Abstract
Understanding the factors that regulate sebum production is important in identifying novel therapeutic targets for acne therapy. Insulin and insulin-like growth factor-1 (IGF-1) stimulate sebaceous gland lipogenesis. IGF-1 increases expression of sterol response element binding protein-1 (SREBP-1), a transcription factor that regulates numerous genes involved in lipid biosynthesis. SREBP-1 expression in turn stimulates lipogenesis in sebocytes. The goal of this study was to identify the intracellular signaling pathway(s) that transduces the lipogenic signal initiated by IGF-1. Sebocytes were treated with IGF-1 and assayed for activation of the PI3- kinase pathway and of the 3 major arms of the MAPK pathway (MAPK/ERK, p38 MAPK and SNK/JNK). IGF-1 activated the MAPK/ERK and PI-3K pathways. Using specific inhibitors of each pathway, we found that the increase in expression of SREBP-1 induced by IGF-1 was blocked in the presence of the PI3-K inhibitor but not in the presence of the MAPK/ERK inhibitor. Furthermore inhibition of the PI3-K pathway also blocked the IGF-1 induced transcription of SREBP target genes and sebocyte lipogenesis. These data indicate that IGF-1 transmits its lipogenic signal in sebocytes through activation of Akt. Specific targeted interruption of this pathway in the sebaceous gland could be a desirable approach to reducing sebum production and improving acne.
INTRODUCTION
Sebum production is a critical factor in the pathogenesis of acne, though the molecular signals involved in sebum production are largely unknown. In order to design rational therapies for acne, it is essential to understand the signaling pathways that drive seborrhea in acne patients. Recent work has shown that insulin-like growth factor-1 (IGF-1) levels reach their peak in adolescents during the growth spurt and then decline, which coincides with the incidence of acne in many individuals (Deplewski and Rosenfield 1999). The same group has also shown that IGF-1 stimulates sebaceous gland lipogenesis (Deplewski and Rosenfield 2000). Lipogenesis is also stimulated by IGF-1 in sebaceous glands grown in organ culture (Downie et al, 2002).
Recent reports indicate a correlation between severity of acne in women and serum IGF-1 levels (Cappel et al, 2005). Furthermore, studies show that IGF-1 increases lipogenesis in the SEB-1 sebocyte model and this increase in lipogenesis is accompanied by an increase in expression of sterol response element binding protein-1 (SREBP-1) mRNA and protein (Smith et al, 2006). Dubbed, “master regulators of lipid homeostasis”, SREBPs have been shown to be regulated by androgens in the hamster ear sebaceous model (Rosignoli et al, 2003; Eberle et al, 2004). In other tissues and cell lines, there have been numerous reports demonstrating an increase in SREBP mRNA and protein in response to a variety of stimuli (Kotzka et al, 1998; Foretz et al, 1999; Chang et al, 2005). There is little doubt that an increase in SREBP protein increases lipogenesis, particularly in the liver (Goldstein et al, 2006). Here we sought to extend these findings to the sebaceous gland.
SREBPs bind sterol response elements (SREs), which are nucleotide sequences found in the promoter regions of several lipogenic genes in the cholesterol and fatty acid biosynthesis pathways. There are three members in the SREBP family: SREBP-1a, SREBP-1c, and SREBP-2. SREBP-1a and 1c are transcribed by alternative start sequences, and the transcripts are each spliced to form an identical protein from the second exon onward. The longer first exon found in SREBP-1a makes it the more potent activator of transcription, and more prone to activate transcription of genes typically controlled by SREBP-2. There is functional overlap between SREBP-1 and SREBP-2, but SREBP-1 typically controls genes in the fatty acid biosynthesis pathway, while SREBP-2 controls transcription of genes associated with cholesterol biosynthesis.(Shimano et al, 1997; Shimano et al, 1999).
The goal of the present study is to determine the molecular signaling pathways by which IGF-1 stimulation of SEB-1 sebocytes increases SREBP-1 mRNA and protein. It has been shown in other cell types that IGF-1 activates both the mitogen-activated protein kinase/c-Jun-N terminal kinase (MAPK/JNK) and the mitogen-activated protein-kinase/extracellular signal-regulated kinase (MAPK/ERK) cascades, in addition to the phosphoinositide 3-kinase (PI3-K) pathway. Furthermore, all three of these pathways have been implicated in activation of SREBP in at least one model system. It seems likely that the molecular pathways governing the SREBPs may be tissue/cell type specific. In this paper we report that IGF-1 activates the PI3-K and MAPK/ERK pathways in SEB-1 sebocytes, but not the MAPK/JNK or MAPK/p38 pathways. Moreover, pharmacological antagonism of the MAPK/ERK pathway has no effect upon the induction of SREBP-1 mRNA, protein, or lipogenesis by IGF-1. Most importantly, induction of the PI3-K pathway by IGF-1 mediates the increase in SREBP-1 mRNA, protein, and lipogenesis in SEB-1 sebocytes. Finally, inhibition of the PI3-K pathway with the inhibitor LY294002 completely blocks the increase in lipogenesis in SEB-1 cells seen in response to IGF-1. These data demonstrate that inhibition of PI3-K signaling in the sebaceous gland decreases lipogenesis and, as such, this approach may represent a novel strategy in the treatment of acne.
