Abstract
BRAF and NRAS are commonly mutated in cancer and represent the most frequent genetic events in malignant melanoma. More recently, a subset of melanomas was shown to overexpress KIT and harbor KIT mutations. Although most gastrointestinal stromal tumors (GISTs) exhibit activating mutations in either KIT or PDGFRA, about 10% of the cases lack mutations in these genes. It is our hypothesis following the melanoma model that mutations in BRAF or NRAS may play a role in wild-type GIST pathogenesis. Alterations in RAS/MEK/ERK pathway may also be involved in development of imatinib resistance in GIST, particularly in tumors lacking secondary KIT or PDGFRA mutations. Imatinib-naive wild-type GISTs from 61 patients, including 15 children and 28 imatinib-resistant tumors without secondary KIT mutations were analyzed. Screening for hot spots mutations in BRAF (exons 11 and 15) and NRAS (exons 2 and 3) was performed. A BRAF exon 15 V600E was identified in 3 of 61 GIST patients, who shared similar clinical features, being 49- to 55-years-old females and having their tumors located in the small bowel. The tumors were strongly KIT immunoreactive and had a high risk of malignancy. An identical V600E BRAF mutation was also identified in one of 28 imatinib resistant GIST lacking a defined mechanism of drug resistance. In conclusion, we identified a primary BRAF V600E mutations in 7% of adult GIST patients, lacking KIT/PDGFRA mutations. The BRAF-mutated GISTs show predilection for small bowel location and high risk of malignancy. A secondary V600E BRAF mutation could represent an alternative mechanism of imatinib resistance. Kinase inhibitors targeting BRAF may be effective therapeutic options in this molecular GIST subset.
INTRODUCTION
Gastrointestinal stromal tumors (GIST) are the most common mesenchymal tumors of the gastrointestinal tract, located mostly in the stomach and small bowel. GISTs express KIT and are thought to arise from a KIT-positive interstitial cell of Cajal (ICC), the pacemaker cells of the GI tract. Although most GISTs show activating mutations in either KIT or PDGFRA, 10–15% of cases have a wild-type genotype for these two oncogenes (Hirota et al., 1998; Rubin et al., 2001; Heinrich et al., 2003a,b). Imatinib mesylate (STI571, Gleevec™, Novartis Pharmaceuticals, Basel, Switzerland), a selective tyrosine kinase inhibitor targeting KITand PDGFRA, is the frontline therapy for metastatic and unresectable GIST patients showing clinical responses in 80% of cases (Demetri et al., 2002). Despite a high rate of response in patients with KIT exon 11 mutated GISTs, the failure rate is significantly higher in patients with a wild type genotype, suggesting an alternative activated pathway not targeted by imatinib therapy (Heinrich et al., 2003a; Debiec-Rychter et al., 2004).
Activating KIT mutations, similar to those described in GISTs, have been reported recently in a subset of acral and mucosal malignant melanomas (Willmore-Payne et al., 2005; Antonescu et al., 2007). These mutations cluster within the juxtamembrane domain of KITand occur in melanomas overexpressing the KIT protein. In contrast, most cutaneous melanomas harbor mutations in either BRAF or NRAS (Cruz et al., 2003; Michaloglou et al., 2007). BRAF mutations are detected in more than half of melanoma cases and their incidence is dependent to UV light exposure, being most common in melanomas arising in skin intermittently exposed to the sun (Curtin et al., 2005). The vast majority of BRAF mutations in melanomas are V600E missense mutations identified in a dominant hot spot, within the kinase domain (Michaloglou et al., 2007). BRAF is a member of the RAF family of serine/threonine protein kinases that are important effectors of RAS activation and is involved in the RAS-RAF-ERK signaling pathway, which connects extracellular signals to transcriptional regulation. BRAF mutations are seen in 7% of all cancers. Apart from melanoma, BRAF mutations are also implicated in the pathogenesis of certain epithelial malignancies, such as papillary thyroid carcinoma, colorectal carcinoma, as well as in some benign/pre-neoplastic lesions, such as melanocytic nevi and serrated colonic polyps (Kebebew et al., 2007; Michaloglou et al., 2007; Minoo et al., 2007).
