Abstract
Ectopic activation of fibroblast growth factor receptor 3 (FGFR3) is associated with several cancers, including multiple myeloma (MM). FGFR3 inhibition in these cells inhibits proliferation and induces apoptosis, validating FGFR3 signaling as a therapeutic target in t(4;14) MM cases. We have identified the PI3K regulatory subunit, p85α, as a novel interactor of FGFR3 by yeast two-hybrid, and confirmed an interaction with both p85α and p85β in mammalian cells. The interaction of FGFR3 with p85 is dependent upon receptor activation. In contrast to the Gab1-mediated association of FGFRs with p85, the FGFR3-p85 interaction we observed requires FGFR3 Y760, previously identified as a PLCγ binding site. The interaction of p85 with FGFR3 does not require PLCγ, suggesting the p85 interaction is direct and independent of PLCγ binding. FGFR3 and p85 proteins also interact in MM cell lines which consistently express p85α and p85β, but not p50 or p55 subunits. siRNA knockdown of p85β in MM cells caused an increased ERK response to FGF2. These data suggest that an endogenous negative regulatory role for the p85-FGFR3 interaction on the Ras/ERK/MAPK pathway may exist in response to FGFR3 activity and identifies a novel therapeutic target for MM.
INTRODUCTION
Fibroblast Growth Factor Receptor 3 (FGFR3) is one of four receptors that mediate the effects of FGFs on diverse cellular processes including proliferation, differentiation and migration (reviewed in 1–3). Ligand activation of the receptor results in phosphorylation of critical tyrosine residues, leading to the activation of multiple signal transduction cascades. Abnormal activation of FGFR3 is directly responsible for human dwarfing syndromes and is reported in association with several cancer types, including bladder cancer and multiple myeloma (MM) (2,4–7).
MM is an incurable malignancy of terminally differentiated B cells, characterized by clonal expansion of plasma cells in the bone marrow. Approximately 50% of intramedullary MM cases involve a chromosomal translocation event between the immunoglobulin heavy chain and one of five recurrent loci, including the 4p16.3 locus, resulting in aberrant expression of FGFR3 (8–10). This translocation event is associated with a particularly poor prognosis, marked by a substantially shortened survival following either conventional or high-dose chemotherapy (11). Roughly 10% of these patients further acquire activating mutations in FGFR3, an additional adverse prognostic factor (12). Inhibition of FGFR3 activity inhibits tumor growth in cell lines and animal models of FGFR3-associated MM (13–17), supporting its therapeutic relevance.
Activation of FGF receptors leads to activation of multiple signaling cascades, including the ERK/MAPK, PLCγ/PKC, PI3K and STAT pathways, all of which have been implicated as contributing to FGFR3-medated transformation (18–23). Signaling through Ras/ERK/MAPK, as monitored by pERK levels, is a required event for manifestation of FGFR3-mediated phenotypes (19,24). Indeed, chemical inhibition of FGFR3 in MM cells results in decreased proliferation and increased apoptosis, which is consistently accompanied by decreased ERK activation (15,17,25,26), suggesting that decreasing ERK activation is a desirable outcome for cancer treatments involving FGFR3. Less is known about the involvement of other downstream events. The PI3K/Akt pathway, which is triggered by FGF and other growth factor receptors and is involved in oncogenic cellular transformation in many cancers (27), may be activated directly, as is the case for the PDGF receptor (28), or indirectly, via the IRS1 and Gab1 adaptor proteins, for the insulin and EGF receptors, respectively (29–31). FGFRs were previously reported to fall in the latter group, activating PI3K indirectly via the Gab1 docking protein (32). The PI3K pathway functions in regulation of cell proliferation and survival, and is implicated in multiple cancer types (33–35). AKT activation (phosphorylation at S473) has been associated with a poor prognosis in many cancers, including those of the hematopoietic system (36,37). Further, PI3K activity is implicated in FGFR3-mediated transformation due to the ability of a PI3K inhibitor, LY294002, to inhibit the growth of ETV6/TEL-FGFR3 transformed Ba/F3 cells (38). In MM cells, PI3K/Akt signaling contributes to cell proliferation and survival, whereas inhibition of PI3K inhibits tumor growth (19,20,23,39–41). Finally, several studies suggest that crosstalk exists between the PI3K and Ras/ERK/MAPK pathways and implicate PI3K activity in the regulation of ERK1/2 activation in a manner dependent on growth factor stimulation (42–46).
We report here that FGFR3 can recruit the p85 regulatory subunit of PI3K independently of the Gab1 adaptor protein in an activation-dependent manner, and further identify FGFR3 Y760 as the critical site involved. The expression of PI3K regulatory isoforms was determined in MM cell lines and the contribution of the PI3K pathway to FGFR3 signaling in MM investigated. Our results suggest that p85 can modulate the ERK response to FGF2 in MM cells and that the interaction of activated FGFR3 with p85 may negatively regulate the downstream activation of ERK.
RESULTS
Identification of a novel interaction between FGFR3 and p85α
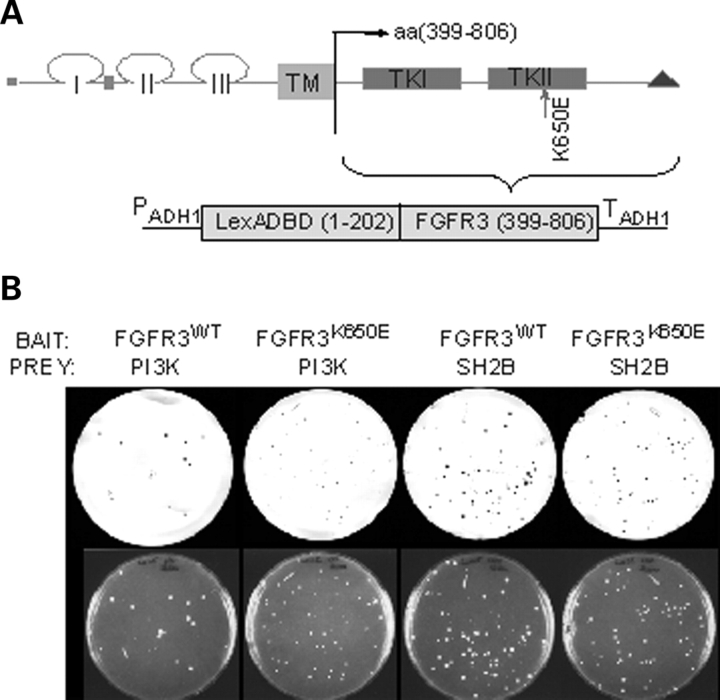
While the molecular components of normal FGFR3 signaling are emerging, a more complete molecular profile of aberrant FGFR3 signaling is necessary to identify effective treatments for FGFR3-associated cancers. In order to identify novel protein interactions of FGFR3 and activated FGFR3 that might lead to the identification of new therapeutic targets, a yeast two-hybrid screen was performed. Previous attempts to use this approach for mutant FGFR3 met with limited success and only a weakly active FGFR3 mutant implicated in a mild from of short-limbed dwarfism, hypochondroplasia (N540K), yielded significant positive clones, allowing for the identification of SH2B as an interacting protein (47). Under the growth conditions used in our studies, expressing the complete cytoplasmic domain of human FGFR3 (amino acids 399–806) with the wild-type (WT) sequence or an activating LYS to GLU mutation at amino acid 650 (K650E) (48) was not toxic. Each was used as bait to screen a primary human chondrocyte cDNA library (Fig. 1A). This library was chosen as FGFR3 is highly expressed in chondrocytes. A pilot screen yielded 87 clones which scored positive on the filter lift β-galactosidase assay, 84 of which were derived using the K650E mutant receptor as bait. One of the strongest interactions observed was identified by sequence analysis as transcript variant 3 of the PI3-kinase (PI3K) regulatory protein, p85α, better known as p50α. This interaction was confirmed in yeast by co-transformation with the isolated library plasmid and the WT or K650E FGFR3 bait plasmid (Fig. 1B), indicating that p85α can interact with both WT and K650E mutant FGFR3 sequences in yeast. No colonies were observed using Lamin C, which does not interact with FGFR3, as a negative control (data not shown).
Figure 1.
FGFR3 and p85 interact in yeast. (A) The cytoplasmic domain of FGFR3 (wild-type or K650E sequences) fused to the LexA DNA-binding domain were used as bait for a yeast two-hybrid screen. (B) L40 yeast expressing FGFR3WT or FGFR3K650E bait plasmids were transformed with PI3K-p85 or SH2B (positive control) prey plasmid as shown and plated on SD/-His/-Leu/-Trp. Colonies were transferred to nitrocellulose filters and subject to X-Gal assay. An interaction with Lamin C (negative control) was not observed for either prey (data not shown).
