Abstract
In Arabidopsis, pericentromeric repeats, retroelements, and silenced rRNA genes are assembled into heterochromatin within nuclear structures known as chromocenters. The mechanisms governing higher-order heterochromatin organization are poorly understood but 24-nt small interfering RNAs (siRNAs) are known to play key roles in heterochromatin formation. Nuclear RNA polymerase IV (Pol IV), RNA-DEPENDENT RNA POLYMERASE 2 (RDR2), and DICER-LIKE 3 (DCL3) are required for biogenesis of 24-nt siRNAs that associate with ARGONAUTE 4 (AGO4). Nuclear RNA polymerase V (Pol V) collaborates with DRD1 (DEFICIENT IN RNA-DEPENDENT DNA METHYLATION 1) to generate transcripts at heterochromatic loci that are hypothesized to bind to siRNA-AGO4 complexes and subsequently recruit the de-novo DNA methylation and/or histone modifying machinery. Here, we report that decondensation of the major pericentromeric repeats and depletion of the heterochromatic mark histone H3 lysine 9 dimethylation at chromocenters occurs specifically in pol V and drd1 mutants. Disruption of pericentromeric repeats condensation is coincident with transcriptional reactivation of specific classes of pericentromeric 180-bp repeats. We further demonstrate that Pol V functions independently of Pol IV, RDR2, and DCL3-mediated siRNA production to affect interphase heterochromatin organization, possibly by involving RNAs that recruit structural or chromatin-modifying proteins.
Keywords: RNA polymerase V, RNA-directed DNA methylation, heterochromatin, centromere, gene expression
INTRODUCTION
During interphase, the genome is differentially organized into regions of decondensed euchromatin, rich in actively transcribed genes, and highly condensed heterochromatin that is less frequently transcribed (Grewal and Moazed, 2003). So-called constitutive heterochromatin remains condensed throughout the cell cycle (Heitz, 1928) and coalesces into higher-order structures, known as chromocenters (Fransz et al., 2002; Soppe et al., 2002). Constitutive heterochromatin is rich in transposons, retrotransposons, and other repetitive DNA elements that are maintained in a transcriptionally repressed state by DNA hypermethylation and/or repressive histone post-translational modifications (Attwood et al., 2002; Bender, 2004; Vongs et al., 1993). The mechanisms by which heterochromatin clusters into chromocenters is mostly unknown; however, mutations that disrupt DNA methylation and Histone H3 lysine 9 dimethylation (H3K9me2) in Arabidopsis thaliana interfere with heterochromatin content and chromocenter formation (Soppe et al., 2002).
Transposons and repetitive elements that comprise constitutive heterochromatin tend to be enriched near centromeres, the highly specialized chromosome domains that are essential for chromosome segregation during mitosis and meiosis. In A. thaliana, centromeric regions include related, but distinct classes of ∼177–180-bp repeats that are tandemly arranged in large clusters (Heslop-Harrison et al., 1999; Round et al., 1997) ranging from 1.4 to 3.3 Mb in length (Haupt et al., 2001). Additional copies, but not long arrays, of ∼180-bp repeats are also found elsewhere in the genome (Lin et al., 1999; Round et al., 1997; The Arabidopsis Genome Initiative, 2000). Interspersed among the centromeric and pericentromeric ∼180-bp repeats are diverse classes of transposable elements and transposon-derived repeats (Hall et al., 2003b; Martinez-Zapater et al., 1986; Thompson et al., 1996).
Although centromeres evolve rapidly and display a remarkable lack of sequence conservation (Henikoff, 2002), a characteristic of their central core is the presence of centromeric Histone H3 (CENH3), known as CENP-A in mammals, centromere identifier (CID) in Drosophila melanogaster, and HTR12 in Arabidopsis (Talbert et al., 2002). These centromere-specific histone H3 variants help mediate the assembly of the kinetochore (Choo, 1997; Guenatri et al., 2004; Sullivan and Karpen, 2004). However, epigenetic modifications of pericentromeric repeats beyond the centromere core also contribute to centromere functions, including sister chromatid cohesion (Bernard et al., 2001; Choo, 2001; Nasmyth et al., 2000).