RESULTS
The MEK inhibitor, PD98059, blocks the activation of MAPK/ ERK signaling by IGF-1, yet has no effect on the induction of SREBP-1 by IGF-1
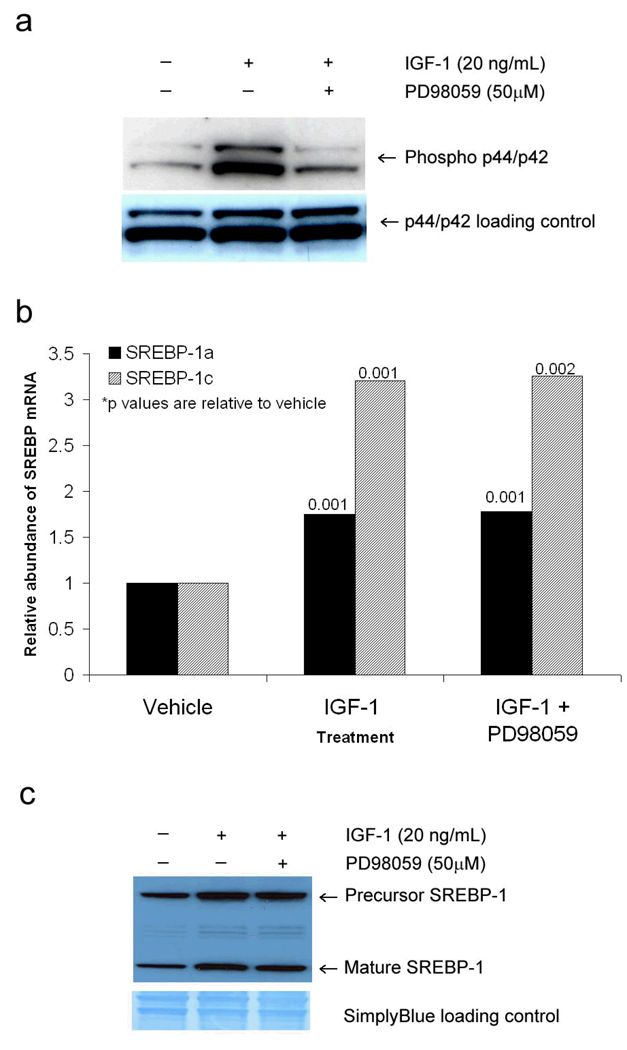
The MAPK/ERK pathway is activated when MEK phosphorylates p44 MAPK or p42 MAPK (ERK 1 and 2, respectively). To determine if IGF-1 mediates its effect on SREBP-1 expression in sebocytes through the MAPK/ERK pathway, SEB-1 cells were treated with and without IGF-1 (20 ng/ml) in the presence and absence of the MEK inhibitor PD98059 (50 µM) and then examined by Western blot for effects on downstream mediators. IGF-1 induced a robust increase in phosphorylated p44/p42 protein in SEB-1 sebocytes after 24 hours (Figure 1). Pretreatment of SEB-1 cells with 50 µM of the MEK inhibitor PD98059 for 30 minutes prior to the addition of IGF-1 blocked activation of this pathway (Figure 1a), indicating that IGF-1 activates the MAPK/ERK pathway in SEB-1 sebocytes. No cytotoxicity was observed with 50 µM PD98059 treatment as determined by cell counts performed at 24 hours (data not shown).
Figure 1. MAPK/ERK pathway is activated by IGF-1 but does affect SREBP-1 expression in SEB-1 cells.
a. Western blot reveals that p44/p42 (ERK1/2) is phosphorylated by IGF-1 in SEB-1 cells and that this stimulation can be blocked by 50 µM PD98059. The phospho p44/p42 blot was stripped and re-probed with an antibody that recognizes total p44/p42 regardless of phosphorylation status to serve as a loading control. Data indicate that IGF-1 activates the MAPK/ERK pathway and that this activation is blocked in the presence of a MEK inhibitor.
b. SEB-1 cells were treated with IGF-1 (20 ng/ml) and/or MEK inhibitor PD98059 (50 µM) and QPCR was performed to quantify SREBP-1a and —1c mRNA. Data are representative of 6 samples per treatment group as analyzed by the REST —XL program. These data indicate that inhibition of the MAPK/ERK pathway does not abrogate the induction of SREBP-1 mRNA by IGF-1.
c. Western blot was used to confirm that the MEK inhibitor PD98059 did not abrogate the induction of SREBP-1 protein by IGF-1 in SEB-1 cells. After being probed for SREBP-1, the membrane was stained with Simply Blue to ensure equal protein loading.
To determine if the MAPK/ERK pathway plays a role in the induction of SREBP-1 by IGF-1, we treated SEB-1 cells with IGF-1 (20ng/ml) and/or PD98059 (50 µM). Addition of the MEK inhibitor failed to block the induction of SREBP-1 mRNA (Figure 1b) or SREBP-1 protein (Figure 1c) by IGF-1, indicating that IGF-1 induced expression of SREBP-1 is not mediated by the MAPK/ERK pathway.
IGF-1 does not activate signaling through the p38 MAPK or the SAPK/JNK pathways in SEB-1 sebocytes at 24 hours
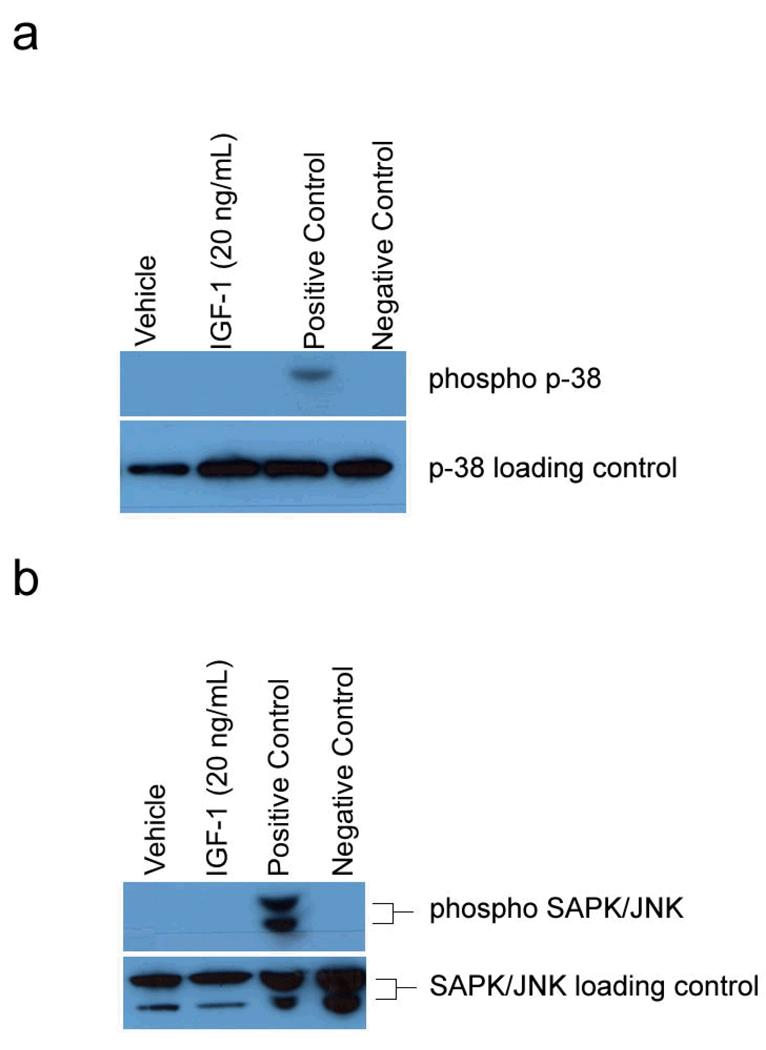
In addition to signaling in cell growth and differentiation, MAP kinases are also involved in signaling pathways involving inflammation and apoptosis. The p38 MAPK and the stress- activated protein kinase (SAPK)/ Jun N-terminal kinase (JNK) are involved in these alternative pathways. To determine if the p38 MAPK or the SAPK/JNK pathways transduce the signal by IGF to increase lipogenesis, Western blots were performed under the same experimental conditions to determine the phosphorylation status of p38 MAPK and SAPK/JNK in SEB-1 sebocytes stimulated with IGF-1. We did not observe activation of either p38 or SAPK/JNK in response to IGF-1 indicating that these pathways are not critical for IGF-1 induced lipogenesis in sebocytes (Figure 2).