FGFR3 interacts with PI3K regulatory proteins in mammalian cells
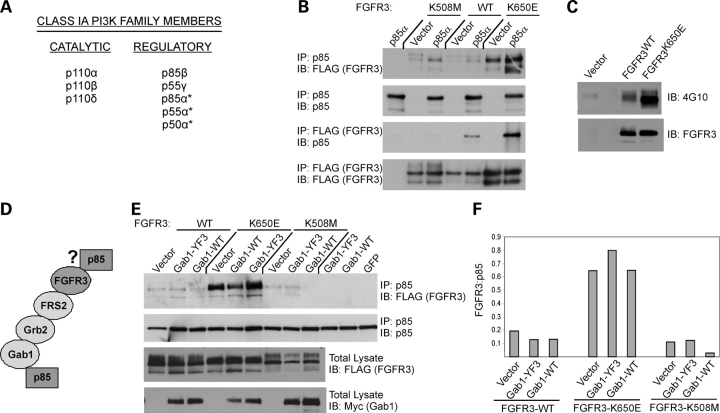
The PI3K proteins are divided into three main classes (e.g. reviewed in 49,50), of which the class IA proteins are activated by receptor tyrosine kinases such as FGFR3. Class IA PI3-kinases are composed of a catalytic subunit (p110α, p110β, p110δ) complexed to one of five regulatory subunits, including p85β, p55γ and the p85α isoforms (p85α, p55α, p50α) (Fig. 2A). The p85α isoforms, encoded by the PIK3R1 gene, contain two SH2 domains, identical in sequence. p85β is structurally similar to p85α, sharing 62% identity at the amino acid level (51), and p55γ shares ∼70% amino acid identity with the SH2 domains and inter-SH2 region of p85α (52).
Figure 2.
FGFR3 interacts with p85 in mammalian cells and does not require Gab1. (A) Class IA PI3K proteins. The asterisks denote isoforms p85α, p55α and p50α, which are encoded by the PIK3R1 gene and are identical in sequence over the SH2 domains. They share 70% amino acid identity with p55γ in this region. p85α and p85β are structurally similar and share 62% amino acid identity. (B) HeLa cells were transiently transfected with FLAG-tagged wild-type (WT), constitutively active (K650E) or kinase-dead (K508M) FGFR3±exogenous p85α (HA-tagged). In the absence of exogenous p85, plasmid vector was co-transfected with FGFR3. Eighteen to twenty-four hours post-transfection, reciprocal co-IPs were performed as indicated from 500 µg total cell lysate. Reduced binding to the kinase-dead form of the receptor is observed following IP with FLAG. (C) HeLa cells were transiently transfected with vector or FLAG-tagged FGFR3, WT or K650E. Cells were harvested 24 h later and 20 µg total cell lysate probed for phosphotyrosine (4G10). Blot was stripped and re-probed for FGFR3. The K650E mutant is strongly phosphorylated and the wild-type receptor exhibits a smaller degree of activation likely due to overexpression. (D) Schematic of proposed interactions between FGFR3 and p85 showing possible direct interaction and indirect adaptor-mediated interaction, the latter of which was identified for FGFR1 (28). (E) HeLa cells were transiently transfected with FLAG-tagged FGFR3, WT, K650E or K508M in combination with wild-type Gab1 or Gab1 defective for p85 binding (YF3). Gab1 constructs were myc-tagged. Where Gab1 or FGFR3 was transfected in the absence of the other, plasmid vector was co-transfected to maintain the total amount of transfected DNA. Eighteen to twenty-four hours post-transfection, reciprocal co-IPs were performed from 500 µg total cell lysate and immunoprecipitates probed for the p85 or FGFR3 as appropriate. IP with p85 is shown. Binding to FGFR3 is observed independent of whether Gab1 is competent for p85 binding or mutated. All blots were stripped and re-probed with the immunoprecipitating antibody to confirm effective IP. Representative of three experiments shown. (F) The western blot in (E) was quantitated using NIH Scion Image and the ratio of FGFR3 to p85 determined.
Having identified p50α as an interacting protein for FGFR3 in the yeast two-hybrid assay, we examined the ability of FGFR3 to interact with several PI3K regulatory proteins in mammalian cells by performing reciprocal co-immunoprecipitations from lysates of HeLa cells transfected with full-length FLAG-tagged FGFR3 in the presence or absence of exogenous HA-tagged p50α and p85α. Three FGFR3 plasmids were used, including the wild-type sequence and the K650E mutation, as well as a third containing a kinase-inactivating K508M mutation. Only the receptor carrying the K650E mutation, which renders FGFR3 constitutively phosphorylated and active (53), showed appreciable interaction with p85α and p50α in both immunoprecipitation orientations (Fig. 2B and data not shown). Some interaction with p85 proteins is observed with the wild-type receptor, particularly in the anti-FLAG IP to pull down FGFR3, likely reflecting receptor phosphorylation characteristic of transient overexpression (Fig. 2C and unpublished results). Endogenous p85α and p85β were also observed to interact in an activation-dependent manner (data not shown). Taken together, these data demonstrate the ability of FGFR3 to interact with multiple PI3K regulatory proteins and suggest a dependence on receptor phosphorylation for these interactions.
The FGFR3 interaction with p85 is not mediated by Gab1
It has been shown for FGFR1 that activation of PI3-kinase signaling can be achieved via an indirect interaction with p85α involving an adaptor protein complex containing FRS2, Grb2 and Gab1 (Fig. 2D). Binding a juxtamembrane region of FGFR1 (residues 412–433) devoid of tyrosine residues in a ligand-independent manner, FRS2 is phosphorylated following receptor stimulation, providing binding sites for Grb2, which subsequently recruits Gab1 via its SH3 domain (32,54,55). Activated Gab1 then directly recruits p85 and activates PI3-kinase. Yeast do not carry homologues of the FRS2, Grb2 or Gab1 adaptor proteins (Saccharomyces Genome Database; http://www.yeastgenome.org), suggesting that the interaction observed between FGFR3 and p85α may not be mediated by this adaptor complex. To determine whether an interaction with Gab1 is required for FGFR3-p85 binding, WT, K650E or K508M FGFR3 constructs were transfected into HeLa cells in combination with either wild-type Gab1 or Gab1-YF3, the latter of which is deficient in p85 binding (56,57). The immunoprecipitation of endogenous p85 was repeated and the precipitates probed for FGFR3. We found that neither wild-type Gab1 nor Gab1-YF3 had any significant effect on the co-immunoprecipitation of FGFR3 with p85α (Fig. 2E and F), indicating that these two proteins can interact independent of Gab1.
FGFR3 interaction with p85 is dependent on FGFR3 Y760
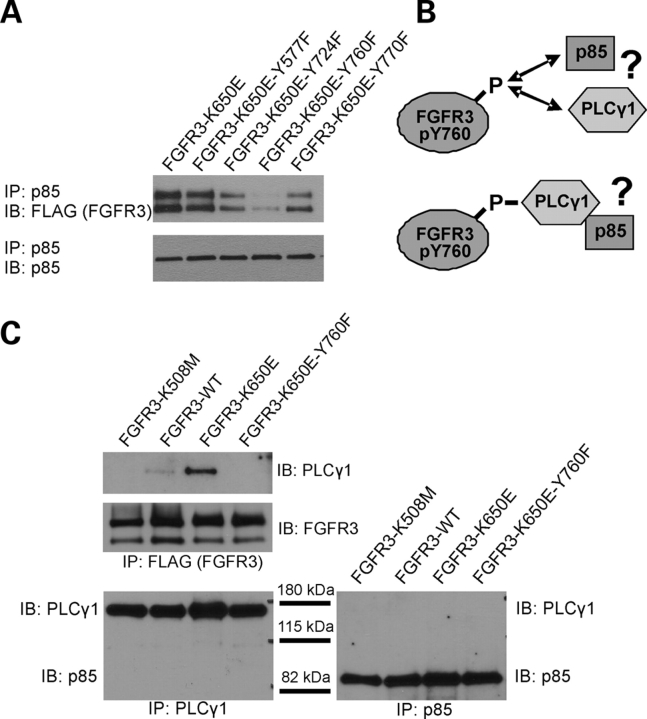
The kinase domain of FGFR3 contains four critical tyrosine residues shown to be important in mediating FGFR3 signals (58). Y724, which appears to be involved in PI3-kinase activation by FGFR3, is within a consensus binding sequence for p85; however, no evidence exists for p85 binding to this residue or its counterpart in FGFR1 (54,58). Y770, in contrast, has been suggested to function as a negative regulator of PI3-kinase activity (58). The counterpart of FGFR3 Y760 in FGFR1 (Y766) is correspondingly implicated in PI3K activation (59). p85 was immunoprecipitated from lysates of HeLa cells transfected with FGFR3 constructs carrying single Y→F mutations at each of the critical tyrosines on the constitutively active K650E background. Evaluation of p85 immunoprecipitates for the presence of FGFR3 indicated that mutation of Y760 greatly reduced binding to p85, and mutation of Y724 less so (Fig. 3A), suggesting Y760 is the residue primarily involved in binding p85.
