (31). However, only seven of the 16 studies (9,10,13,14,16–18) stated that completeness of resection was a criterion for patient selection. In addition, only three studies (10,17,18) placed sufficient importance on patient selection by adhering to consecutive enrollment of patients who had undergone complete resection and received no adjuvant therapy, and only nine studies (9,10,13,14,16–18,22,23) reported having used snap-frozen tissues (Table 1). These points indicate that most of the studies reviewed were based on the use of a convenience sample of patients for whom tissue was available, with limited attention to either patient selection or the collection of important information about them to address specific questions of therapeutic decision making.
Table 1.
Comparison of prognostic gene expression studies*
| Protocols for patient selection and tissue handling |
Model validation and presentation of results |
Addressing medical utility |
|||||||||||
| First author, year (reference) | Training sample size | Validation sample size | Platform | Sample size planning | Appropriate patient selection | Description of patient characteristics | Tissue handling | No resubstitution statistics for the training set | Validation results on external data | Complete model specification | Statistically significant for validation in stages IA and IB | Statistically significant for validation in stage II | Statistically significant improvement over standard risk factors |
| Boutros, 2009 (9) | 147 (92 stage I, 38 stage II, 17 stage III) | 589 (409 stage I, 99 stage II, 81 stage III) | RT-qPCR | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Roepman, 2009 (10) | 103 (72 stage I, 31 stage II) | 69 (45 stage I, 24 stage II) | Microarray | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 |
| Shedden, 2008 (11) | 256 (160 stage I, 49 stage II, 47 stage III) | 186 (119 stage I, 46 stage II, 21 stage III) | Microarray | 1 | 0 | 1 | 0 | 1 | 1 | 0† | 0 | 0 | 0 |
| Sun, 2008 (12) | 86 (ADC: 67 stage I, 19 stage III) 129 (SCC: 73 stage I, 33 stage II, 23 stage III) | 175 (129 stage I, 32 stage II, 14 stage III) | Microarray | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| Skrzypski, 2008 (13) | 66 (SCC: 42 stage I, 22 stage II, 2 stage III) | 26 (22 stage I, 4 stage II) | RT-qPCR | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 |
| Raz, 2008 (14) | 107 (ADC: 70 stage I, 12 stage II, 25 stage III) | No separate validation set | RT-qPCR | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chen, 2007 (15) | 101 (59 stage I or II, 42 stage III) | 146 (109 stage I or II, 37 stage III) | RT-qPCR | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| Lau, 2007 (16) | 147 (92 stage I, 38 stage II, 17 stage III) | 216 (143 stage I, 42 stage II, 15 stage III, 16 stage IV) | RT-qPCR | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 |
| Larsen, 2007 (17) | 51 (SCC: 29 stage I, 15 stage II, 7 stage III) | 58 (SCC: 30 stage I, 14 stage II, 14 stage III) | Microarray | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Larsen, 2007 (18) | 48 (ADC: 46 stage I, 2 stage II) | 95 (ADC: 70 stage I, 17 stage II, 8 stage III) | Microarray | 0 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Guo, 2006 (19) | 86 (ADC: 67 stage I, 19 stage III) | 84 (ADC: 62 stage I, 14 stage II, 8 stage III) | Microarray | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Potti, 2006 (20) | 89 (69 stage I, 14 stage II, 6 stage III) | 109 (70 stage I, 22 stage II, 17 stage III) | Microarray | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1‡ | 0 | 0 |
| Lu, 2006 (21) | 197 (85 stage I, 112 stage IB) | 120 (55 stage IA, 65 stage IB) | Microarray | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
| Raponi, 2006 (22) | 129 (SCC: 73 stage I, 33 stage II, 23 stage III) | 36 (SCC: 25 stage I, 9 stage II, 1 stage III, 1 stage IV) | Microarray | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| Tomida, 2004 (23) | 50 (23 stage I, 11 stage II, 16 stage III) | 6 (stage information not available) | Microarray | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| Beer, 2002 (24) | 86 (ADC: 67 stage I, 19 stage III) | 84 (ADC: 62 stage I, 14 stage II, 8 stage III) | Microarray | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| Total No. of studies | 1 | 3 | 12 | 9 | 7 | 15 | 3 | 2 | 1 | 0 | |||
A score of 1 means that the study complied with the specific evaluation criterion, and a score of 0 means that it did not. ADC = adenocarcinoma; RT-qPCR = real-time quantitative polymerase chain reaction; SCC = squamous cell carcinoma.
For method A reported in the article.
Reported only for stage IA.