Figure 2. IGF-1 does not activate p38 MAPK or SAPK/JNK pathways in SEB-1 cells.
a. A Western blot probing for phosphorylated MAPK/p38 reveals that SEB-1 sebocytes do not phosphorylate p38 in response to IGF-1. C-6 glioma cells treated with anisomycin serve as a positive control for phosphorylated p38, while unstimulated C-6 glioma cells represent the negative control. The phospho p38 blot was stripped and re-probed with an antibody that recognizes total p38 as a loading control.
b. A Western blot probing for phosphorylated SAPK/JNK reveals that SEB-1 sebocytes do not phosphorylate SAPK/JNK in response to IGF-1. Total cell extracts from 293 cells treated with UV light (Cell Signaling Technology) serve as a positive control for phosphorylated JNK, while untreated 293 extracts serve as the negative control. The phospho SAPK/JNK was stripped and re-probed with an antibody that recognizes total SAPK/JNK as a loading control.
The PI3-kinase inhibitor, LY294002, blocks the increase in transcription, translation, and processing of SREBP-1 that are induced by IGF-1
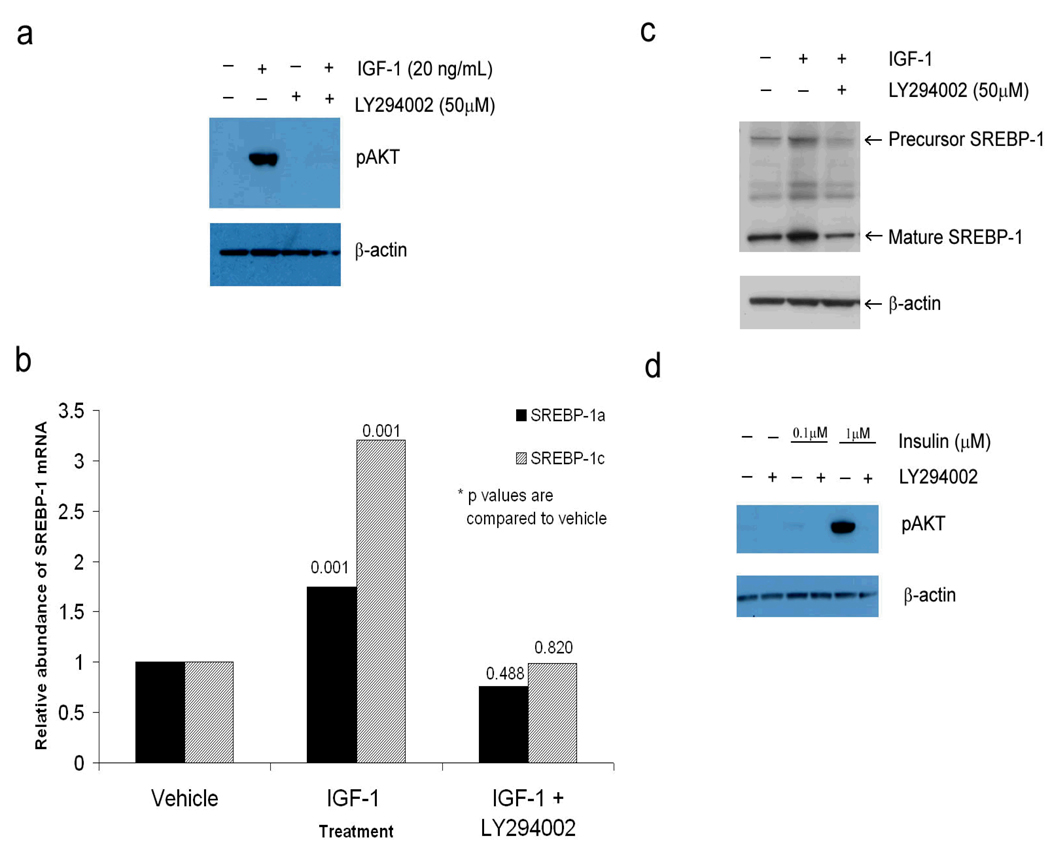
Akt is a critical member of the PI3-K pathway. As such, phosphorylation of Akt is a measure of activation of the PI3-K pathway. To determine if the PI3-K pathway tranduces the signal by IGF to increase lipogenesis, Western blots for pAkt and SREBP-1 were performed in the presence and absence of IGF-1 and the PI3-kinase inhibitor, LY294002. SEB-1 sebocytes were found to have very low amounts of phosphorylated Akt when maintained in medium without serum. However, the addition of IGF-1 (20 ng/ml) to this same medium induced a robust increase in phosphorylated Akt at 24 hours (Figure 3a). Importantly, we demonstrate that this induction of phosphorylated Akt was blocked quite potently by the addition of 50 µM of the PI3-K inhibitor, LY294002 (Figure 3a). No cytotoxicity was observed with 50 µM LY294002 treatment as assessed by viable cell counts after 24 hours of treatment (data not shown).