Figure 3.
p85 interaction with FGFR3 requires Y760 and appears independent of PLCγ binding. (A) HeLa cells were transiently transfected with FLAG-tagged wild type, constitutively active (K650E), or single Y→F mutations on the constitutively active background. Eighteen to twenty-four hours post-transfection, endogenous p85 was IP’d from 500 µg total lysate and the precipitates probed for FGFR3. The Y760F mutation significantly decreases p85 binding to FGFR3. (B) Schematic of possible interactions of p85 and PLCγ with FGFR3 showing independent and possible scenarios. (C) HeLa cells were transfected with FLAG-tagged FGFR3 constructs as indicated. Eighteen to twenty-four hours post-transfection, FGFR3 (FLAG), p85 and PLCγ were immunoprecipitated from 300 µg total lysate and probed as indicated. PLCγ binds active FGFR3 (K650E) and this binding is also decreased by the Y760F mutation. Note that PLCγ and p85 are not pulled down together.
The corresponding residue in FGFR1 (Y766) directly binds PLCγ (60), and has been shown to be required for binding of PLCγ to mouse FGFR3 (Y754) (61) and activation of PLCγ downstream of human FGFR3 (18). Furthermore, PLCγ contains consensus binding sequences for p85, suggesting it may serve as an adaptor for p85 binding to FGF receptors (Fig. 3B). The requirement for Y760 in the interaction of PLCγ was first evaluated with human FGFR3 by reciprocal co-immunoprecipitations in HeLa cells and Y760 is required for PLCγ binding as expected (Fig. 3C and data not shown). We next examined whether p85 and PLCγ bind to FGFR3 independently or whether PLCγ serves as an adaptor for p85 by examining their ability to co-IP with one another in the presence of FGFR3. As shown in Fig. 3C, an interaction between p85 and PLCγ in either direction was undetectable, even in the presence of activating FGFR3 mutations, suggesting that p85 and PLCγ bind independently to FGFR3 in a manner requiring Y760, and are likely to compete for binding.
PI3K p85 proteins are expressed and interact with FGFR3 carrying activating mutations in MM cells
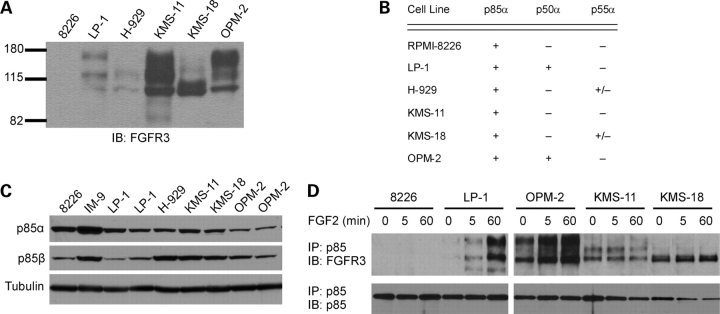
To investigate whether the p85-FGFR3 interaction occurs in MM cells, we first determined which of the five class IA regulatory subunits are expressed in MM cell lines using a combination of western blotting and RT–PCR with isoform-specific antibodies and primers, respectively. Cell lines expressing wild-type FGFR3 were evaluated, as well as lines expressing FGFR3 carrying activating mutations and lines with no FGFR3 expression (Fig. 4A). The regulatory subunits p85α and p85β were detected in all lines evaluated by western analysis, whereas p55γ was consistently lacking (Fig. 4B and C). Since commercial antibodies cannot distinguish between the p50α and p55α isoforms, RT–PCR was used to determine their presence or absence. p50α and p55α were weakly detected in one line each expressing wild-type FGFR3 (LP-1 and H929, respectively) or an activating mutation in FGFR3 (OPM-2 and KMS-18, respectively), suggesting that these isoforms are not consistently expressed and, therefore, are less likely to be relevant to FGFR3-mediated pathogenesis in MM (Fig. 4B). Having identified p85α and p85β as the major regulatory PI3K proteins in MM cells, we next examined the ability of endogenous FGFR3 and p85 to interact in MM cell lines using co-immunoprecipitation. Cell lines expressing FGFR3 with activating mutations (OPM-2, KMS-11 and KMS-18) show an interaction between FGFR3 and p85 with or without ligand (Fig. 4D, detected with pan-p85 antibody). Ligand-dependent FGFR3 interaction with p85 was observed in cells with wild-type FGFR3, although less consistently (Fig. 4D and data not shown). While OPM-2 cells show a robust interaction between FGFR3 and p85, these cells also carry a mutation in the PTEN negative regulator of PI3K (62), rendering these cells unsuitable for studies of FGFR3-mediated PI3K signaling in MM cells. Therefore, we focused on the KMS-18 and KMS-11 MM lines for further studies.
Figure 4.
p85 proteins can interact with FGFR3 in MM cells. (A) FGFR3 expression was confirmed in FGFR3-negative and -positive MM cell lines by western blot of 20 µg total cell lysate. MM lines: 8226, IM-9: FGFR3-negative; LP1: wild-type FGFR3, H929: wild-type FGFR3 + activating Ras mutation; KMS-11: extracellular domain mutant FGFR3Y373C; KMS-18: transmembrane domain mutant FGFR3G384D; OPM-2: intracellular kinase domain mutant FGFR3K650E + PTEN null. (B) PIK3R1 gene products were reversed transcribed from total RNA using a gene-specific primer and individual isoforms amplified using specific primer pairs as described under the experimental procedures. Only p85α is present in every cell line. (C) Expression of the PIK3R2 (p85β) and PIK3R3 (p55γ) gene products was determined by western blot of 20 µg total cell lysate. p85α expression was also examined by western blot. Duplicates represent lines obtained from different sources. Both the α and β forms of p85 are detected. p55γ was not observed (data not shown). (D) Cells were plated at 3×106 per well in six-well plates and serum starved 4 h. Following serum starvation, 10 ng/ml FGF-2 was added for the times indicated. Cells were lysed and p85 immunoprecipitated from 300 µg total cell lysate. Immunoprecipitates were resolved by SDS–PAGE and probed for FGFR3. The blot was stripped and re-probed for p85 to confirm effective IP of all samples.
Knockdown of p85 regulatory isoforms modulates ERK activation in MM cells
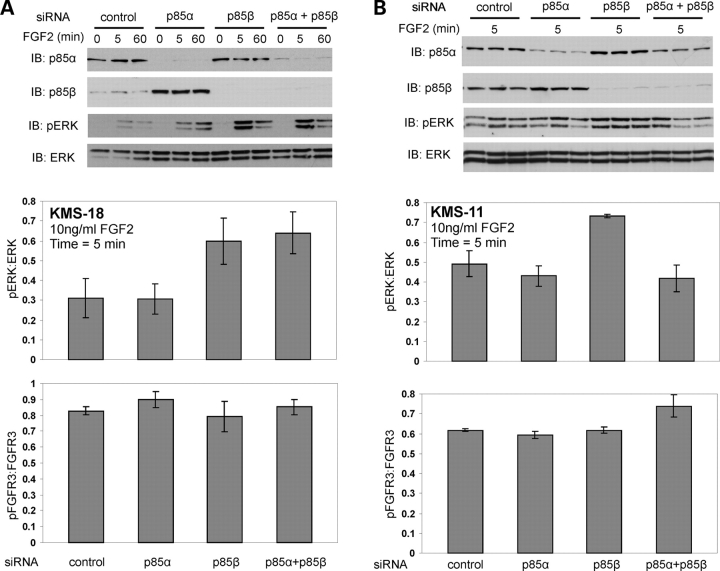
To begin to address the functional consequence of the p85 interaction and better understand the relationship of FGFR3 and p85 in MM, we evaluated the effect of isoform-specific p85 knockdown on the response of the PI3K/AKT and Ras/ERK/MAPK pathways to FGF2 stimulation. We first evaluated AKT phosphorylation, and p85 knockdown had little effect (data not shown). pERK was next evaluated, based upon its important role in FGFR3-mediated phenotypes, and observed that knockdown of p85β in cells expressing constitutively active FGFR3 resulted in an increased phosphorylation of ERK (Fig. 5A and B). In KMS-18 cells, there was no effect on pERK by p85α knockdown, alone or in combination with p85β knockdown (Fig. 5A) at early time points (5 min) when phosphorylation changes are most robust for FGFR3 signaling (Fig. 4D) (53,63). However, in KMS-11 cells, simultaneous knockdown of p85α abrogated the effect of p85β knockdown (Fig. 5B). For both lines, these data suggest that p85β may negatively regulate ERK activity downstream of FGFR3 signaling in MM cells.
Figure 5.
p85 proteins modulate ERK activation in MM cells. (A) Following siRNA knockdown of p85 isoforms in KMS-18 MM cells, cells were serum-starved 4 h and treated with 10 ng/ml FGF-2 for the times indicated. Twenty micrograms total cell lysate was assessed for p85 knockdown as well as phosphorylation of Akt (data not shown) and Erk. Representative of three experiments. The effect of p85 silencing on ERK and FGFR3 phosphorylation at the 5 min time point was quantitated using NIH Scion Image and pERK:ERK and pFGFR3:FGFR3 ratios determined for 5 and 3 replicates, respectively. The mean±SEM is plotted. Knockdown of p85β alone or in combination with p85α resulted in increased phosphorylation of ERK. (B) KMS-11 MM cells were treated as in (A). Data for the 5 min time point is shown and the pERK:ERK ratio was determined for six replicates, the pFGFR3:FGFR3 ratio for three replicates. Knockdown of p85β resulted in increased phosphorylation of ERK.