The most important medical question that needs to be answered by a new prognostic signature in NSCLC is whether it can identify the subset of stage IA patients who might benefit from adjuvant chemotherapy. However, only two studies (20,21) presented validation results for the prognostic signature separately for stage IA patients. Although the stratification results for stage IA patients in the study by Potti et al. (20) look promising, this signature failed to achieve statistical significance in a subsequent independent validation effort by Shedden et al. (11), even for stratifying stage I patients. In the only other study that presented validation results for stage IA samples (21), both the predicted low-risk and high-risk groups achieved 100% 3-year survival.
Most of the studies (9–12,16–18,20–22,24) presented overall validation results for stage I patients. The 3-year overall survival rates for stage I patients in the predicted high- and low-risk groups in the validation datasets of these studies (Table 2) show that some of the signatures succeeded in identifying high-risk stage I patients [eg, (17,18,20,22), studies that reported 40% or less 3-year overall survival for the high-risk group]. However, an evaluation of whether the signature predicted overall survival better than tumor size (ie, using information on whether the tumor stage was IA or IB) and other standard risk factors was not adequately addressed and hence unclear from most of these studies. Only Sun et al. (12) reported a marginal improvement in predictive accuracy for their gene expression signature over tumor size for stage I patients. However, the area under the receiver operating characteristic curve increased only from 0.63 to 0.67.
Table 2.
Three-year overall survival for stage I patients in validation datasets*
| 3-year overall survival (%) |
|||
| First author, year (reference) | No. of samples classified | Predicted low-risk group | Predicted high-risk group |
| Boutros, 2009 (9) | 345 | 75 | 60 |
| Roepman, 2009 (10) | 45 | 90 | 70 |
| Shedden, 2008 (11)† | 63 | 100 (MSK data) | 75 (MSK data) |
| 56 | 100 (CAN/DF data) | 70 (CAN/DF data) | |
| Sun, 2008 (12) | 91 | 75 | 55 |
| Lau, 2007 (16) | 76 | 85 (Harvard data) | 55 (Harvard data) |
| 67 | 75 (Duke data) | 45 (Duke data) | |
| Larsen, 2007 (17) | 58 | 65 | 25 |
| Larsen, 2007 (18) | 30 | 75 [data from Bild et al. (32)] | 40 [data from Bild et al. (32)] |
| Potti, 2006 (20) | 68 | 90‡ | 25‡ |
| Lu, 2006 (21) | 25 | 100 (dataset 6)‡ | 100 (dataset 6)‡ |
| 38 | 100 (dataset 6)§ | 70 (dataset 6)§ | |
| 64 | 95 (dataset 7) | 25 (dataset 7) | |
| Raponi, 2006 (22) | 25 | 60 (SCC data) | 35 (SCC data) |
| 75 (ADC and SCC data) | 35 (ADC and SCC data) | ||
| Beer, 2002 (24) | 62 | 85 | 55 |
The numerical values in this table have been estimated from the Kaplan–Meier survival curves reported in the original publications and hence are only approximate. ADC = adenocarcinoma; CAN/DF = Dana-Farber Cancer Institute; MSK = Memorial Sloan-Kettering Cancer Center; SCC = squamous cell carcinoma.
Values are for method A reported in the article (including covariates).
Classification of stage IA samples.
Classification of stage IB samples.
The recent large multicenter study (11) compared many genomic prognostic models with a model that used clinical covariates alone to predict overall survival in lung cancer patients. The best of the genomic models (identified as method A in the article) provided a statistically significant prognostic gradient for stage I patients in only one of the two validation sets. The authors, however, did not report whether the prognostic gradient for a combined model incorporating gene expression and clinical covariates was statistically significantly greater than that for the model containing only clinical covariates. Also, the clinical information included only age and sex (not tumor size). Separate validation for stage IA and stage IB samples was not addressed in this study.
Identification of the subset of stage IB and stage II patients who are at a low risk of disease recurrence without chemotherapy is also an important medical need. Only the study by Lu et al. (21) presented separate validation results for stage IB patients. The 3-year overall survival was 100% for the low-risk group and 70% for the high-risk group. The study by Roepman et al. (10) was the only one that reported statistical significance of the prognostic signature for validation in stage II samples. The 3-year survival rate for their low-risk stage II group was approximately 90%. These survival estimates, however, were based on very small sample sizes [38 patients in the study by Lu et al. (21) and 24 patients in the study by Roepman et al. (10)], and the authors did not compare the predictive power of their signature with that obtained using standard risk factors. None of the other studies showed results separately for stage IB or stage II samples; however, two studies (15,16) pointed out that the respective signatures did not statistically significantly distinguish prognosis in stage II validation samples. As pointed out in the respective publications, the lack of predictiveness for stage II patients could have resulted from the small number of stage II patients in the samples.