Figure 3. IGF-1 activates the PI3-K/Akt pathway, which mediates the increased expression of SREBP mRNA and protein in SEB-1 sebocytes.
a. Western blot reveals that Akt is phosphorylated by IGF-1 at 24 hours and that this induction is blocked when cells are treated with the PI3-K inhibitor LY294002 for 30 minutes prior to the addition of IGF-1.
b. SEB-1 cells were treated with 50 µM of the PI3-K inhibitor LY294002 and/or 20 ng/mL IGF-1 for 24 hours and QPCR was performed to look for changes in the mRNA for SREBP-1a and —1c. Data are representative of 6 samples per treatment group as analyzed by the REST—XL program. These data indicate that inhibition of the PI3-K pathway with LY294002 abrogates the IGF-1 induced increase in expression of mRNA for both SREBP-1a and –1c.
c. Western blot probing for SREBP-1 was performed in SEB-1 cells in the presence or absence of IGF-1 (20 ng/ml) or the PI3-K inhibitor LY294002. β-actin was used as the loading control. Data indicate that the precursor and mature forms of the SREBP-1 protein are not induced by IGF-1 in the presence of the PI-3K inhibitor, which supports the QPCR data above.
d. Previous studies indicate that 1µM insulin (which can act through the IGF-1 receptor) increases SREBP-1 expression whereas lower doses of insulin (100 nM) did not. Western blots were performed with 2 doses of insulin in the presence and absence of the PI3-K inhibitor to test the hypothesis that Akt is activated by high dose insulin, but not low dose insulin. Data indicate that 1µM insulin induces a robust increase in pAkt that is blocked in the presence of the PI3-K inhibitor. In contrast, negligible activation of Akt is noted with 100 nM insulin. These data suggest that although lipogenesis can be stimulated by activation of the insulin receptor or the IGF-1 receptor in SEB-1 sebocytes, different intracellular signaling pathways are involved.
We then treated SEB-1 cells with LY294002 and/or IGF-1 to determine if Akt activation was essential for the induction of SREBP-1 mRNA and protein by IGF-1 (20 ng/ml) observed in previous studies (Smith, Cong et al, 2006). We found that inhibition of the PI3-K/Akt pathway by LY294002 blocked the induction of SREBP-1 mRNA and protein by IGF-1 (Figures 3b and 3c).
In our previous study we noted that insulin (100 nM) induced a modest increase in lipogenesis in SEB-1 cells but it did not increase SREBP expression (Smith, Cong et al, 2006). We then sought to determine if there were differences in the level of Akt activation in SEB-1 based on the dose of insulin. We chose 100 nM that acts through the insulin receptor and 1µM, which is high enough to activate the IGF-1 receptor in addition to the insulin receptor. We found that 100 nM insulin, unlike 1 µM insulin, does not activate Akt, which may account for its lack of effect on SREBP expression (Figure 3d).
IGF-1 induces lipogenesis in SEB-1 sebocytes via the PI3-K pathway
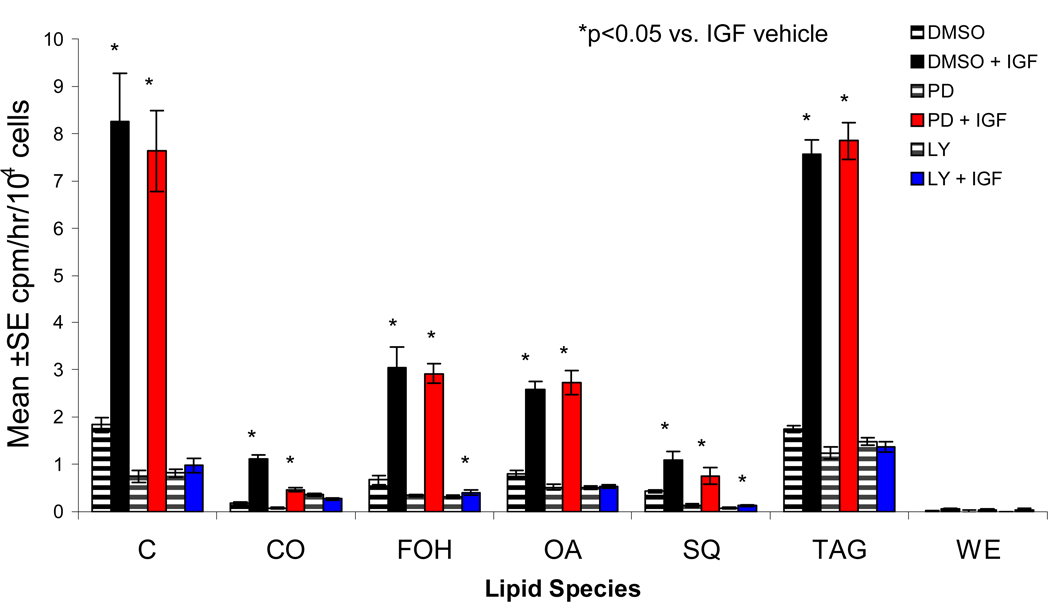
Since both the PI3-K and MAPK/ERK pathways are activated in SEB-1 cells by IGF-1, we next wanted to dissect the contribution of each of these signaling pathways to the increase in lipogenesis noted in response to IGF-1 treatment. We know that the MEK inhibitor PD98059 has no effect on SREBP activation by IGF-1. Keeping in line with our central hypothesis that SREBPs are important mediators of lipid metabolism in SEB-1 cells, we hypothesized that treatment of SEB-1 cells with a MAPK inhibitor would reduce lipogenesis to a much lesser degree than treatment with a PI3-K inhibitor. The 14C acetate incorporation assay was performed on SEB-1 cells treated with IGF-1 alone, or with the addition of PD98059 or LY294002 in conjunction with IGF-1. IGF-1 increased lipogenesis as we have shown previously (Smith, Cong et al, 2006). Additionally, treatment with MAPK inhibitor PD98059 had no effect on this induction of lipogenesis (Figure 4). However, when cells were treated with LY294002, IGF-1 failed to induce lipogenesis, as cells in this treatment group produce a lipid profile nearly identical to the group that received no IGF. From this we conclude that IGF-1 induces lipogenesis in SEB-1 cells via the PI3-K pathway (Figure 4).
Figure 4. IGF-1 induces lipogenesis in SEB-1 sebocytes through activation of the PI3-K pathway.