DISCUSSION
Here we show that FGFR3 can interact with the p85 regulatory isoform of PI3K through a direct interaction at a site distinct from that mediating a Gab1-dependent, indirect interaction, suggesting an additional level of PI3K regulation. Surprisingly, the data presented shows that FGF2-stimulated phosphorylation of ERK is increased following p85β isoform-specific knockdown in MM cells expressing an activated form of FGFR3, suggesting a distinct inhibitory axis that may serve as a cellular attempt to prevent unregulated ERK activation. These findings have broad implications to understanding tumorigenesis and the relatively poor prognosis for patients having FGFR3-associated MM.
p85 interacts with FGFR3 in an activation-dependent manner and requires Y760
The PI3K/AKT pathway can be activated by growth factor receptors, which may involve direct recruitment of PI3K, as for the PDGF and EGF (ErbB3) receptors, or indirectly through association with the IRS or Gab adaptor proteins (e.g. insulin receptor and EGFR, respectively) (29–31,64). With respect to the FGF receptors, FGFR1 can recruit PI3K indirectly via an FRS2/Grb2/Gab1 adaptor protein complex (32). Previous yeast two-hybrid data indicates that FGFR1 can also recruit PI3K via binding of p85α and p85β (54). Similarly, FGFR3 has been found to interact with p50α (this report) and with p85β, in a parallel study (D.D., unpublished results) by yeast two-hybrid. A search of the yeast database (Saccharaomyces Genome Database; http://www.yeastgenome.org) indicates that yeast lack homologues of the FRS2, Grb2 and Gab1 proteins, suggesting that the interaction observed in yeast represents a direct interaction between FGFR3 and PI3K. However, this interaction was not verified in mammalian cells and to date, evidence of direct interaction between an FGF receptor and PI3K in a non-yeast system has been solely provided for Xenopus FGFR (54,65). We find that FGFR3 can associate with PI3K regulatory proteins in mammalian cells. This interaction appears to be dependent on receptor activation since the constitutively active receptor shows significant interaction in HeLa cells, whereas ligand was required to observe a strong interaction with wild-type receptor (Fig. 2A, data not shown). Similarly, in MM cells, interaction with FGFR3 and p85 was only observed constitutively in lines expressing receptors with activating mutations and upon ligand addition in lines carrying wild-type receptor (Fig. 4D). The interaction of p85 with wild-type FGFR3 in the absence of ligand that we observe in yeast, and to a lesser extent in HeLa cells, is likely due to overexpression of the receptor which produces some autophosphorylation in the absence of ligand (Fig. 2C and unpublished observations).
Our data suggest that FGFR3 can associate with p85 independently of the Gab1 adaptor protein (32) and appears to require Y760 of FGFR3, as a mutant of Gab1 that cannot bind p85 (56,57) did not alter the level of p85 that associates with FGFR3. Using single Y→F at tyrosines known to be critical to transducing FGFR3 signals (58), we find that mutation of Y760 and, to a lesser extent, Y724 (Fig. 3A), significantly reduces co-immunoprecipitation of FGFR3 with p85. The corresponding residue for Y760 in human FGFR1 (Y766) is a binding site for PLCγ (60), and this site in mouse FGFR3 (Y754) is also implicated in PLCγ binding (61). Mutation of Y760 in human FGFR3 significantly attenuates PLCγ activation in Ba/F3 cells (18) and we confirm this site is also required for binding of PLCγ in our studies. However, p85 and PLCγ, while both capable of binding activated FGFR3, do not co-immunoprecipitate with one another in the presence of the activated receptor. We interpret these data to indicate that p85 and PLCγ can both bind activated FGFR3 directly and potentially in a competitive or mutually exclusive manner at Y760, and influence downstream signaling. The use of distinct residues for potentially different outcomes of PI3K recruitment is intriguing, particularly if recruitment of specific regulators of PI3K signaling to different sites is associated with either differential regulation of PI3K activation and downstream effects, or competes for binding of other signaling molecules such as PLCγ. In support of this possibility, while p85 can bind to activated FGFR3, results from an in vitro FGFR3 kinase assay indicate that p85 is not a substrate of FGFR3 (data not shown), suggesting that when p85 directly binds receptor, binding does not lead to phosphorylation of p85 and activation of downstream pathways, such as AKT. However, the presence of p85 in complex with the FGFR3 catalytic subunit inhibits the ability of FGFR3 to phosphorylate STAT1 in the kinase assay (unpublished results), suggesting that the primary role for this interaction may be to modulate the phosphorylation of other FGFR3 targets, such as STAT1 or PLCγ. This possibility will be explored in future studies.
Knockdown of p85 isoforms moduates ERK activation in MM cells
The Ras/ERK/MAPK and PI3K/AKT pathways are key regulators of cell growth and survival frequently altered in cancer, including FGFR3-related cancers such as MM. Dominant negative proteins of the Ras/MAPK pathway inhibit the transforming potential of FGFR3, implicated in 10–20% of MM cases (19). Similarly, PI3K signaling is implicated in mediating the transforming property of the TEL-FGFR3 fusion protein found in a peripheral T cell lymphoma that progressed to acute myelogenous leukemia (38), and chemical inhibition of PI3K has been shown to abolish the proliferative effect of combined FGF and IL-6 stimulation in MM cells (66).
Here, we show that p85α and p85β are the predominant PI3K regulatory proteins expressed in MM cell lines, and each are capable of interacting with activated FGFR3 in these cells. Treatment with FGF2 results in AKT and ERK phosphorylation, with the level and kinetics of phosphorylation varying between the different cell lines (data not shown and (67)). However, isoform-specific knockdown of p85β, but not p85α, in the KMS-18 and KMS-11 lines specifically resulted in increased phosphorylation of ERK in response to FGF2 treatment (Fig. 5A and B and data not shown). It is not clear why dual p85α appears to abrogate p85β knockdown in KMS11 cells. However, differential effects are consistent with results for c-kit signaling in hematopoietic stem/progenitor cells (HSC/Ps), where genetic disruption of p85α, but not p85β, dramatically reduces proliferation and colony formation of HSC/Ps carrying the activating D814V mutation of c-kit present in some AML cases (68). While p85α and p85β share significant structural homology in the two SH2 domains, they show differences in the N-terminal SH3 domain, the latter of which accounts for some of the differential affects of the p85 isoforms in the reported system. A similar difference could be occurring in the MM cells studied here, explaining the stronger effect with p85β.
The increase in ERK phosphorylation elicited by p85 inhibition was unexpected since several reports implicate PI3K activity in the enhancement of ERK activation when crosstalk between these two pathways is observed (reviewed in 44,45). In contrast, other studies implicate an AKT-dependent phosphorylation of Raf, resulting in abrogation of Raf action on downstream substrates and causing decreased ERK phosphorylation (69–71). Interestingly, there is precedence for this type of effect in chondrocytes, where addition of constitutively active AKT partially reversed some of the pathological effects of FGFR3 in these cells (72). Given the protective effects of PI3K inhibitors in MM cell lines, the results suggest that the direct p85 interaction may be distinct and that PI3K signaling may be solely modulated by the indirect interaction and activation via the Gab1 cascade. Extensive evidence indicates that the Ras/ERK/MAPK pathway is critical to the oncogenic potential of FGFR3 activity and to MM pathogenesis. Activating mutations in FGFR3 do not occur in the same MM cells as activating mutations in Ras, suggesting they may have a similar role in MM progression (19). Indeed, inhibition of the Ras/ERK/MAPK pathway significantly reduces the transforming potential of FGFR3 (19) and small molecule inhibitors of FGFR3 which successfully attenuate MM phenotypes result in decreased phosphorylation of ERK (15,17,25,26). Recently, Kang et al. (21) reported that the direct ERK substrate, RSK2, mediates growth and survival of FGFR3-expressing MM cells, including the KMS-11 line.