Until recently, centromeric regions were not thought to be actively transcribed. However, genes are present within centromeric regions (Appelgren et al., 2003; Kanellopoulou et al., 2005) and transcripts that traverse pericentromeric repeats contribute to heterochromatin formation in these regions (Gaubatz and Cutler, 1990; Nakano et al., 2003; Topp et al., 2004). As was first shown in Schizosacharomyces pombe, pericentromeric transcripts play at least two roles, serving as precursors for short interfering RNA (siRNA) production and serving as the targets for siRNA action via the RNA interference (RNAi) pathway (Hall et al., 2003a; Fukagawa et al., 2004; Volpe et al., 2002). In this pathway, dsRNAs generated by bidirectional transcription, or by RNA-dependent RNA polymerases acting on single-stranded primary transcripts, are cleaved by one or more Dicer endonucleases to produce siRNAs (Hannon, 2002). The siRNAs are then incorporated into an Argonaute protein that comprises the core of an RNA-induced silencing complex (RISC) knows as the RITS (RNA induced transcriptional silencing) complex (Noma et al., 2004). The siRNAs then guide the RITS complex to homologous loci by base-pairing with nascent RNA transcripts, promoting the recruitment of histone methyltransferases and deacetylases that bring about heterochromatin formation and transcriptional silencing of a pericentromeric repeats subset (Noma et al., 2004; Verdel et al., 2004; Volpe et al., 2003). Consequently, mutations in the RNAi pathway disrupt heterochromatin formation and gene silencing within the pericentromeric regions. Moreover, the association with centromeres of the Rad21/Scc1 regulatory subunit of cohesin is lost in RNAi mutants, causing defects in chromosome cohesion and aberrant chromosome segregation. However, CENH3 association at the centromere core is not disrupted in RNAi-deficient mutants (Fukagawa et al., 2004; Hall et al., 2003a; Kanellopoulou et al., 2005; Volpe et al., 2003).
Like fission yeast, A. thaliana makes use of siRNAs to direct DNA methylation and silencing of retroelements, transposons, and other repeats that are enriched in the pericentromeric regions (Zaratiegui et al., 2007). This RNA-directed DNA methylation (RdDM) process involves two plant-specific nuclear RNA polymerases, Pol IV and Pol V (formerly Pol IVa and Pol IVb, respectively) (Herr et al., 2005; Kanno et al., 2005; Onodera et al., 2005), that are structurally distinct and functionally non-redundant. Pol IV collaborates with RDR2 (RNA DEPENDENT RNA POLYMERASE 2) to generate double-stranded RNAs that are processed by DCL3 (DICER-LIKE 3) into 24-nt siRNAs (Xie et al., 2004; Chan et al., 2004) that bind to AGO4 (ARGONAUTE 4). Analogous to the fission yeast RITS complex, AGO4–RISC is thought to target heterochromatic modifications at loci complementary to 24-nt siRNAs (Qi et al., 2006; Zilberman et al., 2004). In a parallel process, Pol V generates transcripts at target loci (Wierzbicki et al., 2008) with the help of DRD1, a SWI2/SNF2-like chromatin remodeler (Kanno et al., 2004). Recent evidence suggests that siRNA–AGO4 complexes bind to Pol V transcripts (Wierzbicki et al., 2008), ultimately guiding the de novo DNA methyltransferase DRM2 (Chan et al., 2004) and histone modifying complexes to the target loci.
In this study, we tested components of the A. thaliana siRNA-directed DNA methylation pathway for effects on heterochromatin organization in chromocenters. Our study reveals that loss of Pol V or DRD1 function disrupts heterochromatin organization during interphase, which correlates with the transcriptional derepression of specific pericentromeric repeats. By contrast, loss of Pol IV, RDR2, or DCL3 functions, which are all required for 24-nt siRNA biogenesis, does not cause major disruptions in chromocenter organization. Our data suggest that Pol V contributes to heterochromatin formation and pericentromeric repeat silencing via a previously unrecognized ability to function outside of the currently defined siRNA-dependent DNA methylation pathway.