Most of the studies presented validation results on data that were not used for developing the predictive signatures (Table 1). Four studies (15,20,22,24) developed signatures that were subsequently independently evaluated by other authors. Only the signature reported by Beer et al. (24) provided a statistically significant difference in outcome of the low-risk vs high-risk group on independent validation by Sun et al. (12). However, the signature was not statistically significantly prognostic after adjustment for clinical covariates. This validation study of the Beer et al. signature by Sun et al. (12) also included all stages of disease and reported no separate analysis of stage I or stage II patients. Sun et al. (12) also attempted to validate the signature reported by Raponi et al. (22), which provided nearly statistically significant differences (P = .09) in outcome among the predicted risk groups after adjusting for clinical covariates. However, this validation study also included patients from all stages and again, no separate validation of stage I or stage II patients was reported.
Shedden et al. (11) attempted unsuccessfully to validate the signatures reported by Chen et al. (15) and Potti et al. (20). In neither case was there convincing evidence that the signatures alone provided statistically significant risk discrimination for stage I or stage II patients. Shedden et al. (11) reported that the signature of Chen et al. (15), when combined with clinical covariates, provided statistically significant risk discrimination for one of their validation sets of stage I patients. However, in this case, the model with clinical covariates (age and sex) alone gave statistically significant discrimination, and no evidence was presented that the signature added statistically significant prognostic power to the clinical covariates.
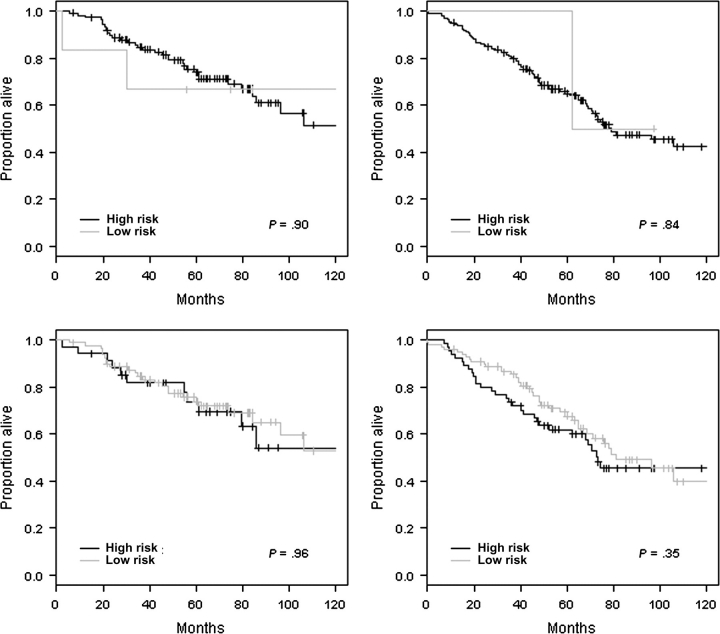
The studies by Shedden et al. (11) and Sun et al. (12) were the only attempts at independent validation of prognostic signatures reported by others. Such attempts at independent assessment of signatures are difficult because the prognostic models are often not fully specified in the original publications; in most cases, only the list of statistically significant genes is provided. A predictive signature is not just a gene list. To enable independent confirmation of a prognostic signature, all other aspects of the predictive model, such as weights and cut points, should also be reported. Only three of the 16 studies we reviewed presented fully specified models (Table 1). It is interesting that these three studies were RT-qPCR studies with simple three- to five-gene prognostic models. Two of these studies (15,16) specified the normalization and preprocessing steps used to apply their prognostic models to microarray data. We attempted to independently assess the prognostic signatures reported in (15) and (16) for stage IA and stage IB samples using the data of Shedden et al. (11). However, in our validation study, the signatures did not demonstrate statistically significant differences in outcome among the predicted risk groups (Figure 1; Supplementary Methods, available online).
Figure 1.
Independent validation of gene expression–based prognostic signatures on stage IA (left) and stage IB (right) samples obtained from the datasets reported by Shedden et al. (11). Kaplan–Meier survival curves for the five-gene signature reported by Chen et al. (15) (top panels) and for the three-gene signature reported by Lau et al. (16) (bottom panels). The P values (two-sided) are from the log-rank test. Tick marks indicate censored observations. Each patient was classified into the high- or low-risk group as outlined in Chen et al. (15) and Lau et al. (16). Further details on methodology for this independent validation study are given in Supplementary Material (available online).