How then does one reconcile the apparently contradictory results showing that ERK and PI3K activation appear necessary for FGFR3-mediated MM pathogenesis while p85 knockdown enhances acute ERK activation. Further, how might the direct interaction of FGFR3 with p85 impact MM. Since FGFRs can activate both ERK and PI3K both through binding of FRS2 to Grb2 where, respectively, Grb2 interacts with Sos guanine nucleotide exchange factor via its amino-terminal SH3 domain and with Gab1 via its C-terminal SH3 domain and this binding can occur simultaneously (55), it is likely that the indirect interaction and phosphorylation of p85 via FRS2 and Gab1 activates a canonical pathway involved in promoting tumorigenesis. In contrast, the direct interaction of FGFR3 at Y760 does not appear to cause p85 phosphorylation in vitro and may instead represent a negative compensatory axis to keep signaling in check to prevent cancer. In MM, where constitutive activation of aberrantly expressed FGFR3 following chromosomal rearrangement can occur, MM may be a consequence in part of an inability of this and other normally compensatory pathways (73) to keep signaling in check, possibly because constitutive activation overcomes the ability of p85 to negatively regulate ERK activation. Alternatively, some other non-canonical pathway may become activated in MM via the direct p85 interaction. Since Y760 also mediates an interaction with PLCγ (PLCγ/diacylglycerol-mediated activation of the RasGRP1 exchange factor (reviewed in 74)), it is possible that when p85β levels are decreased, PLCγ has increased access to this site and the enhanced PLCγ binding causes increased activation of PLCγ and subsequently ERK. Indeed, in preliminary studies, we observe that silencing of p85β results in increased phosphorylation of PLCγ following FGF2 treatment of KMS-18 and KMS-11 MM lines (data not shown). Further, as described earlier, phosphorylation of a second FGFR3 substrate, STAT1, was also inhibited in vitro in the presence of p85 (unpublished results), suggesting that several pathways could be impacted by the p85 interaction. Precedence exists for both direct and indirect binding sites for p85 and for dual regulatory functions of receptor binding partners. The hepatocyte growth factor receptor (Met) binds p85 both directly and indirectly via Gab1 (75) and a balance exists for various domains to mediate tumor progression activity of Met/Ras signaling, suggesting that selectivity for specific adaptor protein involvement may be critical for tumorigenesis (75). Further, Gab1 binding elicits opposing effects in IGF-1 signaling. IGF-1 activates PI3K to promote myogenic differentiation and as stated earlier, Gab1 can activate PI3K-AKT signaling through association of phosphorylated p85 downstream of various growth factors. However, upon IGF-1 activation, Gab1 can associate with SHP2 to activate ERK1/2, which inhibits myogenic differentiation, suggesting an inhibitory axis for IGF-I-activation as well (76). Therefore, future studies will involve investigating levels and turnover of various proteins (e.g. Gab1, PLCγ) in MM samples to determine whether abundance may influence positive and negative regulatory axes of the FGFR3 signaling pathway.
The results presented here suggest that in FGFR3-associated MM lines, p85 is involved in a negative regulatory axis of ERK signaling, possibly by directly competing with PLCγ or STATs for binding at Y760, representing a novel regulatory process that may be overcome by expression of constitutively active FGFR3. These findings have therapeutic significance in that delivery of peptide mimics of the specific p85 domain that interacts with Y760 and potentially inhibits PLCγ binding could prevent deleterious signaling from ectopic and constitutively active FGFR3 expression. This is similar to that shown for a peptide developed for the region surrounding the FGFR high-affinity binding site for PLCγ (77). For MM, it is conceivable that FGFR3 activation may lead to two outcomes involving p85—an interaction via the adaptor protein Gab1 with subsequent activation of PI3K that contributes to pathogenesis and can be targeted with PI3K inhibitors, and the direct interaction, which acts as an endogenous competitive inhibitor to prevent overactivation of ERK. Attempts to inhibit ERK directly and in combination with FGFR3 inhibitors are likely to be protective. However, mimicking the p85 interacting peptide may also provide a therapeutic strategy to dampen ERK phosphorylation originating from FGFR3 signaling with fewer side effects than general ERK inhibitors. Future studies will identify the p85 domain involved and develop peptide mimics to distinguish between effects specifically derived from this interaction versus the indirect pathway and to test the contribution of this interaction to MM and aberrant proliferation of plasma cells ectopically expressing activated FGFR3.
MATERIALS AND METHODS
Reagents
Antibodies to FGFR3 (B-9) and ERK1 were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA) and the phospho-ERK antibody from Cell Signaling Technology, Inc. (Danvers, MA). Pan p85 antibody was from Upstate Cell Signaling Solutions (Lake Placid, NY) or from Abcam (Cambridge, MA); isoform-specific antibodies to p85α, p85β and p55γ were also from Abcam. Antibody to the FLAG epitope tag was obtained from Sigma (St Louis, MO) and recombinant human FGF2 was from R&D Systems (Minneapolis, MN). siRNA to p85α and p85β (both ON-TARGETplus SMARTpool), as well as control siRNA, were purchased from Dharmacon (Lafayette, CO).
Plasmids
The C-terminally FLAG-tagged wild-type and constitutively active (K650E) FGFR3 vectors have been described previously (78). FLAG-tagged single Y→F mutations on the K650E background were prepared in the same manner by PCR amplification and subcloning from the ‘Single F’ mutants described in Hart et al. (58). HA-tagged human p85α was a kind gift from Dr Jon Backer (Albert Einstein College of Medicine, Bronx, NY) and the wild-type and YF3 Gab1 constructs were generously provided by Dr Patrick Raynal (Toulouse, France). The GFP control was pEGFP-N1 (Clontech, Mountain View, CA).
Yeast two-hybrid screen
A yeast two-hybrid screen was performed using the cytoplasmic domain of wild-type or constitutively active (K650E) human FGFR3 fused to the LexA DNA-binding domain in the pBTM116 plasmid. These were used to screen a human chondrocyte cDNA library, expressed as fusions to the Gal4 activation domain in pACT2 (BD Biosciences Clontech, Palo Alto, CA). Transformants were selected for 3–4 days at 30°C on medium containing 6.7% yeast nitrogen base (BD Biosciences, Franklin Lakes, NJ), 2% glucose, 2% bacto agar (BD Biosciences) and complete amino acids (all from Sigma, St Louis, MO) except histidine, leucine and tryptophan. The resulting colonies were subjected to colony filter lift assay and tested for β-galactosidase activity according to previously published protocol (79).
Cell culture and FGF2 treatment
HeLa cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS. Human MM cell lines that do not express (IM-9, RPMI-08226), express wild-type (NCI-H929, LP-1 or express mutant [OPM-2 (K650E), KMS-11 (Y373C), KMS-18 (G384D)] FGFR3 were obtained from American Type Tissue Culture [ATCC; Manassas, VA (IM-9, NCI-H929)], German Collection of Microorganisms and Cell Cultures [DSMZ; Braunschweig, Germany (RPMI-8226, OPM-2, LP-1)], Dr Guido Tricot (OPM-2 and KMS-11) and Dr P. Leif Bergsagel (KMS-18). MM cells were maintained in RPMI 1640 media supplemented with 10% fetal bovine serum (both from Invitrogen, Carlsbad, CA). For FGF2 treatment, cells were serum-starved 4 h in RPMI 1640 without supplement. Following serum starvation, FGF2 was added to a final concentration of 10 ng/ml for the times indicated. Experiments were performed in triplicate.
Transfections
Transient transfection of HeLa cells was achieved using Lipofectamine 2000 (Invitrogen). Cells were plated at 50% confluence in six-well plates with fresh growth medium, and the following day, washed twice with serum-free medium then incubated with 10 µg DNA in Optimem medium (Invitrogen). After 4 h at 37°C, complete growth medium was added. The following day, cells were harvested for immunoprecipitation and immunoblot analysis. Transfection of MM cells with siRNA was performed using the Nucleofector system (Amaxa Biosystems, Germany). Following the manufacturer’s procedure, 5 × 106 cells were suspended in 100 µl Solution V containing 10 µg siRNA and pulsed once under program T-16 (KMS-18) or program O-10 (KMS-11). Cells were subsequently cultured in 5 ml complete growth medium and 48 h later treated with FGF2 as described earlier.
RT–PCR
Total RNA was isolated from 1 × 107 MM cells using the RNeasy midi kit (Qiagen, Valencia, CA). One microgram total RNA was used for cDNA synthesis, primed with the gene-specific primer 5′-GAGATTCATTCCGGTAGTGG-3′ which recognizes all three isoforms of the PIK3R1 gene (p85α, p55α and p50α) and carried out using the Superscript II RT Kit (Invitrogen). Isoform-specific expression was determined by PCR using the primer above coupled with one of the following, as published (80): 5′-CCGTTGAAATGCATAACCTGC-3′ (p50α), 5′-ATTGTGGCACAGACTTGATG-3′ (p55α), 5′-ATTCTCAGCAGCCAGCTCTG-3′ (p85α) or 5′-ACTACTGTAGCCAACAACGG-3′ (all isoforms). All primer pairs were intron-spanning and a no RT control was included. PCR was performed for 25 cycles as follows: denaturation at 94°C for 25 s, annealing at a primer-specific temperature for 35 s and extension at 72°C for 30 s.