The 14C incorporation assay was performed on SEB-1 cells treated with IGF-1 (20 ng/ml) and/or 50 µM of a pharmacological inhibitor of PI3-K (LY294002) or MAPK/ERK (PD98059). IGF-1 induces a robust increase in lipogenesis (black striped bars vs. solid black bars). Interestingly, addition of 50 µM PD98059 to cells treated with IGF-1 has no effect on the IGF-1 induced lipogenesis (striped red bars vs. solid red bars). Most importantly, the addition of 50 µM PI3-K inhibitor LY294002 blocks any induction of lipogenesis when cells are stimulated with IGF-1 (blue striped bars vs. blue solid bars). All samples within an experiment were done in triplicate, and each experiment was performed at least three separate times. Data were analyzed by ANOVA and considered significant if a p-value of <0.05 was observed compared to the DMSO vehicle treated control group. Abbreviations used in this figure are: C=cholesterol; CO=cholesterol oleate; FOH=fatty alcohols; OA=oleic acid; SQ=squalene; TAG=triglycerides; WE=wax esters.
Analysis of transcriptional targets of SREBPs
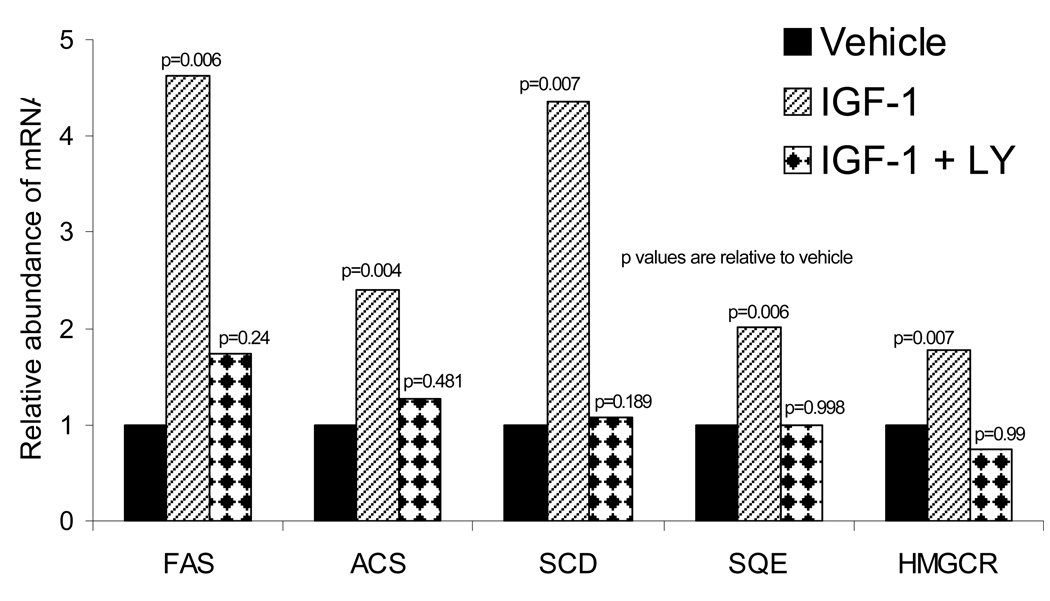
We next sought to determine if the increase in nuclear SREBP protein was accompanied by a corresponding increase in the mRNA levels of SREBP transcriptional targets. Following 14 hours of IGF-1 treatment (20 ng/mL), SEB-1 RNA was isolated and QPCR was performed to determine the relative amounts of fatty acid synthase (FAS), acyl-CoA synthetase (ACS), stearoyl-CoA desaturase (SCD), squalene epoxidase (SQE) and HMG-CoA reductase (HMGCR). Transcription of each of these genes has been shown to be activated by SREBP-1 in other models (Shimano, Horton et al, 1997; Ikeda et al, 2001; Sakakura et al, 2001; Sone et al, 2002; Chang, Wang et al, 2005; Murphy et al, 2006). Here we show that IGF-1 increases mRNA of each of these five genes, and further, this effect was attenuated in the presence of the PI3-K inhibitor LY294002. (Figure 5).
Figure 5. Inhibition of the PI3-K pathway in SEB-1 sebocytes blocked the induction of SREBP target genes by IGF-1.
SEB-1 cells were treated with IGF-1 (20ng/ml) in the presence or absence of 50 µM of the PI3-K inhibitor LY294002 and QPCR was performed. IGF-1 induces a robust increase in the expression of SREBP target genes that is blocked in the presence of the PI3-K inhibitor (which blocks SREBP induction). FAS, ACS, and SCD are traditional targets of SREBP-1, as they are involved in fatty acid biosynthesis. SQE and HMGCR are involved in cholesterol biosynthesis. Data are representative of 5 samples per treatment group as analyzed by the REST program. The following abbreviations are used: FAS=fatty acid synthase; ACS=Acyl-CoA synthetase; SCD=stearoyl-CoA desaturase; SQE=squalene epoxidase; HMGCR=HMG-CoA reductase.
DISCUSSION
It is well established in the literature that expression of SREBP-1 is increased in a variety of cell lines in response to a litany of growth factors (Nadeau et al, 2004). Previous work in the SEB-1 sebaceous cell model system has shown that IGF-1 increases both expression of SREBP-1 protein and lipogenesis (Smith, Cong et al, 2006). Recent reports indicate that SREBP-1 and other lipogenic factors are also expressed in sebaceous glands and the SZ95 sebocyte cell line. (Harrison et al, 2007). There are conflicting reports as to which molecular pathway(s) are involved in transmitting the growth factor signal that eventually leads to induction of the SREBPs in other cell systems (Nadeau, Leitner et al, 2004). In this study, we sought to determine which pathway(s) is critical for the induction of SREBP-1 protein by IGF-1 in SEB-1 sebocytes, and furthermore, to determine which pathway(s) is important for the increase in lipogenesis observed when SEB-1 cells are stimulated with IGF-1. An understanding of the signaling mechanisms that regulate the production of sebaceous lipids can lead to the identification of potential therapeutic target sites to reduce sebum production and improve acne. In this regard, we studied the role of the PI3-K pathway and the 3 major arms of the MAPK pathway (p38, SAPK/JNK, and p44/p42) in mediating the lipogenic signal induced by IGF-1.
There are cell-type specific pathways that mediate signals from growth factor receptors and that regulate the expression of SREBP and lipogenesis (Nadeau, Leitner et al, 2004). The p38 MAPK and the SAPK/JNK pathways are typically associated with cell stress and are variably activated by growth factors. In fact, activation of the IGF-1 receptor can actually downregulate both pathways . Conversely, in the H292 pulmonary epithelial cancer cell line, keratinocyte growth factor (KGF) activates the SAPK/JNK pathway to increase SREBP protein and lipogenesis (Chang, Wang et al, 2005). Our data indicate that neither the p38 MAPK nor the SAPK/JNK pathways are activated in SEB-1 cells in response to IGF-1 stimulation.