In developing predictive models that use data in which the number of variables is much greater than the number of samples, it is essential to separate the data used for model development from the data used for model evaluation (33). Statistics that are computed by using the same data for model development and evaluation are called “resubstitution” statistics. The separation between the Kaplan–Meier survival curves for low- and high-risk patients of the training set used for model development is an example of a resubstitution statistic. Even though the enormous bias involved in presenting such resubstitution statistics has been repeatedly emphasized (25,33), presentation of resubstitution statistics has again emerged as an area of concern in our analysis, with nine studies (9,13–19,21) presenting such biased survival curves. We conducted a small simulation study to demonstrate the bias involved in presenting resubstitution-based estimates of prediction accuracy for prognostic models. Full details on the methodology for this simulation study are provided in the Supplementary Methods (available online). Our simulation studies show that even with completely random gene expression profiles, a prognostic model can always be developed that provides excellent associations with survival time for the training set. The poor predictive power of the model in such cases is revealed only when applied to independent validation data (Figure 2).
Figure 2.

Kaplan–Meier survival estimates for the simulation study. *Prediction accuracy for the training and validation datasets with random gene expression profiles. For this simulation, survival data on 129 patients were obtained from Bild et al. (32). For each patient, 5000 random numbers obtained from the standard normal distribution formed the gene expression profile. This master dataset was divided randomly into training and validation sets. A model predicting survival based on gene expression was developed from the training data. This model was used to classify survival risk group for patients in the training set and the validation set. The Kaplan–Meier curves show the proportion alive (vertical axis) vs time in months (horizontal axis) for predicted high-risk (black line) and low-risk (gray line) groups. Tick marks indicate censored observations. The P values are two-sided and are from the log-rank test. The Kaplan–Meier survival curves for the training set are “resubstitution” estimates because the same data are used to develop the model and to test it. Additional details of the simulation methodology are provided in the Supplementary Methods (available online).
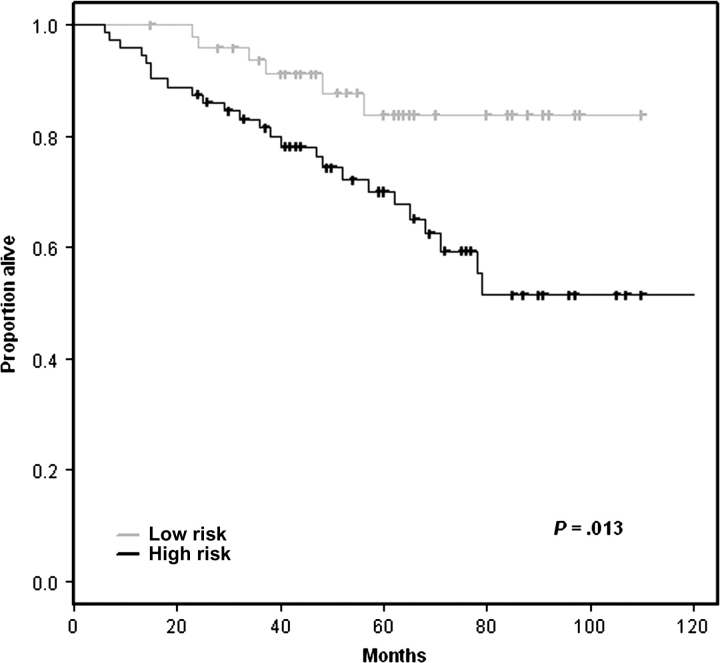
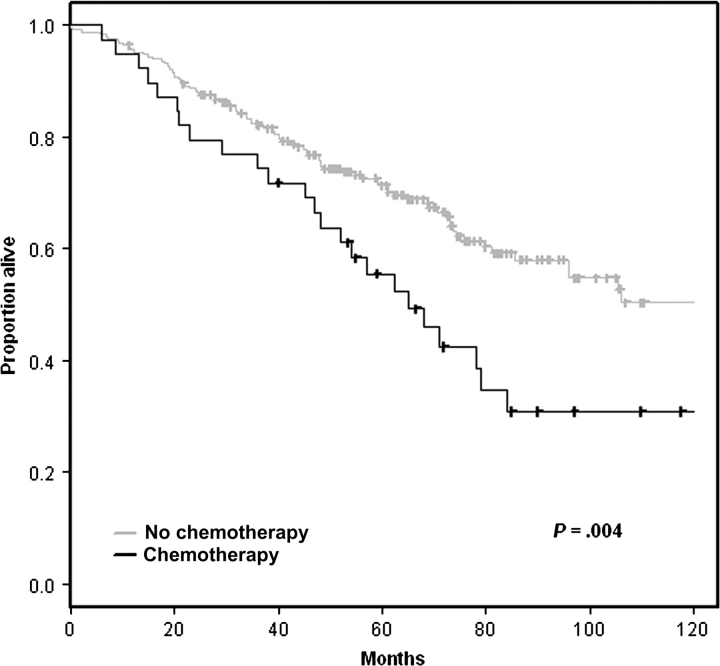
None of the 16 studies reviewed adequately addressed the question of the predictive power that could be attained by using easily measurable clinicopathological factors for stage I samples. We attempted to analyze the predictive power of clinicopathological factors for stage I samples by using the training data from Shedden et al. (11). We developed a predictive model based on age, tumor stage (IA vs IB), and adjuvant chemotherapy (received vs not received) for stage I patients [the study by Shedden et al. (11) was among those studies that did not exclude patients receiving adjuvant chemotherapy]. Full details on the methods used for this study are provided in Supplementary Methods (available online). Statistically significant separation of the risk groups (P = .013) was obtained for the test datasets using this model (Figure 3). An unexpected finding from this analysis was the poorer outcome for stage I patients who received adjuvant chemotherapy (Table 3, Figure 4). The poorer outcome for stage I patients receiving adjuvant chemotherapy is probably because “adjuvant chemotherapy” acts as a surrogate variable for risk factors that are being used by clinicians to select stage I patients for chemotherapy. These risk factors unfortunately were neither recorded nor analyzed in the publications reviewed. Our analysis also emphasizes again the importance of establishing appropriate patient selection criteria for studies of gene expression–based prognostic signatures. Because the objective of such studies is to identify patients for adjuvant chemotherapy, they should be restricted to patients who do not receive adjuvant chemotherapy.
Figure 3.
Evaluation of the prognostic models for stage I samples developed using clinical information alone (Table 3) on the Memorial Sloan-Kettering Cancer Center (MSK) and Dana-Farber Cancer Institute (CAN/DF) test datasets of Shedden et al. (11). The (two-sided) P value is from the log-rank test. Tick marks indicate censored observations. Each patient was classified into the high- or low-risk group based on whether his predicted risk score was greater or less than the median risk score for the training set. Further details on the methodology for developing these prognostic models are given in Supplementary Methods (available online).