Immunoprecipitation and immunoblot analysis
Cells were washed briefly with cold PBS containing 0.5 mm sodium orthovanadate and lysed in cold lysis buffer [50 mm HEPES (pH 7.5), 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, 10% glycerol, 1% Triton X-100 and protease inhibitors (10 µg/ml leupeptin, 10 µg/ml aprotinin, 1 mm PMSF and 0.5 mm sodium orthovanadate)] (81). Cell debris was removed by centrifugation and protein concentration determined using the Bradford colorimetric assay (Bio-Rad, Hercules, CA). Equal amounts of total protein were boiled 5 min in SDS sample buffer then resolved on a 10% polyacrylamide gel, transferred to nitrocellulose and probed with antibody as indicated. For immunoprecipitation, 200–500 µg total protein was pre-cleared 1 h with protein G PLUS-agarose (Santa Cruz) in 500 µl immunoprecipitation buffer [50 mm HEPES (pH 7.5), 50 mm NaCl, 10% glycerol, 1% Triton X-100 and protease inhibitors as indicated for the lysis buffer] (81). Antibody was added and following an overnight incubation at 4°C, immunocomplexes were recovered by incubation with protein G PLUS-agarose. The beads were washed three times with immunoprecipitation buffer, resuspended in SDS sample buffer and processed as earlier for total cell lysate. Quantitation of western blots was performed using NIH Scion Image.
FUNDING
This work was supported by the Multiple Myeloma Research Foundation (L.M.T. and W.R.W.), Yang Sheng Tang USA (L.M.T. and W.R.W.), Winnick Family Research Scholar’s award (W.R.W.) and the Ministry of Education, Youth and Sports of the Czech Republic (MSM0021622430; P.K.).
ACKNOWLEDGEMENTS
We would like to thank Ya-Zhen Zhu and Katalin Illes for technical assistance. We would also like to thank Dr Joan S. Steffan and the Sandmeyer and Nomura laboratories at UCI for helpful advice and reagents. HA-tagged human p85α was a kind gift from Dr Jon Backer (Albert Einstein College of Medicine, Bronx, NY), the wild-type and YF3 Gab1 constructs were generously provided by Dr Patrick Raynal (Toulouse, France) and the p55α plasmid was kindly provided by Dr Anne Hamburger at the University of Maryland School of Medicine, Baltimore, MD.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Eswarakumar V.P., Lax I., Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 2.L’Hote C.G., Knowles M.A. Cell responses to FGFR3 signalling: growth, differentiation and apoptosis. Exp. Cell Res. 2005;304:417–431. doi: 10.1016/j.yexcr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Thisse B., Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev. Biol. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Chaffer C.L., Dopheide B., Savagner P., Thompson E.W., Williams E.D. Aberrant fibroblast growth factor receptor signaling in bladder and other cancers. Differentiation. 2007;75:831–842. doi: 10.1111/j.1432-0436.2007.00210.x. [DOI] [PubMed] [Google Scholar]
- 5.Ornitz D.M., Marie P.J. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002;16:1446–1465. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- 6.Ronchetti D., Greco A., Compasso S., Colombo G., Dell’Era P., Otsuki T., Lombardi L., Neri A. Deregulated FGFR3 mutants in multiple myeloma cell lines with t(4;14): comparative analysis of Y373C, K650E and the novel G384D mutations. Oncogene. 2001;20:3553–3562. doi: 10.1038/sj.onc.1204465. [DOI] [PubMed] [Google Scholar]
- 7.Webster M.K., Donoghue D.J. FGFR activation in skeletal disorders: too much of a good thing. Trends Genet. 1997;13:178–182. doi: 10.1016/s0168-9525(97)01131-1. [DOI] [PubMed] [Google Scholar]
- 8.Bergsagel P.L., Kuehl W.M. Molecular pathogenesis and a consequent classification of multiple myeloma. J. Clin. Oncol. 2005;23:6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 9.Chesi M., Nardini E., Brents L.A., Schrock E., Ried T., Kuehl W.M., Bergsagel P.L. Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat. Genet. 1997;16:260–264. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richelda R., Ronchetti D., Baldini L., Cro L., Viggiano L., Marzella R., Rocchi M., Otsuki T., Lombardi L., Maiolo A.T., et al. A novel chromosomal translocation t(4; 14)(p16.3; q32) in multiple myeloma involves the fibroblast growth-factor receptor 3 gene. Blood. 1997;90:4062–4070. [PubMed] [Google Scholar]
- 11.Moreau P., Facon T., Leleu X., Morineau N., Huyghe P., Harousseau J.L., Bataille R., Avet-Loiseau H. Recurrent 14q32 translocations determine the prognosis of multiple myeloma, especially in patients receiving intensive chemotherapy. Blood. 2002;100:1579–1583. doi: 10.1182/blood-2002-03-0749. [DOI] [PubMed] [Google Scholar]
- 12.Intini D., Baldini L., Fabris S., Lombardi L., Ciceri G., Maiolo A.T., Neri A. Analysis of FGFR3 gene mutations in multiple myeloma patients with t(4;14) Br. J. Haematol. 2001;114:362–364. doi: 10.1046/j.1365-2141.2001.02957.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen J., Lee B.H., Williams I.R., Kutok J.L., Mitsiades C.S., Duclos N., Cohen S., Adelsperger J., Okabe R., Coburn A., et al. FGFR3 as a therapeutic target of the small molecule inhibitor PKC412 in hematopoietic malignancies. Oncogene. 2005;24:8259–8267. doi: 10.1038/sj.onc.1208989. [DOI] [PubMed] [Google Scholar]
- 14.Trudel S., Ely S., Farooqi Y., Affer M., Robbiani D.F., Chesi M., Bergsagel P.L. Inhibition of fibroblast growth factor receptor 3 induces differentiation and apoptosis in t(4;14) myeloma. Blood. 2004;103:3521–3528. doi: 10.1182/blood-2003-10-3650. [DOI] [PubMed] [Google Scholar]
- 15.Trudel S., Li Z.H., Wei E., Wiesmann M., Chang H., Chen C., Reece D., Heise C., Stewart A.K. CHIR-258, a novel, multitargeted tyrosine kinase inhibitor for the potential treatment of t(4;14) multiple myeloma. Blood. 2005;105:2941–2948. doi: 10.1182/blood-2004-10-3913. [DOI] [PubMed] [Google Scholar]
- 16.Trudel S., Stewart A.K., Rom E., Wei E., Li Z.H., Kotzer S., Chumakov I., Singer Y., Chang H., Liang S.B., et al. The inhibitory anti-FGFR3 antibody, PRO-001, is cytotoxic to t(4;14) multiple myeloma cells. Blood. 2006;107:4039–4046. doi: 10.1182/blood-2005-10-4179. [DOI] [PubMed] [Google Scholar]
- 17.Xin X., Abrams T.J., Hollenbach P.W., Rendahl K.G., Tang Y., Oei Y.A., Embry M.G., Swinarski D.E., Garrett E.N., Pryer N.K., et al. CHIR-258 is efficacious in a newly developed fibroblast growth factor receptor 3-expressing orthotopic multiple myeloma model in mice. Clin. Cancer Res. 2006;12:4908–4915. doi: 10.1158/1078-0432.CCR-06-0957. [DOI] [PubMed] [Google Scholar]
- 18.Chen J., Williams I.R., Lee B.H., Duclos N., Huntly B.J., Donoghue D.J., Gilliland D.G. Constitutively activated FGFR3 mutants signal through PLCgamma-dependent and -independent pathways for hematopoietic transformation. Blood. 2005;106:328–337. doi: 10.1182/blood-2004-09-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chesi M., Brents L.A., Ely S.A., Bais C., Robbiani D.F., Mesri E.A., Kuehl W.M., Bergsagel P.L. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood. 2001;97:729–736. doi: 10.1182/blood.v97.3.729. [DOI] [PubMed] [Google Scholar]
- 20.Harvey R.D., Lonial S. PI3 kinase/AKT pathway as a therapeutic target in multiple myeloma. Future Oncol. 2007;3:639–647. doi: 10.2217/14796694.3.6.639. [DOI] [PubMed] [Google Scholar]
- 21.Kang S., Dong S., Gu T.L., Guo A., Cohen M.S., Lonial S., Khoury H.J., Fabbro D., Gilliland D.G., Bergsagel P.L., et al. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell. 2007;12:201–214. doi: 10.1016/j.ccr.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollett J.B., Trudel S., Stern D., Li Z.H., Stewart A.K. Overexpression of the myeloma-associated oncogene fibroblast growth factor receptor 3 confers dexamethasone resistance. Blood. 2002;100:3819–3821. doi: 10.1182/blood-2002-02-0608. [DOI] [PubMed] [Google Scholar]