Our hypothesis is that activating BRAF and/or NRAS mutations may play a role in the pathogenesis of GISTs lacking an identifiable mechanism of KIT or PDGFRA activation. We further postulate that the acquisition of secondary BRAF or NRAS mutations may be involved in the mechanism of imatinib resistance in GIST.
MATERIALS AND METHODS
Patient Selection and Clinicopathologic Features
Patients with a diagnosis of GIST were identified from the Memorial Sloan-Kettering Cancer Center sarcoma database. Patient demographics, treatment data, and follow up information were obtained from chart review. The pathologic diagnosis was confirmed using standard H&E staining and immunoreactivity for CD117 (DAKO Corp., Carpinteria, California; 1:500) on formalin fixed paraffin embedded tissue. In case no. 4 other immunohistochemical studies were performed, including antibodies for PDGFRA (LabVision; 1:500), desmin (DAKO, 1:50) and myogenin (Ventana Medical Systems, Inc, Tucson, Arizona; prediluted). In addition, the protein expression status for p16 and PTEN was examined by immunohistochemistry using prediluted antibodies from Ventana Medical Systems, Inc, Tucson, Arizona. Pediatric and adult wild-type GISTs from imatinib-naïve patients were included in the study. Also selected for analysis were imatinib resistant GISTs lacking a defined mechanism of resistance, such as the presence of a secondary mutation in KIT/PDGFRA. The study was approved by the Institutional Review Board.
KIT/PDGFRA, BRAF, and NRAS Genotyping
Genomic DNA was isolated either from snap-frozen tumor tissue in 48 samples or from paraffin-embedded tissue in 41, as described previously (Antonescu et al., 2003). Adequate DNA for mutational analysis was obtained in all 89 samples which were then tested for the known sites of KIT (exons 9, 11, 13, 14, and 17) and PDGFRA (exons 12, 14, and 18) mutations. Primer sequences and annealing temperatures were as described (Antonescu et al., 2003, Agaram et al., 2007). Direct sequencing of PCR products was performed for all exons tested and each ABI sequence was compared to the NCBI human KIT and PDGFRA gene sequences.
BRAF exons 11 and 15 and NRAS exons 2 and 3 were screened for mutations by Denaturing High Pressure Liquid Chromatography (DHPLC) using the WAVE System (Transgenomic, Inc., Omaha, Nebraska). The elution temperature for each amplicon was calculated using the WAVEMAKER™ software. The PCR products of cases with DHPLC profiles indicating mutations were further confirmed by direct sequencing in half of the wild-type sequences as well as all mutated samples.
One case (case no. 4) was also subjected to FISH analysis for the presence of KIT or PDGFRA gene copy alterations, using paraffin sections from pre- and post-imatinib samples. The KIT probes used were two overlapping BAC clones: CTD-3180G20 and RP11-722F21 (Invitrogen), labeled by nick-translation with Spectrum Green (Vysis, Abbott Laboratories, Illinois). The PDGFRA probe used included 2 BAC clones that spanned about 290 kb around the gene: RP11-117E8 and RP11-231C18. A chromosome 4 centromeric probe labeled with Spectrum Orange (CEP 4, Vysis) was used as reference.
RESULTS
Eighty-nine samples from 87 patients were selected for the molecular analysis. From the untreated group, there were 61 wild-type GIST patients, including 15 children (≤18 years of age), 5 young adults (>18, ≤30 years), and 41 adults. The remaining 26 patients were selected from the imatinib resistance group which lacked secondary mutations in KIT or PDGFRA.