Table 3.
Cox regression analysis of association between patient characteristics and overall survival for stage I samples*
| Characteristic | HR of death (95% CI) | P† |
| Age, continuous | 1.03 (1.01 to 1.06) | .006 |
| Stage (IB vs IA) | 1.67 (1.07 to 2.60) | .02 |
| Adjuvant chemotherapy (Yes vs No) | 1.76 (0.88 to 3.52) | .11 |
The regression model was built using the University of Michigan Cancer Center and the Moffitt Cancer Center (HLM) training data reported by Shedden et al. (11). More details on the methods are given in the supplementary information (available online). CI = confidence interval; HR = hazard ratio.
Two-sided Wald test.
Figure 4.
Effect of adjuvant chemotherapy on survival. These survival curves are based on the combined University of Michigan Cancer Center and Moffitt Cancer Center (HLM), Memorial Sloan-Kettering Cancer Center (MSK), and Dana-Farber Cancer Institute (CAN/DF) datasets reported in Shedden et al. (11). Tick marks indicate censored observations. The (two-sided) P value is from the log-rank test.
On the basis of observations made during this review and considering previous publications on analysis and reporting recommendations for microarray studies (25,34), we present a set of design, analysis, and reporting practice guidelines for prognostic gene expression studies, with a focus on NSCLC (Table 4).
Table 4.
Guidelines for prognostic factor studies in NSCLC*
| Section | Guidelines |
| Introduction | Clear statement of objectives. Clinically important objectives include: |
| 1. Through gene expression profiling, identification of a high-risk subgroup of stage IA NSCLC patients who might benefit from adjuvant chemotherapy. | |
| 2. Through gene expression profiling, further stratification of stage IB NSCLC patients who did not receive chemotherapy to improve adjuvant treatment decisions. | |
| 3. Through gene expression profiling, identification of stage II NSCLC patients who have low risk of recurrence without chemotherapy. | |
| Data collection and reporting | 1. Patient selection and sample size should be carefully planned based on the intended use of the prognostic signature to be developed. |
| 2. Characteristics of the study patients, how they were selected, and inclusion/exclusion criteria should be fully explained. | |
| 3. Data on treatment and all standard risk factors for the patients in the study should be available and reported. | |
| 4. Adequate description of the protocols for procurement of tissues and gene expression assays should be provided. | |
| 5. All raw data should be made publicly available. | |
| 6. If reusing data collected for a previous study, strict care must be taken to ensure that it meets the objectives of the new study. | |
| Statistical analysis and presentation of results | 1. When the endpoint is time to death or recurrence, data should not be binary transformed. |
| 2. Full details of the analysis method must be provided, including normalization procedures, gene filtering methods, variable selection and model building technique(s), handling of missing data, any cutoffs used, and rationale behind the cutoffs. | |
| 3. To demonstrate robustness of the signature, it must be validated on at least one completely independent dataset. | |
| 4. Resubstitution statistics for the training should not be presented. | |
| 5. To demonstrate medical utility for the new signature, the minimal set of results that need to be shown for all validation data separately for stages IA, IB, and II are: | |
| (a) Kaplan–Meier plots showing the risk stratification using the new signature. This requires the specification of a cutoff for defining the high- and low-risk groups, which should be fully specified using the training set data alone. | |
| (b) Positive predictive value, negative predictive values, and receiver operating characteristic curves to test whether the new signature is a statistically significantly better predictor of survival than a combination of age and other standard risk factors. Hazard ratios and regression coefficients from multivariable analysis or their P values are insufficient in this regard. | |
| 6. Full details of the final prognostic model(s) should be reported so that others can use it to classify patients in independent datasets. | |
| Discussion | The utility of the new signature as compared with standard risk factors must be clearly addressed including the limitations of the study. |
NSCLC = non–small cell lung cancer.