Numerous reports, particularly by groups using skeletal myoctyes, have found the SREBPs to be activated by the p42/p44 (ERK1/2) arm of the MAPK pathway in response to insulin (Kotzka et al, 2000; Nadeau, Leitner et al, 2004). When myocytes were treated with insulin in the presence of PD98059, an inhibitor of the p42/p44 MAPK pathway, the increase in expression of SREBP-1 was blocked. However, in adipocytes, - the converse was true wherein the induction of SREBP-1 by insulin was blocked in the presence of the PI3-K inhibitor, LY294002, but not in the presence of the MAPK/ERK inhibitor, PD98059. As expected, SEB-1 sebocytes stimulated with IGF-1 display a robust activation of the p44/p42 MAPK/ERK pathway (Figure 1) and also activation of the PI3-K pathway (Figure 3). As activation of SREBP proteins has been shown to increase transcription of several lipogenic genes (Rawson 2003), we hypothesized that one or both pathways are responsible for the increase in SREBP and ultimately, the increase in lipogenesis that we have observed previously with IGF treatment (Smith, Cong et al, 2006).
As far as SREBP activation is concerned, we found that inhibition of the p44/p42 MAPK pathway with PD98059 had no effect on SREBP-1a or SREBP-1c mRNA, the translation of SREBP-1 protein, or the processing of the protein into the mature form (Figures 1b & c). Here we establish that MAPK activation is not necessary for IGF-1 induced activation of SREBPs in SEB-1 sebocytes. Importantly, inhibition of the PI3-K pathway with LY294002 blocked transcription of SREBP-1a and 1c mRNA, translation of SREBP-1 protein, and also decreased the amount of the mature SREBP-1 protein found in the nucleus of IGF-1 stimulated SEB-1 cells (Figure 3). This clearly implicates the PI3-K/Akt pathway as the pathway by which IGF-1 mediates the increase in SREBP-1 activity in SEB-1 sebocytes. Additionally, earlier work done in our laboratory has shown that low dose insulin (100nM) induces a modest increase in lipogenesis but does not increase SREBP-1 expression unlike IGF-1 or high dose insulin (1 µM) which induce SREBP-1 expression and a robust increase in lipogenesis (Smith, Cong et al, 2006). Low dose insulin acts solely through the insulin receptor whereas IGF-1 and high doses of insulin stimulate both the insulin and the IGF-1 receptor. Here we provide a molecular explanation for the differences in the induction of lipogenesis by these hormones. In SEB-1 sebocytes, IGF-1 and high dose insulin activate the PI3-K pathway leading to an increase in SREBP expression and lipogenesis whereas 100 nM insulin does not activate the PI3-K pathway as evidenced by a lack of significant increase in phosphorylated Akt (Figure 3d). These data suggest that although lipogenesis can be stimulated by activation of the insulin receptor or the IGF-1 receptor in SEB-1 sebocytes, different intracellular signaling pathways are involved that mediate signals from these cell surface receptors.
Recent reports indicate that activation of Akt is involved in the transport of the SREBP cleavage activating protein (SCAP)/SREBP complex from the endoplasmic reticulum to the Golgi (Du et al, 2006). This is a major regulatory step in SREBP activity. Our data indicate that IGF-1 increases the amount of cleaved (mature) SREBP protein and that this increase in mature protein can be blocked in the presence of the PI3-K inhibitor (Figure 3c). This suggests that, in addition to possible transcriptional/translational control, Akt activation may also affect SREBP processing in SEB-1 sebocytes. Additional experiments are needed to test this hypothesis.
Akt activation has been shown to increase the expression of lipogenic genes (Porstmann et al, 2005). Microarray analysis of gene expression was compared between retinal pigment epithelial cells that express a constitutively activated Akt construct and wild type cells. Data indicated increased expression of numerous genes involved in sterol and lipid metabolism in the cells expressing activated Akt (Porstmann, Griffiths et al, 2005). Additional experiments revealed that these gene changes were mediated by an increase in the expression of SREBP-1a and SREBP-1 c (Porstmann, Griffiths et al, 2005). These data are supported by our observation that inhibition of Akt activation blocks the increase in expression of lipogenic genes induced by SREBP-1.
After establishing that activation of the PI3-K pathway by IGF-1 leads to an increase in mRNA for SREBP-1 and its target genes, we sought to determine the role of the PI3-K pathway in lipogenesis in SEB-1 sebocytes. Lipid production in sebocytes is a key predictive endpoint for drugs that may be of benefit in acne. Sebum, the lipid product of sebocytes, consists of primarily of cholesterol, triglycerides, and wax esters. Here we show that treatment with the PI3-K inhibitor LY294002 blocks the robust induction of lipogenesis noted in response to IGF- treatment for all lipids assayed. In contrast, although IGF-1 activated the MAPK/ERK pathway in SEB-1, inhibition of the later pathway had no effect on the IGF-1 induced lipogenesis in SEB1 cells (Figure 4). These data provide a rationale for investigation of members of the PI3-K pathway as drug targets to decrease sebum production and improve acne. Topical application of a specific PI3-K isoform would be desirable since the broad inhibition of PI3-K by an oral agent renders it undesirable as a systemic therapy.
The PI3-K enzyme consists of a p110 and a p85 subunit, each of which has several isoforms. The p110α isoform is activated in response to insulin and mediates the metabolic effects of insulin including glucose uptake (Knight et al, 2006). Whether this isoform mediates the same effects in sebocytes remains to be determined. Research is underway to develop small molecule drugs to target several p110 isoforms. As it has been recently learned that p110α is frequently mutated in human tumors, this class will come to the forefront of cancer research (Samuels et al, 2004). Progress in this area could be utilized to develop a topical inhibitor of PI3-K to reduce sebum production. While the pathogenesis of many diseases may be attributable to PI3-K signaling, acne may be easier to treat with small molecules inhibitors due to the accessibility of the sebaceous gland by topical application, particularly lipophilic molecules.