BRAFV600E Mutations are Present in a Subset of Small Bowel KIT/PDGFRA Wild-Type GISTs
Three (4%) of 61 KIT/PDGFRA wild-type GISTs showed a BRAF exon 15 V600E mutation, while none were detected in BRAF exon 11. In one of these three BRAF mutation positive patients, normal tissue was available for molecular analysis, which did not display the V600E, thus excluding the possibility of a germline mutation. The three patients with BRAF-mutated GISTs shared similar clinical findings, being middle-aged females (range of 49–55 years) with tumors located in the small bowel (Table 1). Microscopically, the three tumors had a predominantly spindle cell morphology and in two cases there was a small epithelioid component as well (Figs. 1A and 1B). In one case the tumor showed areas of sclerosis and dystrophic calcification (Fig. 1C). Tumors were diffusely immunoreactive for CD117 (Fig. 1D). There were no distinctive histological features that would distinguish this subset from the more commonly KIT-mutated tumors. The mitotic count ranged from 5 to 90 mitoses/50 high power fields (HPFs). These three patients were considered to have a high risk of malignancy GIST either based on tumor size (≥10 cm), high mitotic count (≥10 MF/50 HPFs) or both. One patient (Patient no. 1), who developed peritoneal metastasis, died of disease 18 months following diagnosis and the remaining two are with no evidence of disease (NED) at 9 and 13 months. BRAF mutations were not detected in any of the 15 pediatric and 5 young adult GISTs.
TABLE 1.
Clinical and Pathologic Findings in BRAF mutated GIST patients
| Age/Sex | Primary Tumor Size (cm) |
Primary Tumor Site |
MF/50 HPF |
Stage at presentation |
CD117 | PTEN | P16 | LFU/mo | |
|---|---|---|---|---|---|---|---|---|---|
| 1a | 52/F | 10 | SB | 90 | Periton Mets | P | P | N | DOD/18 |
| 2 | 55/F | 10 | SB | 5 | Primary | P | NA | NA | NED/9 |
| 3 | 49/F | 9 | SB | 50 | Primary | P | P | P | NED/13 |
| 4b | 66/M | 20 | Stomach | 68 | Primary | N | N | N | NED/32 |
F, female; M, male; cm, centimeters; SB, small bowel; periton mets, peritoneal metastases; MF, mitotic figures; HPF, high power fields; P, positive; N, negative; DOD, dead of disease; NED, no evidence of disease; AWD, alive with disease; NA, not available, LFU, last follow-up; mo, months.
Genotyping on normal tissue as well.
Imatinib resistant GIST.
Figure 1.
Pathologic findings in BRAF mutated GIST, showing a predominantly monomorphic spindle cell morphology (A: patient no.1, HE, ×200), with only focal epithelioid features and high mitotic activity (B: patient no. 1, HE ×400); one tumor showed focal areas of dystrophic calcification (C: patient 4, HE, ×100). Tumors were diffusely immunoreactive for CD117 (D: patient no. 2, ×200). Additional findings included: loss of P16 protein expression while PTEN expression was maintained (E, F: patient no. 1; ×200), as compared to patient no. 3 who had maintained P16 protein expression (G, ×200) and to patient no. 4 who lost PTEN (H, ×200).
BRAF Mutation is a Potential Alternative Mechanism of Imatinib Resistance in GIST
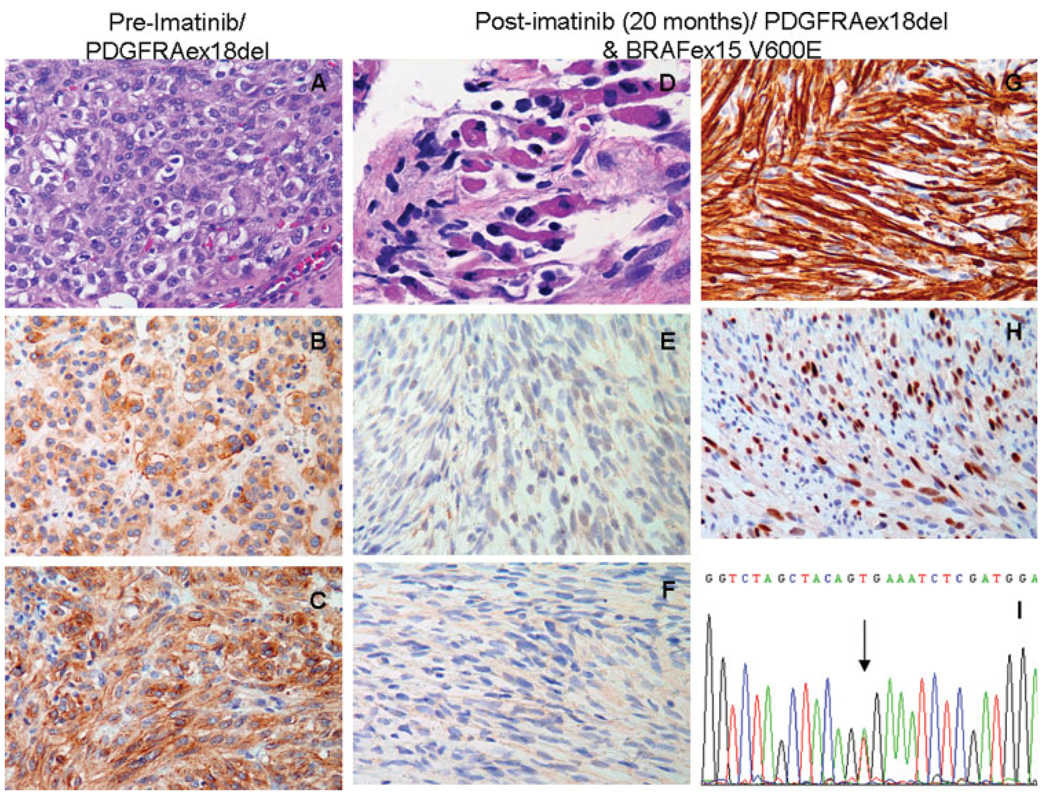