Discussion
Our review of published studies reporting the development of gene expression–based prognostic signatures for NSCLC found little evidence that any of the signatures are ready for clinical application. The review also showed that many of the studies contain serious problems, starting from unfocused design and continuing through inadequate analysis and biased reporting. These points are further elaborated in the discussions below.
Critical points that need to be clearly addressed by studies reporting prognostic signatures from gene expression data are the statistical validation and reproducibility of the signatures and their actual medical utility. Rigorous validation and demonstration of reproducibility are important in any data-derived measurement and especially so in situations where the number of variables is much larger than the number of samples, which is the case with gene expression data. In addition, to be broadly accepted by the medical community, the new prognostic signature should address current medical questions and show good predictive power over and above the risk factors that are part of existing treatment guidelines. In the case of NSCLC, a new prognostic signature should show that it can successfully identify stage IA and stage IB patients who might benefit from, and stage IB and stage II patients who could be spared, adjuvant chemotherapy.
It was evident from our review that although some of the earlier prognostic factor studies were plagued by small sample size and insufficient independent validation (7,23), large-scale collaborative efforts and accumulation of data in multiple laboratories have, to some extent, alleviated these problems. However, the fact that most studies now attempt to validate their signature in sufficiently large independent datasets does not imply that recent studies are without problems. Specific problems include a lack of clear specification of therapeutically relevant objectives, inappropriate patient selection, poor documentation of important prognostic factors, presentation of biased resubstitution statistics, and lack of a demonstration of medical utility for the resulting signature.
None of the studies reviewed were successful in showing clear usefulness for the gene expression signatures over and above the known risk factors. In fact, most of the risk factors outlined by NCCN (4) were not addressed or even measured in any of the studies. Most importantly, the number of studies that demonstrated that the new signature is helpful in making improved treatment decisions in disease stages IA, IB, or II was almost nil (Table 1). Even though Potti et al. (20) reported excellent discriminatory power for their signature on stage IA patients, this result was not reproduced in an independent validation study on a different dataset (11).
Even though covariates such as tumor size, surgical margins, and adjuvant chemotherapy are associated with survival (Figure 4), most of the reviewed studies did not take these variables into consideration during the selection of patients or make any attempt to adjust for the effects of these variables when reporting on the statistical significance of their prognostic signature. Hence, we again emphasize the fact that care must be taken to collect and use as much clinical information about patients as possible when developing prognostic signatures.
Another common problem that we observed during our review was the failure to completely specify a prognostic model that uses gene expression data. Without reporting a completely specified model, validation by independent investigators is not possible (35). We noted that most often, authors only specify the list of contributing genes in the model, which is not sufficient. Full model specification should include unambiguous documentation of all array preprocessing steps, a list of genes whose expression was statistically significantly associated with outcome, their weights in the multivariate model or complete decision trees, and cutoffs used for defining the risk groups. We noted that it is the cutoffs that are frequently ignored in the documentation.
It is surprising that despite validating the prognostic signature on independent data, many authors continue to present biased resubstitution statistics for their training set. This approach, taken together with the fact that superiority over standard risk factors was not clearly demonstrated by any reported signature, conveys an overly optimistic picture of the value of gene expression signatures. Our independent assessment of two previously reported signatures (15,16) failed to demonstrate a statistically significant discrimination between predicted high- and low-risk groups when evaluated on a different set of stage IA and stage IB samples (Figure 1) and further confirmed the overoptimistic and nonrobust results reported in many prognostic signature studies.