- 23.Younes H., Leleu X., Hatjiharissi E., Moreau A.S., Hideshima T., Richardson P., Anderson K.C., Ghobrial I.M. Targeting the phosphatidylinositol 3-kinase pathway in multiple myeloma. Clin. Cancer Res. 2007;13:3771–3775. doi: 10.1158/1078-0432.CCR-06-2921. [DOI] [PubMed] [Google Scholar]
- 24.Murakami S., Balmes G., McKinney S., Zhang Z., Givol D., de Crombrugghe B. Constitutive activation of MEK1 in chondrocytes causes Stat1-independent achondroplasia-like dwarfism and rescues the Fgfr3-deficient mouse phenotype. Genes Dev. 2004;18:290–305. doi: 10.1101/gad.1179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grand E.K., Chase A.J., Heath C., Rahemtulla A., Cross N.C. Targeting FGFR3 in multiple myeloma: inhibition of t(4;14)-positive cells by SU5402 and PD173074. Leukemia. 2004;18:962–966. doi: 10.1038/sj.leu.2403347. [DOI] [PubMed] [Google Scholar]
- 26.Paterson J.L., Li Z., Wen X.Y., Masih-Khan E., Chang H., Pollett J.B., Trudel S., Stewart A.K. Preclinical studies of fibroblast growth factor receptor 3 as a therapeutic target in multiple myeloma. Br. J. Haematol. 2004;124:595–603. doi: 10.1111/j.1365-2141.2004.04814.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhao L., Vogt P.K. Class I PI3K in oncogenic cellular transformation. Oncogene. 2008;27:5486–5496. doi: 10.1038/onc.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piccione E., Case R.D., Domchek S.M., Hu P., Chaudhuri M., Backer J.M., Schlessinger J., Shoelson S.E. Phosphatidylinositol 3-kinase p85 SH2 domain specificity defined by direct phosphopeptide/SH2 domain binding. Biochemistry. 1993;32:3197–3202. doi: 10.1021/bi00064a001. [DOI] [PubMed] [Google Scholar]
- 29.Holgado-Madruga M., Emlet D.R., Moscatello D.K., Godwin A.K., Wong A.J. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature. 1996;379:560–564. doi: 10.1038/379560a0. [DOI] [PubMed] [Google Scholar]
- 30.Rodrigues G.A., Falasca M., Zhang Z., Ong S.H., Schlessinger J. A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol. Cell. Biol. 2000;20:1448–1459. doi: 10.1128/mcb.20.4.1448-1459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yenush L., White M.F. The IRS-signalling system during insulin and cytokine action. Bioessays. 1997;19:491–500. doi: 10.1002/bies.950190608. [DOI] [PubMed] [Google Scholar]
- 32.Ong S.H., Hadari Y.R., Gotoh N., Guy G.R., Schlessinger J., Lax I. Stimulation of phosphatidylinositol 3-kinase by fibroblast growth factor receptors is mediated by coordinated recruitment of multiple docking proteins. Proc. Natl Acad. Sci. USA. 2001;98:6074–6079. doi: 10.1073/pnas.111114298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia Z., Kumar A., Marques M., Cortes I., Carrera A.C. Phosphoinositide 3-kinase controls early and late events in mammalian cell division. EMBO J. 2006;25:655–661. doi: 10.1038/sj.emboj.7600967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song G., Ouyang G., Bao S. The activation of Akt/PKB signaling pathway and cell survival. J. Cell Mol. Med. 2005;9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogt P.K., Kang S., Elsliger M.A., Gymnopoulos M. Cancer-specific mutations in phosphatidylinositol 3-kinase. Trends Biochem. Sci. 2007;32:342–349. doi: 10.1016/j.tibs.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 36.LoPiccolo J., Granville C.A., Gills J.J., Dennis P.A. Targeting Akt in cancer therapy. Anticancer Drugs. 2007;18:861–874. doi: 10.1097/CAD.0b013e3280cc2c6f. [DOI] [PubMed] [Google Scholar]
- 37.Min Y.H., Cheong J.W., Kim J.Y., Eom J.I., Lee S.T., Hahn J.S., Ko Y.W., Lee M.H. Cytoplasmic mislocalization of p27Kip1 protein is associated with constitutive phosphorylation of Akt or protein kinase B and poor prognosis in acute myelogenous leukemia. Cancer Res. 2004;64:5225–5231. doi: 10.1158/0008-5472.CAN-04-0174. [DOI] [PubMed] [Google Scholar]
- 38.Maeda T., Yagasaki F., Ishikawa M., Takahashi N., Bessho M. Transforming property of TEL-FGFR3 mediated through PI3-K in a T-cell lymphoma that subsequently progressed to AML. Blood. 2005;105:2115–2123. doi: 10.1182/blood-2003-12-4290. [DOI] [PubMed] [Google Scholar]
- 39.Menu E., Kooijman R., Van Valckenborgh E., Asosingh K., Bakkus M., Van Camp B., Vanderkerken K. Specific roles for the PI3K and the MEK-ERK pathway in IGF-1-stimulated chemotaxis, VEGF secretion and proliferation of multiple myeloma cells: study in the 5T33MM model. Br. J. Cancer. 2004;90:1076–1083. doi: 10.1038/sj.bjc.6601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pene F., Claessens Y.E., Muller O., Viguie F., Mayeux P., Dreyfus F., Lacombe C., Bouscary D. Role of the phosphatidylinositol 3-kinase/Akt and mTOR/P70S6-kinase pathways in the proliferation and apoptosis in multiple myeloma. Oncogene. 2002;21:6587–6597. doi: 10.1038/sj.onc.1205923. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J., Choi Y., Mavromatis B., Lichtenstein A., Li W. Preferential killing of PTEN-null myelomas by PI3K inhibitors through Akt pathway. Oncogene. 2003;22:6289–6295. doi: 10.1038/sj.onc.1206718. [DOI] [PubMed] [Google Scholar]
- 42.Downward J. Role of phosphoinositide-3-OH kinase in Ras signaling. Adv. Second Messenger Phosphoprotein Res. 1997;31:1–10. doi: 10.1016/s1040-7952(97)80004-3. [DOI] [PubMed] [Google Scholar]
- 43.Liu L., Xie Y., Lou L. PI3K is required for insulin-stimulated but not EGF-stimulated ERK1/2 activation. Eur. J. Cell Biol. 2006;85:367–374. doi: 10.1016/j.ejcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Sebolt-Leopold J.S., Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat. Rev. Cancer. 2004;4:937–947. doi: 10.1038/nrc1503. [DOI] [PubMed] [Google Scholar]
- 45.Vivanco I., Sawyers C.L. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 46.Wang Q., Zhou Y., Wang X., Evers B.M. Glycogen synthase kinase-3 is a negative regulator of extracellular signal-regulated kinase. Oncogene. 2006;25:43–50. doi: 10.1038/sj.onc.1209004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong M., Wang C.S., Donoghue D.J. Interaction of fibroblast growth factor receptor 3 and the adapter protein SH2-B. A role in STAT5 activation. J. Biol. Chem. 2002;277:15962–15970. doi: 10.1074/jbc.M102777200. [DOI] [PubMed] [Google Scholar]
- 48.Tavormina P.L., Shiang R., Thompson L.M., Zhu Y.Z., Wilkin D.J., Lachman R.S., Wilcox W.R., Rimoin D.L., Cohn D.H., Wasmuth J.J. Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nat. Genet. 1995;9:321–328. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- 49.Anderson K.E., Jackson S.P. Class I phosphoinositide 3-kinases. Int. J. Biochem. Cell Biol. 2003;35:1028–1033. doi: 10.1016/s1357-2725(02)00270-4. [DOI] [PubMed] [Google Scholar]
- 50.Vanhaesebroeck B., Ali K., Bilancio A., Geering B., Foukas L.C. Signalling by PI3K isoforms: insights from gene-targeted mice. Trends Biochem. Sci. 2005;30:194–204. doi: 10.1016/j.tibs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Otsu M., Hiles I., Gout I., Fry M.J., Ruiz-Larrea F., Panayotou G., Thompson A., Dhand R., Hsuan J., Totty N., et al. Characterization of two 85 kd proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes, and PI3-kinase. Cell. 1991;65:91–104. doi: 10.1016/0092-8674(91)90411-q. [DOI] [PubMed] [Google Scholar]
- 52.Pons S., Asano T., Glasheen E., Miralpeix M., Zhang Y., Fisher T.L., Myers M.G., Jr, Sun X.J., White M.F. The structure and function of p55PIK reveal a new regulatory subunit for phosphatidylinositol 3-kinase. Mol. Cell. Biol. 1995;15:4453–4465. doi: 10.1128/mcb.15.8.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raffioni S., Zhu Y.Z., Bradshaw R.A., Thompson L.M. Effect of transmembrane and kinase domain mutations on fibroblast growth factor receptor 3 chimera signaling in PC12 cells. A model for the control of receptor tyrosine kinase activation. J. Biol. Chem. 1998;273:35250–35259. doi: 10.1074/jbc.273.52.35250. [DOI] [PubMed] [Google Scholar]
- 54.Hu Y., Fang X., Dunham S.M., Prada C., Stachowiak E.K., Stachowiak M.K. 90-kDa ribosomal S6 kinase is a direct target for the nuclear fibroblast growth factor receptor 1 (FGFR1): role in FGFR1 signaling. J. Biol. Chem. 2004;279:29325–29335. doi: 10.1074/jbc.M311144200. [DOI] [PubMed] [Google Scholar]