We identified 28 GIST tumors from 26 imatinib resistant patients, lacking an identifiable mechanism of treatment failure. All except one patient had tumors harboring a primary mutation in either KIT or PDGFRA and developed secondary resistance to imatinib after an initial response. The only patient with a wild-type genotype showed primary resistance to imatinib. A BRAF V600E mutation was identified in 1 of the 28 samples tested. This patient is a 66-years-old man with a primary high risk gastric GIST measuring 20 cm and a high mitotic count of 68 MF/HPFs. The tumor displayed a pure epithelioid morphology and showed diffuse staining for both KIT and PDGFRA by immunohistochemistry (Figs. 2A–2C). Mutation analysis of the primary tumor revealed a heterozygous PDGFRA exon 18 (4 amino acid deletion 842–845) mutation and absence of BRAF or NRAS mutations. Following resection of the primary tumor, he was treated with imatinib for 20 months for metastatic peritoneal disease. On resection of the peritoneal nodules, despite a good overall response with large areas of fibrosis, there were microscopic foci of viable and proliferating tumor cells, in keeping with a resistant phenotype. However, these foci did not resemble the pre-imatinib morphology and were composed of a spindle cell proliferation lacking both KIT and PDGFRA expression (Figs. 2D–2F). Moreover, these imatinib-resistant areas closely resembled an embryonal rhabdomyosarcoma phenotype, with spindle cells containing abundant eosinophilic cytoplasmic and cross-striations, which were diffusely positive for desmin and myogenin (Figs. 2G and 2H), markers of skeletal muscle differentiation. Mutational analysis from DNA obtained from micro-dissection of this area showed the presence of a BRAF V600E mutation in addition to the primary PDGFRA exon 18 deletion (Fig. 2I). No mutations were identified in BRAF exon 11 in any of the imatinib-resistant patients tested.
Figure 2.
Pre-imatinib pathologic findings revealed a high risk primary gastric GIST with epithelioid morphology (A, HE, ×200) and diffuse reactivity for CD117 (B, ×200) and PDGFRA (C, ×200). The post-imatinib resection for peritoneal metastasis showed foci of viable spindle cells with abundant eosinophilic cytoplasm and strap cells, resembling an embryonal rhabdomyosarcoma phenotype (D, HE, ×400), which was negative for CD117, PDGFRA, (E,F, ×200), but strongly positive for skeletal muscle markers (G, desmin, ×200; H, myogenin, ×200). The mutation analysis of this sample revealed in addition to a PDGFRA exon 18 deletion, a BRAF exon 15 V600E substitution (I, ABI sequence).
This case was also subjected to FISH analysis for the presence of KIT or PDGFRA gene copy alterations, using paraffin sections from pre- and post-imatinib samples. This analysis is particularly relevant since loss of protein expression for both KIT and PDGFRA was seen in the post-imatinib sample, suggesting a possible large deletion encompassing both genes. However, there were no abnormalities seen by FISH, with two copies of KIT and PDGFRA probes in both the pre and post-treatment samples tested (data not shown).