Prognostic factor studies need to be designed with a focus on the intended use. We have proposed a set of guidelines to aid the design and analysis of prognostic factor studies in NSCLC (Table 4). These guidelines are the result of observations made during this review and in the past (25,34) about good practices for the design and analysis of microarray studies. Although some of the points in our guidelines appear to be specific to NSCLC, they can, in fact, be translated to any therapeutic area once the intended use of the prognostic factor has been clearly identified.
One of the difficulties in evaluating whether a proposed prognostic gene expression signature has greater medical utility than established prognostic variables is the lack of a widely accepted multivariate prognostic model for early-stage lung cancer. In the now-popular Adjuvant! Online program in breast cancer, the risk estimate with and without therapy is evaluated based on multiple easily measured known risk factors (36). Developing such a model for NSCLC based on rigorous multivariate modeling of the NCCN prognostic factors would help to establish the prognostic power that can be achieved without gene expression data for patients with a given stage of disease, as illustrated by our Cox regression modeling of associations between clinical variables and survival in stage I patients (Table 3). Such models need to be evaluated on larger datasets and should include other important clinical covariates. The risk estimates thus obtained could then serve as the baseline for comparing gene expression–based prognostic factors. However, in the development of these new prognostic signatures, the focus must always be on the clinical validity and medical utility of the prognostic signature.
Clinical validity of a prognostic signature implies demonstrating that the test result correlates with clinical outcome. Medical utility of a prognostic signature means that the test result is actionable, leading to patient benefit. The ultimate test of clinical validity for a prognostic signature is its performance in a prospective clinical trial. Two such trials are already being conducted for breast cancer: the Trial Assigning Individualized Options for Treatment (Rx) or TAILORx, which tests the 21-gene Oncotype DX assay (37), and the Microarray In Node-negative and 1 to 3 positive lymph node Disease may Avoid ChemoTherapy or MINDACT trial, which uses Adjuvant! and a 70-gene profile, MammaPrint (38). The CALGB 30506 trial was recently initiated in lung cancer to clinically test the lung metagene prognostic signature (20). The objectives of this trial are to 1) determine the potential survival benefit of adjuvant chemotherapy in stage I NSCLC; 2) determine the potential survival benefit of adjuvant chemotherapy in predicted high-risk stage I NSCLC patients; and 3) determine the survival difference between the predicted high- and low-risk groups who are not given adjuvant chemotherapy. Even if chemotherapy is found to be beneficial in stage I patients predicted to be high risk by the model, presumably, it will be important to establish that the patients who benefit could not have been identified based on tumor size and other standard risk factors.
Regardless of clinical validation, unless a new prognostic signature provides additional risk stratification within the stage and risk factor groupings on which current treatment guidelines are based, its broad acceptance in medical practice is unlikely. From our review, it is clear that medical utility for any of the reported prognostic signatures has not yet been convincingly demonstrated. We hope that future research in this important field will strive to move away from being another exercise in clinical correlation to one that truly makes an impact on widespread medical practice.
Funding
The authors received no external funding for this study.
Supplementary Material
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer Statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Tanoue LT. Staging of non-small cell lung cancer. Semin Respir Crit Care Med. 2008;29(3):248–260. doi: 10.1055/s-2008-1076745. [DOI] [PubMed] [Google Scholar]
- 3.Tsuboi M, Ohira T, Saji H, et al. The present status of postoperative adjuvant chemotherapy for completely resected non-small cell lung cancer. Ann Thorac Cardiovasc Surg. 2007;13(2):73–77. [PubMed] [Google Scholar]
- 4.NCCN Clinical Practice Guidelines in Oncology™Non-Small Cell Lung Cancer V.2. 2009 http://www.nccn.org/professionals/physician_gls/PDF/nscl.pdf. Accessed April 22, 2009. [Google Scholar]
- 5.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98(24):13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98(24):13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wigle DA, Jurisica I, Radulovich N, et al. Molecular profiling of non-small cell lung cancer and correlation with disease-free survival. Cancer Res. 2002;62(11):3005–3008. [PubMed] [Google Scholar]
- 8.Kratz JR, Jablons DM. Genomic prognostic models in early-stage lung cancer. Clin Lung Cancer. 2009;10(3):151–157. doi: 10.3816/CLC.2009.n.021. [DOI] [PubMed] [Google Scholar]