Building on our previous work that showed IGF-1 induces lipogenesis in SEB-1 sebocytes, here we have shown that 1) The PI3-K molecular signaling pathway transduces the IGF-1 signal, resulting in an increase in lipogenesis, 2) The PI3-K pathway is essential for IGF-1 induced lipogenesis, and 3) The increase in SREBP-1 nuclear protein is accompanied by an increase in the transcription of several lipogenic genes downstream of SREBP-1. As the p110α subunit of the PI3-K molecule has recently been shown to mediate the insulin signal, it is plausible that the small molecules designed to inhibit this isoform of the protein could be useful in reducing sebaceous gland lipogenesis and improving acne.
EXPERIMENTAL PROCEDURES
Cell culture and treatments
SEB-1 (passage 22–27) SV40 immortalized human sebocytes were grown to confluence in all experiments unless stated otherwise and were cultured in standard medium consisting of Dulbecco’s Modified Eagle’s Medium (DMEM) with 5.5 mM glucose/Ham’s F-12 3:1, fetal bovine serum 2.5%, adenine 1.8 × 10−4M, hydrocortisone 0.4µg/mL, insulin 10ng/mL, epidermal growth factor 3ng/mL, and cholera toxin 1.2×10−10M (Thiboutot et al, 2003).
For treatments with IGF-1, SEB-1 cells were plated at 7.6 × 105 in a 100mm dish, or 1.3 × 105 in a 35mm dish and grown for 6 days. On the sixth day, media was removed; cells were washed twice with PBS, and given DMEM containing 5.5mM glucose. IGF-1 was added 1:1000 in 0.1M acetic acid plus 0.1% BSA for 24 hours. MEK inhibitor PD98059 (50 µM) was used as a pharmacological inhibitor of the MAPK/ERK pathway, and cells were pretreated with the compound 30 minutes prior to stimulation with 20 ng/ml IGF-1. Likewise, the cell permeable PI3-K inhibitor, LY294002, at a dose of 50 µM was also administered 30 minutes prior to IGF-1 treatment. Both compounds were suspended in DMSO and the final concentration of DMSO in the media was 0.1%. Dose response experiments (10 µM, 20 µM and 50 µM) with each inhibitor were conducted to determine the optimal concentration that inhibited phosphorylation of Akt (LY294002) or ERK (PD98059). For each inhibitor, optimal inhibition was obtained with the 50 µM concentration as has been commonly reported in other cell systems.
Lipogenesis Assay
The incorporation of 14C-acetate into lipids was used as a measure of lipogenesis as described previously (Smith, Cong et al, 2006). Briefly, SEB-1 cells were grown to confluence in a 35mm dish and treated as described above. Cells were trypsinized and resuspended in a solution containing 1µCi 14C-acetate (New England Nuclear, Boston, MA) in DMEM, and incubated for 2 hours at 37°C with agitation. Following the incubation, lipids were extracted twice with ethyl ether and non-radioactive carrier lipids. The solvents were evaporated under a stream of nitrogen gas, and lipids were taken up in petroleum ether and separated by thin layer chromatography. Lipids spots were visualized, excised, and radioactivity in each spot was quantified in a liquid scintillation counter. All samples within an experiment were done in triplicate, and each experiment was performed at least three separate times. Data were analyzed by ANOVA and considered significant if a p-value of <0.05 was observed compared to the vehicle treated control group.
Western Blot
Protein was extracted using the Ne-Per kit (Pierce, Rockford, IL). Both the precursor and mature SREBP-1 protein was found in the nuclear lysate as described previously (Smith, Cong et al, 2006). Twenty-five micrograms of protein was run on a 4–12% Bis-Tris NuPage polyacrylamide gel (Invitrogen, Carlsbad, CA). Protein was then transferred to a polyvinylidene fluoride membrane (PVDF) and probed using standard methods. The SREBP-1 (K-10) antibody was purchased from Santa Cruz (Santa Cruz, CA). In samples that were probed for phospho-JNK, JNK, phospho-p38, p38, phospho-ERK, ERK, or phospho-Akt, cytoplasmic protein lysates were used and the antibody was purchased from Cell Signaling Technology (Boston, MA). Membranes were probed with an anti-actin antibody or stained with SimplyBlue (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions to ensure even protein loading.
C-6 glioma cells treated with anisomycin were used as a positive control for phosphorylated p38, while unstimulated C-6 glioma cells were the negative control. Total cell extracts from 293 cells treated with UV light served as a positive control for phosphorylated JNK, while normal 293 extracts served as the negative control. All control cell extracts were purchased from Cell Signaling Technology.
Quantitative RT-PCR
For QPCR, cells were grown in standard medium for 6 days. On the sixth day, cells were washed twice with phosphate-buffered saline and treated with 20 ng/mL IGF-1 and the appropriate inhibitor/vehicle in serum-free DMEM for 14 hours. Total RNA was isolated and complementary DNA was generated from 4.2 µg RNA/reaction primed with oligo-dT using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). QPCR was performed using the Brilliant SYBR Green QPCR Core Reagent Kit in an Mx4000 Multiplex Quantitative PCR System (Stratagene, La Jolla, CA). TATA-binding protein was used as a reference gene. The following primer sequences were used: TATA-binding protein upstream (GenBank number NM_003194) 5’c acg gca ctg att ttc agt tct, TATA-binding protein downstream 5’ttc ttg ctg cca gtc tgg act, SREBP-1a upstream (GenBank number NM_004176) 5’gct gct gac cga cat cga a, SREBP-1c upstream (GenBank number NM_001005291 5’gga gcc atg gat tgc act tt, and SREBP-1a, c downstream 5’tca aat agg cca ggg aag tca. Primer pairs for fatty acid synthase (product number Hs00188012_m1), acyl-CoA synthetase short-chain family member 2 (Hs002318766_m1), stearoyl-CoA desaturase (delta-9-desaturase) (Hs00748952_s1), squalene epoxidase (Hs00162288_m1), and HMG-CoA reductase (Hs00168352_m1) were purchased from ABI (Foster City, CA). Data were analyzed using the REST-XL© program (Pfaffl et al, 2002). A p-value <0.05 was considered significant.
Acknowledgement
This work was supported by NIH NIAMS R01AR047820 to D.M.T. and the Jake Gittlen Cancer Foundation. The authors thank Amanda Nelson, PhD for assistance with manuscript preparation.