No frozen tissue was available from any of the BRAF mutation positive GISTs to assess the phosphorylation status of downstream targets, MEK and ERK by Western blotting.
NRAS Mutations do not Appear to be Involved in GIST Pathogenesis
In 56 untreated wild-type GIST patients (15 children, 5 young adult, and 36 adults) and 26 imatinib-resistant patients without secondary KIT or PDGFRA mutations adequate material was available for detecting mutations in NRAS exons 2 and 3. However no NRAS mutations were identified in any of these GIST subsets.
CDKN2A (p16) Alterations may be Associated with BRAF Mutations in GISTs
We analyzed the protein expression status of both p16 and PTEN as examined by immunohistochemistry in 3 of the 4 BRAF mutated GISTs with available material. In two of the three cases tested cases, there was lack of P16 staining, while PTEN expression was retained (Figs. 1E–1H).
DISCUSSION
Melanoma is the prototypical example in which BRAF mutations have been detected in ~50% of the patients and NRAS in about 15%, with the two mutations typically being mutually exclusive (Brose et al., 2002; Gorden et al., 2003). More recently, activating KIT mutations were identified in a small subset of melanomas lacking BRAF mutations (Curtin et al., 2006). As the progenitor cells for both melanoma and GIST, i.e., the melanocyte and the ICC, are KIT-expressing and KIT-dependent during embryologic development, it is possible that GIST pathogenesis may follow similar oncogenic signaling pathways observed in melanomas. Thus our working hypothesis was that alterations in the RAS-RAF-MEK signaling cascade may occur in GIST lacking identifiable mutations in KIT and PDGFRA. Furthermore, half of the imatinib-resistant GIST patients lack an identifiable mechanism of resistance, such as a secondary KIT mutation or KIT gene amplification (Antonescu et al., 2005; Debiec-Rychter et al., 2005; Heinrich et al., 2006). In this subset the acquisition of mutations in other signaling pathways, which are not targeted by imatinib, may occur. Thus our second hypothesis was that BRAF or NRAS mutations, commonly involved in other cancer types, may trigger an alternate mechanism of imatinib resistance in GIST patients.
To our knowledge this is the first report identifying BRAF mutations in either imatinib-naïve or imatinib-resistant GISTs. Although detected only in a minority of KIT/PDGFRA wild-type GISTs, BRAF mutations appear to be associated with a distinct clinicopathologic phenotype, occurring in middle-aged females and being preferentially located in the small bowel. Furthermore, the three tumors showed pathologic features consistent with a high risk of malignancy, as defined either by large tumor size or high mitotic count or both. KIT protein was consistently overexpressed in these cases and there were no other distinctive features microscopically that would differentiate them from KIT mutated GISTs. The mechanism through which BRAF activating mutations in GISTs may affect KIT signaling remains unclear. Similar to other tumor types where BRAF mutations are more commonly observed, the mutation in GIST was present in the exon 15 V600E hot spot. No other mutations were identified in the BRAF exon 11 or NRAS exons 2 and 3.
BRAF is a member of the RAF family of serine/threonine protein kinases that are important effectors of RAS activation and is involved in the RAS-RAF-ERK signaling pathway, which connects extracellular signals to transcriptional regulation. Activated RAF proteins phosphorylate MEK1 and 2 (MAPKKs), which in turn phosphorylate ERK1 and 2 (MAPKs), leading to the activation of several cytoplasmic and nuclear targets, including transcription factors such as ETS11, c-JUN, and c-MYC. Thus the MEK-ERK effector pathway is commonly dysregulated in cancer, often through gain-of-function mutations of either RAS or RAF family members (Davies et al., 2002). The RAS/MAP kinase pathway is activated by many receptor tyrosine kinases, including KIT. The role of oncogenic KIT signaling on ERK1/2 activation has been recently evaluated by our group in a KITV558Δ/+ GIST mouse model, produced by a knock-in strategy introducing a KIT exon 11-activating mutation into the mouse genome (Rossi et al., 2006). Although untreated tumors show consistent activation of ERK1/2 phosphorylation, there is no effect of imatinib treatment on ERK activation (Rossi et al., 2006). Thus BRAF mutated GIST patients may be similarly resistant to imatinib inhibition.