- 9.Boutros PC, Lau SK, Pintilie M, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A. 2009;106(8):2824–2828. doi: 10.1073/pnas.0809444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roepman P, Jassem J, Smit EF, et al. An immune response enriched 72-gene prognostic profile for early-stage non-small-cell lung cancer. Clin Cancer Res. 2009;15(1):284–290. doi: 10.1158/1078-0432.CCR-08-1258. [DOI] [PubMed] [Google Scholar]
- 11.Shedden K, Taylor JM, Enkemann SA, et al. Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14(8):822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Z, Wigle DA, Yang P. Non-overlapping and non-cell-type-specific gene expression signatures predict lung cancer survival. J Clin Oncol. 2008;26(6):877–883. doi: 10.1200/JCO.2007.13.1516. [DOI] [PubMed] [Google Scholar]
- 13.Skrzypski M, Jassem E, Taron M, et al. Three-gene expression signature predicts survival in early-stage squamous cell carcinoma of the lung. Clin Cancer Res. 2008;14(15):4794–4799. doi: 10.1158/1078-0432.CCR-08-0576. [DOI] [PubMed] [Google Scholar]
- 14.Raz DJ, Ray MR, Kim JY, et al. A multigene assay is prognostic of survival in patients with early-stage lung adenocarcinoma. Clin Cancer Res. 2008;14(17):5565–5570. doi: 10.1158/1078-0432.CCR-08-0544. [DOI] [PubMed] [Google Scholar]
- 15.Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356(1):11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 16.Lau SK, Boutros PC, Pintilie M, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol. 2007;25(35):5562–5569. doi: 10.1200/JCO.2007.12.0352. [DOI] [PubMed] [Google Scholar]
- 17.Larsen JE, Pavey SJ, Passmore LH, et al. Expression profiling defines a recurrence signature in lung squamous cell carcinoma. Carcinogenesis. 2007;28(3):760–766. doi: 10.1093/carcin/bgl207. [DOI] [PubMed] [Google Scholar]
- 18.Larsen JE, Pavey SJ, Passmore LH, Bowman RV, Hayward NK, Fong KM. Gene expression signature predicts recurrence in lung adenocarcinoma. Clin Cancer Res. 2007;13(10):2946–2954. doi: 10.1158/1078-0432.CCR-06-2525. [DOI] [PubMed] [Google Scholar]
- 19.Guo L, Ma Y, Ward R, Castranova V, Shi X, Qian Y. Constructing molecular classifiers for the accurate prognosis of lung adenocarcinoma. Clin Cancer Res. 2006;12(11, pt 1):3344–3354. doi: 10.1158/1078-0432.CCR-05-2336. [DOI] [PubMed] [Google Scholar]
- 20.Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355(6):570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 21.Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3(12):3. doi: 10.1371/journal.pmed.0030467. e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raponi M, Zhang Y, Yu J, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66(15):7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 23.Tomida S, Koshikawa K, Yatabe Y, et al. Gene expression-based, individualized outcome prediction for surgically treated lung cancer patients. Oncogene. 2004;23(31):5360–5370. doi: 10.1038/sj.onc.1207697. [DOI] [PubMed] [Google Scholar]
- 24.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 25.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99(2):147–157. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 26.Tumor Analysis Best Practices Working Group. Expression profiling—best practices for data generation and interpretation in clinical trials. Nat Rev Genet. 2004;5(3):229–237. doi: 10.1038/nrg1297. [DOI] [PubMed] [Google Scholar]
- 27.Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. Am J Epidemiol. 2004;159(9):882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- 28.Kattan MW. Evaluating a new marker's predictive contribution. Clin Cancer Res. 2004;10(3):822–824. doi: 10.1158/1078-0432.ccr-03-0061. [DOI] [PubMed] [Google Scholar]
- 29.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 30.Moskowitz CS, Pepe MS. Quantifying and comparing the accuracy of binary biomarkers when predicting a failure time outcome. Stat Med. 2004;23(10):1555–1570. doi: 10.1002/sim.1747. [DOI] [PubMed] [Google Scholar]
- 31.Pfannschmidt J, Muley T, Bulzebruck H, Hoffmann H, Dienemann H. Prognostic assessment after surgical resection for non-small cell lung cancer: experiences in 2083 patients. Lung Cancer. 2007;55(3):371–377. doi: 10.1016/j.lungcan.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 32.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 33.Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95(1):14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 34.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23(36):9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 35.Kostka D, Spang R. Microarray based diagnosis profits from better documentation of gene expression signatures. PLoS Comput Biol. 2008;4(2):4. doi: 10.1371/journal.pcbi.0040022. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravdin PM, Siminoff LA, Davis GJ, et al. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol. 2001;19(4):980–991. doi: 10.1200/JCO.2001.19.4.980. [DOI] [PubMed] [Google Scholar]
- 37.Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol. 2008;26(5):721–728. doi: 10.1200/JCO.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 38.Cardoso F, Van’t Veer L, Rutgers E, Loi S, Mook S, Piccart-Gebhart MJ. Clinical application of the 70-gene profile: the MINDACT trial. J Clin Oncol. 2008;26(5):729–735. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.