- 55.Ong S.H., Guy G.R., Hadari Y.R., Laks S., Gotoh N., Schlessinger J., Lax I. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol. Cell. Biol. 2000;20:979–989. doi: 10.1128/mcb.20.3.979-989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montagner A., Yart A., Dance M., Perret B., Salles J.P., Raynal P. A novel role for Gab1 and SHP2 in epidermal growth factor-induced Ras activation. J. Biol. Chem. 2005;280:5350–5360. doi: 10.1074/jbc.M410012200. [DOI] [PubMed] [Google Scholar]
- 57.Yart A., Laffargue M., Mayeux P., Chretien S., Peres C., Tonks N., Roche S., Payrastre B., Chap H., Raynal P. A critical role for phosphoinositide 3-kinase upstream of Gab1 and SHP2 in the activation of ras and mitogen-activated protein kinases by epidermal growth factor. J. Biol. Chem. 2001;276:8856–8864. doi: 10.1074/jbc.M006966200. [DOI] [PubMed] [Google Scholar]
- 58.Hart K.C., Robertson S.C., Donoghue D.J. Identification of tyrosine residues in constitutively activated fibroblast growth factor receptor 3 involved in mitogenesis, Stat activation, and phosphatidylinositol 3-kinase activation. Mol. Biol. Cell. 2001;12:931–942. doi: 10.1091/mbc.12.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cross M.J., Hodgkin M.N., Roberts S., Landgren E., Wakelam M.J., Claesson-Welsh L. Tyrosine 766 in the fibroblast growth factor receptor-1 is required for FGF-stimulation of phospholipase C, phospholipase D, phospholipase A(2), phosphoinositide 3-kinase and cytoskeletal reorganisation in porcine aortic endothelial cells. J. Cell Sci. 2000;113:643–651. doi: 10.1242/jcs.113.4.643. [DOI] [PubMed] [Google Scholar]
- 60.Mohammadi M., Honegger A.M., Rotin D., Fischer R., Bellot F., Li W., Dionne C.A., Jaye M., Rubinstein M., Schlessinger J. A tyrosine-phosphorylated carboxy-terminal peptide of the fibroblast growth factor receptor (Flg) is a binding site for the SH2 domain of phospholipase C-gamma 1. Mol. Cell Biol. 1991;11:5068–5078. doi: 10.1128/mcb.11.10.5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lievens P.M., Roncador A., Liboi E. K644E/M FGFR3 mutants activate Erk1/2 from the endoplasmic reticulum through FRS2 alpha and PLC gamma-independent pathways. J. Mol. Biol. 2006;357:783–792. doi: 10.1016/j.jmb.2006.01.058. [DOI] [PubMed] [Google Scholar]
- 62.Hyun T., Yam A., Pece S., Xie X., Zhang J., Miki T., Gutkind J.S., Li W. Loss of PTEN expression leading to high Akt activation in human multiple myelomas. Blood. 2000;96:3560–3568. [PubMed] [Google Scholar]
- 63.Thompson L.M., Raffioni S., Wasmuth J.J., Bradshaw R.A. Chimeras of the native form or achondroplasia mutant (G375C) of human fibroblast growth factor receptor 3 induce ligand-dependent differentiation of PC12 cells. Mol. Cell. Biol. 1997;17:4169–4177. doi: 10.1128/mcb.17.7.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suenaga A., Takada N., Hatakeyama M., Ichikawa M., Yu X., Tomii K., Okimoto N., Futatsugi N., Narumi T., Shirouzu M., et al. Novel mechanism of interaction of p85 subunit of phosphatidylinositol 3-kinase and ErbB3 receptor-derived phosphotyrosyl peptides. J. Biol. Chem. 2005;280:1321–1326. doi: 10.1074/jbc.M410436200. [DOI] [PubMed] [Google Scholar]
- 65.Ryan P.J., Paterno G.D., Gillespie L.L. Identification of phosphorylated proteins associated with the fibroblast growth factor receptor type I during early Xenopus development. Biochem. Biophys. Res. Commun. 1998;244:763–767. doi: 10.1006/bbrc.1998.8326. [DOI] [PubMed] [Google Scholar]
- 66.Ishikawa H., Tsuyama N., Liu S., Abroun S., Li F.J., Otsuyama K., Zheng X., Ma Z., Maki Y., Iqbal M.S., et al. Accelerated proliferation of myeloma cells by interleukin-6 cooperating with fibroblast growth factor receptor 3-mediated signals. Oncogene. 2005;24:6328–6332. doi: 10.1038/sj.onc.1208782. [DOI] [PubMed] [Google Scholar]
- 67.Krejci P., Mekikian P.B., Wilcox W.R. The fibroblast growth factors in multiple myeloma. Leukemia. 2006;20:1165–1168. doi: 10.1038/sj.leu.2404202. [DOI] [PubMed] [Google Scholar]
- 68.Munugalavadla V., Sims E.C., Borneo J., Chan R.J., Kapur R. Genetic and pharmacologic evidence implicating the p85 alpha, but not p85 beta, regulatory subunit of PI3K and Rac2 GTPase in regulating oncogenic KIT-induced transformation in acute myeloid leukemia and systemic mastocytosis. Blood. 2007;110:1612–1620. doi: 10.1182/blood-2006-10-053058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guan K.L., Figueroa C., Brtva T.R., Zhu T., Taylor J., Barber T.D., Vojtek A.B. Negative regulation of the serine/threonine kinase B-Raf by Akt. J. Biol. Chem. 2000;275:27354–27359. doi: 10.1074/jbc.M004371200. [DOI] [PubMed] [Google Scholar]
- 70.Moelling K., Schad K., Bosse M., Zimmermann S., Schweneker M. Regulation of Raf-Akt Cross-talk. Biol. Chem. 2002;277:31099–31106. doi: 10.1074/jbc.M111974200. [DOI] [PubMed] [Google Scholar]
- 71.Rommel C., Clarke B.A., Zimmermann S., Nunez L., Rossman R., Reid K., Moelling K., Yancopoulos G.D., Glass D.J. Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science. 1999;286:1738–1741. doi: 10.1126/science.286.5445.1738. [DOI] [PubMed] [Google Scholar]
- 72.Priore R., Dailey L., Basilico C. Downregulation of Akt activity contributes to the growth arrest induced by FGF in chondrocytes. J. Cell Physiol. 2006;207:800–808. doi: 10.1002/jcp.20620. [DOI] [PubMed] [Google Scholar]
- 73.Hanafusa H., Torii S., Yasunaga T., Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat. Cell Biol. 2002;4:850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- 74.McKay M.M., Morrison D.K. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 75.Seiden-Long I., Navab R., Shih W., Li M., Chow J., Zhu C.Q., Radulovich N., Saucier C., Tsao M.S. Gab1 but not Grb2 mediates tumor progression in Met overexpressing colorectal cancer cells. Carcinogenesis. 2008;29:647–655. doi: 10.1093/carcin/bgn009. [DOI] [PubMed] [Google Scholar]
- 76.Koyama T., Nakaoka Y., Fujio Y., Hirota H., Nishida K., Sugiyama S., Okamoto K., Yamauchi-Takihara K., Yoshimura M., Mochizuki S., et al. Interaction of scaffolding adaptor protein Gab1 with tyrosine phosphatase SHP2 negatively regulates IGF-I-dependent myogenic differentiation via the ERK1/2 signaling pathway. J. Biol. Chem. 2008;283:24234–24244. doi: 10.1074/jbc.M803907200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hall H., Williams E.J., Moore S.E., Walsh F.S., Prochiantz A., Doherty P. Inhibition of FGF-stimulated phosphatidylinositol hydrolysis and neurite outgrowth by a cell-membrane permeable phosphopeptide. Curr. Biol. 1996;6:580–587. doi: 10.1016/s0960-9822(02)00544-4. [DOI] [PubMed] [Google Scholar]
- 78.Krejci P., Masri B., Salazar L., Farrington-Rock C., Prats H., Thompson L.M., Wilcox W.R. Bisindolylmaleimide I suppresses fibroblast growth factor-mediated activation of Erk MAP kinase in chondrocytes by preventing Shp2 association with the Frs2 and Gab1 adaptor proteins. J. Biol. Chem. 2007;282:2929–2936. doi: 10.1074/jbc.M606144200. [DOI] [PubMed] [Google Scholar]
- 79.Vojtek A.B., Hollenberg S.M., Cooper J.A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 80.Lefai E., Roques M., Vega N., Laville M., Vidal H. Expression of the splice variants of the p85alpha regulatory subunit of phosphoinositide 3-kinase in muscle and adipose tissue of healthy subjects and type 2 diabetic patients. Biochem. J. 2001;360:117–126. doi: 10.1042/0264-6021:3600117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bonofiglio D., Gabriele S., Aquila S., Catalano S., Gentile M., Middea E., Giordano F., Ando S. Estrogen receptor alpha binds to peroxisome proliferator-activated receptor response element and negatively interferes with peroxisome proliferator-activated receptor gamma signaling in breast cancer cells. Clin. Cancer Res. 2005;11:6139–6147. doi: 10.1158/1078-0432.CCR-04-2453. [DOI] [PubMed] [Google Scholar]