BRAF mutations are known to occur in melanocytic nevi and other benign or precursor lesions like colonic serrated polyps. This observation raises the possibility that BRAF mutations are an early event in the tumor development and subsequent gene alterations may be required for tumor progression to a malignant phenotype. Additional alterations in tumor suppressor genes, such as CDKN2A or PTEN have been shown to play a synergistic role along with the existing BRAF mutations, necessary for tumor development and/or progression (Daniotti et al., 2004; Curtin et al., 2005; Michaloglou et al., 2007). Melanocytic nevi typically show increased levels of CDKN2A, whereas most melanomas lose P16 expression (Curtin et al., 2005). CDKN2A encodes a 16 kDa nuclear protein (P16INK4A or P16) that specifically binds to the cyclin-dependent kinases 4 (CDK4) and 6 (CDK6), which are involved in the G1 checkpoint of the mammalian cell cycle. The preserved P16 function in melanocytic nevi maintains a stable cell cycle arrest even in the presence of a BRAF mutation, while loss of P16 leads to development of melanoma. PTEN mutations have also been implicated along with BRAF mutations in a subset of melanomas. PTEN encodes for a lipid and protein phosphatase and regulates intracellular levels of the lipid phosphatidylinositol phosphate (PIP3), involved in the control of apoptosis via AKT (Bonneau and Longy, 2000). In our study, loss of P16 expression by immunohistochemistry was noted in two of the BRAF mutated GISTs. PTEN expression was maintained in two of the three cases tested.
The most frequent mechanism of imatinib-resistance in GIST is the acquisition of secondary mutations either in KIT or PDGFRA which are detected in half of the patients. These 2nd site mutations occur after a longer period of imatinib therapy (median 27 months), compared with resistant tumors without secondary mutations (median 14.5 months) and are typically seen in patients with KIT exon 11 mutated GISTs (Antonescu et al., 2005). Our patient with a high risk gastric GIST showed expression of both KIT and PDGFRA proteins in the primary tumor and direct sequencing demonstrated a primary PDGFRA exon 18 deletion in the absence of BRAF or NRAS mutations. The imatinib resistant tumor was resected after 20 months of imatinib therapy and showed not only loss of KIT and PDGFRA protein expression, but trans-differentiation into a rhabdomyosarcoma phenotype. These foci showed a high proliferation rate and diffuse expression for skeletal muscle markers, such as desmin and myogenin. A similar phenotypic switch or dedifferentiation phenomenon, with loss of KIT expression and acquisition of aberrant lines of differentiation has been previously reported in rare cases of imatinib-resistant patients, but thus far was not attributed to a particular genotype (Pauwels et al., 2005).
The detection of BRAF mutations in GIST patients is highly relevant clinically. First, it identifies a subset of KIT/PDGFRA wild type GIST patients who are less sensitive to imatinib and could benefit from the inhibition of alternative pathways. Second, it has been recently shown that mutation in BRAF is associated with enhanced and selective sensitivity to MEK inhibition when compared to “wild-type” cell or cells harboring RAS mutations. This MEK dependency was noted in BRAF mutant cells regardless of tissue lineage (Solit et al., 2006). Thus newer compounds targeting BRAF mutations and the RAS-RAF-MEK pathway now under study in clinical trials in melanoma or other tumor types, may show efficacy in BRAF-mutated GIST patients. Sorafenib, an inhibitor of RAF, VEGFR and PDGFR has been studied in melanoma and thyroid cell lines and has shown an inhibitory effect (Haluska et al., 2007). In in vitro studies modeling imatinib resistant GIST patients carrying the gatekeeper KIT T670I secondary mutation, sorafenib showed the highest potency in inhibiting cell proliferation and inducing apoptosis compared to other kinase inhibitors (Guo et al., 2007). Similar results have been obtained using the MEK inhibitor CI-1040 (Solit et al., 2006). Heat shock protein 90 (HSP90) inhibitors (geldanamycin or 17AAG) have also shown to be effective in BRAF mutants based on in vitro studies (Sharp and Workman, 2006).