Abbreviations
- IGF-1
Insulin-like growth factor-1
- SREBP
sterol response element binding protein
- MAPK
mitogen-activated protein kinase
- JNK
c-Jun-N terminal kinase
- ERK
extracellular signal-regulated kinase
- MEK
MAPK/ERK kinase
- SAPK
stress-activated protein kinase
- PI3-K
phosphoinositide 3-kinase
Footnotes
Conflict of Interest
The authors state no conflict of interest.
REFERENCES
- Cappel M, Mauger D, et al. Correlation between serum levels of insulin-like growth factor 1, dehydroepiandrosterone sulfate, and dihydrotestosterone and acne lesion counts in adult women. Arch Dermatol. 2005;141(3):333–338. doi: 10.1001/archderm.141.3.333. [DOI] [PubMed] [Google Scholar]
- Chang Y, Wang J, et al. KGF induces lipogenic genes through a PI3K and JNK/SREBP-1 pathway in H292 cells. J Lipid Res. 2005;46(12):2624–2635. doi: 10.1194/jlr.M500154-JLR200. [DOI] [PubMed] [Google Scholar]
- Deplewski D, Rosenfield RL. Growth hormone and insulin-like growth factors have different effects on sebaceous cell growth and differentiation. Endocrinology. 1999;140(9):4089–4094. doi: 10.1210/endo.140.9.6957. [DOI] [PubMed] [Google Scholar]
- Deplewski D, Rosenfield RL. Role of hormones in pilosebaceous unit development. Endocr Rev. 2000;21(4):363–392. doi: 10.1210/edrv.21.4.0404. [DOI] [PubMed] [Google Scholar]
- Downie MM, Sanders DA, et al. Modeling the remission of individual acne lesions in vitro. Br J Dermatol. 2002;147(5):869–878. doi: 10.1046/j.1365-2133.2002.04946.x. [DOI] [PubMed] [Google Scholar]
- Du X, Kristiana I, et al. Mol Biol Cell. 6. Vol. 17. 2006. Involvement of Akt in ER-to-Golgi transport of SCAP/SREBP: a link between a key cell proliferative pathway and membrane synthesis; pp. 2735–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle D, Hegarty B, et al. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86(11):839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Foretz M, Pacot C, et al. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol Cell Biol. 1999;19(5):3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, et al. Protein sensors for membrane sterols. Cell. 2006;124(1):35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Harrison W, Bull J, et al. Expression of lipogenic factors galectin-12, resistin, SREBP-1 and SCD in human sebaceous glands and cultured sebocytes. J Invest Dermatol online March. 2007;15 doi: 10.1038/sj.jid.5700743. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Yamamoto J, et al. Transcriptional regulation of the murine acetyl-CoA synthetase 1 gene through multiple clustered binding sites for sterol regulatory element-binding proteins and a single neighboring site for Sp1. J Biol Chem. 2001;276(36):34259–34269. doi: 10.1074/jbc.M103848200. [DOI] [PubMed] [Google Scholar]
- Knight ZA, Gonzalez B, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125(4):733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzka J, Muller-Wieland D, et al. ADD1/SREBP-1c mediates insulin-induced gene expression linked to the MAP kinase pathway. Biochem Biophys Res Commun. 1998;249(2):375–379. doi: 10.1006/bbrc.1998.9161. [DOI] [PubMed] [Google Scholar]
- Kotzka J, Muller-Wieland D, et al. Sterol regulatory element binding proteins (SREBP)-1a and SREBP-2 are linked to the MAP-kinase cascade. J Lipid Res. 2000;41(1):99–108. [PubMed] [Google Scholar]
- Murphy C, Ledmyr H, et al. Promoter analysis of the murine squalene epoxidase gene. Identification of a 205 bp homing region regulated by both SREBP'S and NF-Y. Biochim Biophys Acta. 2006;1761(10):1213–1227. doi: 10.1016/j.bbalip.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Nadeau KJ, Leitner JW, et al. Insulin regulation of sterol regulatory element-binding protein-1 expression in L-6 muscle cells and 3T3 L1 adipocytes. J Biol Chem. 2004;279(33):34380–34387. doi: 10.1074/jbc.M403596200. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, et al. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T, Griffiths B, et al. Oncogene. Vol. 24. 2005. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP; pp. 6465–6481. [DOI] [PubMed] [Google Scholar]
- Rawson RB. The SREBP pathway--insights from Insigs and insects. Nat Rev Mol Cell Biol. 2003;4(8):631–640. doi: 10.1038/nrm1174. [DOI] [PubMed] [Google Scholar]
- Rosignoli C, Nicolas JC, et al. Involvement of the SREBP pathway in the mode of action of androgens in sebaceous glands in vivo. Exp Dermatol. 2003;12(4):480–489. doi: 10.1034/j.1600-0625.2003.00014.x. [DOI] [PubMed] [Google Scholar]
- Sakakura Y, Shimano H, et al. Biochem Biophys Res Commun. 1. Vol. 286. 2001. Sterol regulatory element-binding proteins induce an entire pathway of cholesterol synthesis; pp. 176–183. [DOI] [PubMed] [Google Scholar]
- Samuels Y, Wang Z, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- Shimano H, Horton JD, et al. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99(5):846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Yahagi N, et al. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J Biol Chem. 1999;274(50):35832–35839. doi: 10.1074/jbc.274.50.35832. [DOI] [PubMed] [Google Scholar]
- Smith TM, Cong Z, et al. Insulin-like growth factor-1 induces lipid production in human SEB-1 sebocytes via sterol response element-binding protein-1. J Invest Dermatol. 2006;126(6):1226–1232. doi: 10.1038/sj.jid.5700278. [DOI] [PubMed] [Google Scholar]
- Sone H, Shimano H, et al. Acetyl-coenzyme A synthetase is a lipogenic enzyme controlled by SREBP-1 and energy status. Am J Physiol Endocrinol Metab. 2002;282(1):E222–E230. doi: 10.1152/ajpendo.00189.2001. [DOI] [PubMed] [Google Scholar]
- Thiboutot D, Jabara S, et al. Human skin is a steroidogenic tissue: steroidogenic enzymes and cofactors are expressed in epidermis, normal sebocytes, and an immortalized sebocyte cell line (SEB-1) J Invest Dermatol. 2003;120(6):905–914. doi: 10.1046/j.1523-1747.2003.12244.x. [DOI] [PubMed] [Google Scholar]