In summary, this is the first report detecting BRAF mutations in imatinib-naive or imatinib-resistant GISTs. They occur in a small subset of intestinal high risk GISTs lacking KIT/PDGFRA mutations. This finding delineates a new molecular group of patients who may benefit from selective BRAF inhibitors, as an alternative therapeutic option to imatinib. Furthermore, the acquisition of BRAF mutations may play a role in triggering imatinib- resistance in GIST patients and may induce a KIT-negative trans-differentiation phenotype in these tumors. Since our observations were made on a limited number of cases, additional cases are thus required to confirm that BRAF mutated GIST indeed show a distinct clinicopathologic phenotype.
Acknowledgments
Supported by: ACS MRSG CCE-106841, P01CA47179, Life Raft Group, GIST Cancer Research Fund, Shuman Family Fund for GIST Research, CA102613, CA102774, HL/DK55748.
REFERENCES
- Agaram NP, Besmer P, Wong GC, Guo T, Socci ND, Maki RG, DeSantis D, Brennan MF, Singer S, DeMatteo RP, Antonescu CR. Pathologic and molecular heterogeneity in imatinib-stable or imatinib-responsive gastrointestinal stromal tumors. Clin Cancer Res. 2007;13:170–181. doi: 10.1158/1078-0432.CCR-06-1508. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Sommer G, Sarran L, Tschernyavsky SJ, Riedel E, Woodruff JM, Robson M, Maki R, Brennan MF, Ladanyi M, DeMatteo RP, Besmer P. Association of KIT exon 9 mutations with nongastric primary site and aggressive behavior: KIT mutation analysis and clinical correlates of 120 gastrointestinal stromal tumors. Clin Cancer Res. 2003;9:3329–3337. [PubMed] [Google Scholar]
- Antonescu CR, Besmer P, Guo T, Arkun K, Hom G, Koryotowski B, Leversha MA, Jeffrey PD, Desantis D, Singer S, Brennan MF, Maki RG, DeMatteo RP. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res. 2005;11:4182–41190. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Busam KJ, Francone TD, Wong GC, Guo T, Agaram NP, Besmer P, Jungbluth A, Gimbel M, Chen CT, Veach D, Clarkson BD, Paty PB, Weiser MR. L576P KIT mutation in anal melanomas correlates with KIT protein expression and is sensitive to specific kinase inhibition. Int J Cancer. 2007;121:257–264. doi: 10.1002/ijc.22681. [DOI] [PubMed] [Google Scholar]
- Bonneau D, Longy M. Mutations of the human PTEN gene. Hum Mutat. 2000;16:109–122. doi: 10.1002/1098-1004(200008)16:2<109::AID-HUMU3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, Roth JA, Albelda SM, Davies H, Cox C, Brignell G, Stephens P, Futreal PA, Wooster R, Stratton MR, Weber BL. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- Cruz F, III, Rubin BP, Wilson D, Town A, Schroeder A, Haley A, Bainbridge T, Heinrich MC, Corless CL. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res. 2003;63:5761–5766. [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, LeBoit PE, Pinkel D, Bastian BC. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- Daniotti M, Oggionni M, Ranzani T, Vallacchi V, Campi V, Di Stasi D, Torre GD, Perrone F, Luoni C, Suardi S, Frattini M, Pilotti S, Anichini A, Tragni G, Parmiani G, Pierotti MA, Rodolfo M. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene. 2004;23:5968–5977. doi: 10.1038/sj.onc.1207780. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Debiec-Rychter M, Dumez H, Judson I, Wasag B, Verweij J, Brown M, Dimitrijevic S, Sciot R, Stul M, Vranck H, Scurr M, Hagemeijer A, van Glabbeke M, van Oosterom AT. Use of c-KIT/PDGFRA mutational analysis to predict the clinical response to imatinib in patients with advanced gastrointestinal stromal tumours entered on phase I and II studies of the EORTC Soft Tissue and Bone Sarcoma Group. Eur J Cancer. 2004;40:689–695. doi: 10.1016/j.ejca.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, Fletcher JA, Silverman SG, Silberman SL, Capdeville R, Kiese B, Peng B, Dimitrijevic S, Druker BJ, Corless C, Fletcher CD, Joensuu H. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- Gorden A, Osman I, Gai W, He D, Huang W, Davidson A, Houghton AN, Busam K, Polsky D. Analysis of BRAF and N-RAS mutations in metastatic melanoma tissues. Cancer Res. 2003;63:3955–3957. [PubMed] [Google Scholar]
- Guo T, Agaram NP, Wong GC, Hom G, D’Adamo D, Maki RG, Schwartz GK, Veach D, Clarkson BD, Singer S, DeMatteo RP, Besmer P, Antonescu CR. Sorafenib inhibits the imatinib-resistant KITT670I gatekeeper mutation in gastrointestinal stromal tumor. Clin Cancer Res. 2007;13:4874–4881. doi: 10.1158/1078-0432.CCR-07-0484. [DOI] [PubMed] [Google Scholar]
- Haluska F, Pemberton T, Ibrahim N, Kalinsky K. The RTK/RAS/BRAF/PI3K pathways in melanoma: Biology, small molecule inhibitors, and potential applications. Semin Oncol. 2007;34:546–554. doi: 10.1053/j.seminoncol.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ, Kiese B, Eisenberg B, Roberts PJ, Singer S, Fletcher CD, Silberman S, Dimitrijevic S, Fletcher JA. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003a;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, Demetri GD, Fletcher CD, Fletcher JA. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003b;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Blanke CD, Demetri GD, Joensuu H, Roberts PJ, Eisenberg BL, von Mehren M, Fletcher CD, Sandau K, McDougall K, Ou WB, Chen CJ, Fletcher JA. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006;24:4764–4774. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Muhammad Tunio G, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- Kebebew E, Weng J, Bauer J, Ranvier G, Clark OH, Duh QY, Shibru D, Bastian B, Griffin A. The prevalence and prognostic value of BRAF mutation in thyroid cancer. Ann Surg. 2007;246:466–470. doi: 10.1097/SLA.0b013e318148563d. discussion 470–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Mooi WJ, Peeper DS. BRAF(E600) in benign and malignant human tumours. Oncogene. 2008;27:877–895. doi: 10.1038/sj.onc.1210704. [DOI] [PubMed] [Google Scholar]
- Minoo P, Moyer MP, Jass JR. Role of BRAF-V600E in the serrated pathway of colorectal tumourigenesis. J Pathol. 2007;212:124–133. doi: 10.1002/path.2160. [DOI] [PubMed] [Google Scholar]
- Pauwels P, Debiec-Rychter M, Stul M, De Wever I, Van Oosterom AT, Sciot R. Changing phenotype of gastrointestinal stromal tumours under imatinib mesylate treatment: a potential diagnostic pitfall. Histopathology. 2005;47:41–47. doi: 10.1111/j.1365-2559.2005.02179.x. [DOI] [PubMed] [Google Scholar]
- Rossi F, Ehlers I, Agosti V, Socci ND, Viale A, Sommer G, Yozgat Y, Manova K, Antonescu CR, Besmer P. Oncogenic Kit signaling and therapeutic intervention in a mouse model of gastrointestinal stromal tumor. Proc Natl Acad Sci USA. 2006;103:12843–12848. doi: 10.1073/pnas.0511076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R, Hibbard MK, Chen CJ, Xiao S, Tuveson DA, Demetri GD, Fletcher CD, Fletcher JA. KITactivation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res. 2001;61:8118–8121. [PubMed] [Google Scholar]
- Sharp S, Workman P. Inhibitors of the HSP90 molecular chaperone: Current status. Adv Cancer Res. 2006;95:323–348. doi: 10.1016/S0065-230X(06)95009-X. [DOI] [PubMed] [Google Scholar]
- Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, Golub TR, Sebolt-Leopold J, Sellers WR, Rosen N. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmore-Payne C, Holden JA, Tripp S, Layfield LJ. Human malignant melanoma: Detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum Pathol. 2005;36:486–493. doi: 10.1016/j.humpath.2005.03.015. [DOI] [PubMed] [Google Scholar]