RESULTS
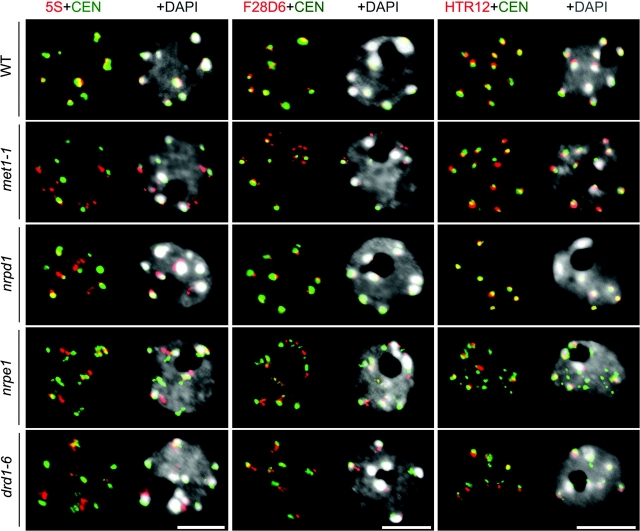
Nuclear Localization of Pericentromeric Repeats Is Disrupted in pol V and drd1 Mutants
We demonstrated previously that the gene encoding the second largest subunit of both Pol IV and Pol V, NRPD2/NRPE2, is necessary for the compaction and association of pericentromeric 5S rRNA genes within chromocenters (Onodera et al., 2005). However, it has been unclear whether Pol IV, Pol V or both enzymes are needed for chromatin condensation or whether other proteins of the siRNA pathway are also involved. To address these questions, we compared wild-type and mutant plants for the co-localization of heterochromatic repeats within chromocenters using DNA fluorescence in-situ hybridization (DNA-FISH). In wild-type nuclei, 180-bp centromere repeats (Figure 1, green signals) partially co-localize with 5S genes, with abundant repeat classes present within pericentromeric BAC clone F28D6, and with the centromeric Histone H3, HTR12, within chromocenters that stain intensively with the fluorescent DNA-binding dye, DAPI (Figure 1, top row). In met1 (DNA METHYLTRANSFERASE 1) mutants defective for the major CG maintenance methyltransferase (Kakutani et al., 1995), the centromere repeats and HTR12 remain co-localized with the chromocenters, as in wild-type cells. In contrast, while we observed only a subtle decondensation of 5S gene loci, F28D6 repeats are significantly decondensed such that they no longer co-localize precisely with chromocenters (Figure 1; Supplemental Tables 1–3). Interestingly, the Pol IV mutant, nrpd1, causes a small degree of 5S gene decondensation but has no significant effect on F28D6 or HTR12 co-localization with 180-bp repeats or chromocenters. However, in nrpe1 or drd1 mutants, 180-bp centromeric repeats, 5S gene repeats, and F28D6 repeats are all decondensed and no longer co-localize precisely with chromocenters (Figure 1 and Supplemental Tables 1 and 2). Like nrpd1 mutants, rdr2 and dcl3 mutants that disrupt 24-nt siRNA biogenesis have only subtle effects on pericentromeric repeat condensation (Supplemental Figure 1 and Supplemental Tables 1 and 2).
Figure 1.
Major Pericentromere Repeat Organization at Interphase Is Disrupted in nrpe1 and drd1 But Not nrpd1 Mutants.
DNA-FISH of 5S genes (red) and 180-bp centromeric repeats (green) (left panel).
Organization pattern of pericentromeric regions analyzed by FISH using the 180-bp centromere repeat (green) and the BAC F28D6 (red) DNA probes (middle panel).
Immunostaining followed by DNA-FISH performed respectively with an antibody recognizing the A. thaliana centromeric histone H3, HTR12 (red), and 180-bp centromeric repeats (green) (right panel). In all panels, DNA was visualized by DAPI (gray) and size bars indicate 5 μm.
Despite the pericentromeric region decompaction apparent in nrpe1 and drd1 mutants, it is noteworthy that the centromere core is unaltered such that HTR12 signals remain at the chromocenters, similar to wild-type cells (Figure 1). We conclude that nrpe1 and drd1 are required for the association of pericentromeric heterochromatic repeats with chromocenters but are not required for the deposition of HTR12 at the centromere core.
Heterochromatin Content Is Reduced in pol V and drd1 Mutants
DNA methyltransferase MET1, which functions primarily in the maintenance of methylation at CG motifs (Kakutani et al., 1995), and DDM1 (DECREASE IN DNA METHYLATION 1), a SWI2/SNF2 chromatin remodeler required for the maintenance of cytosine methylation in multiple sequence contexts (Vongs et al., 1993), are both required for the organization of pericentromeric heterochromatin within chromocenters (Soppe et al., 2002). As a consequence, heterochromatin content in interphase nuclei visualized by DAPI staining is diminished in met1 and ddm1 mutants (Soppe et al., 2002). Using the approach of Soppe et al. (2002), we determined the fraction of nuclear area that is occupied by chromocenters. In nrpe1, nrpd2/nrpe2 and drd1 mutants, heterochromatic content is strongly reduced, similarly to met1 and ddm1 mutants (Figure 2 and Supplemental Figure 2). By contrast, chromocenter content in nuclei of the Pol IV mutant nrpd1 resembles that of wild-type cells (ecotypes Col and Ler).
Figure 2.
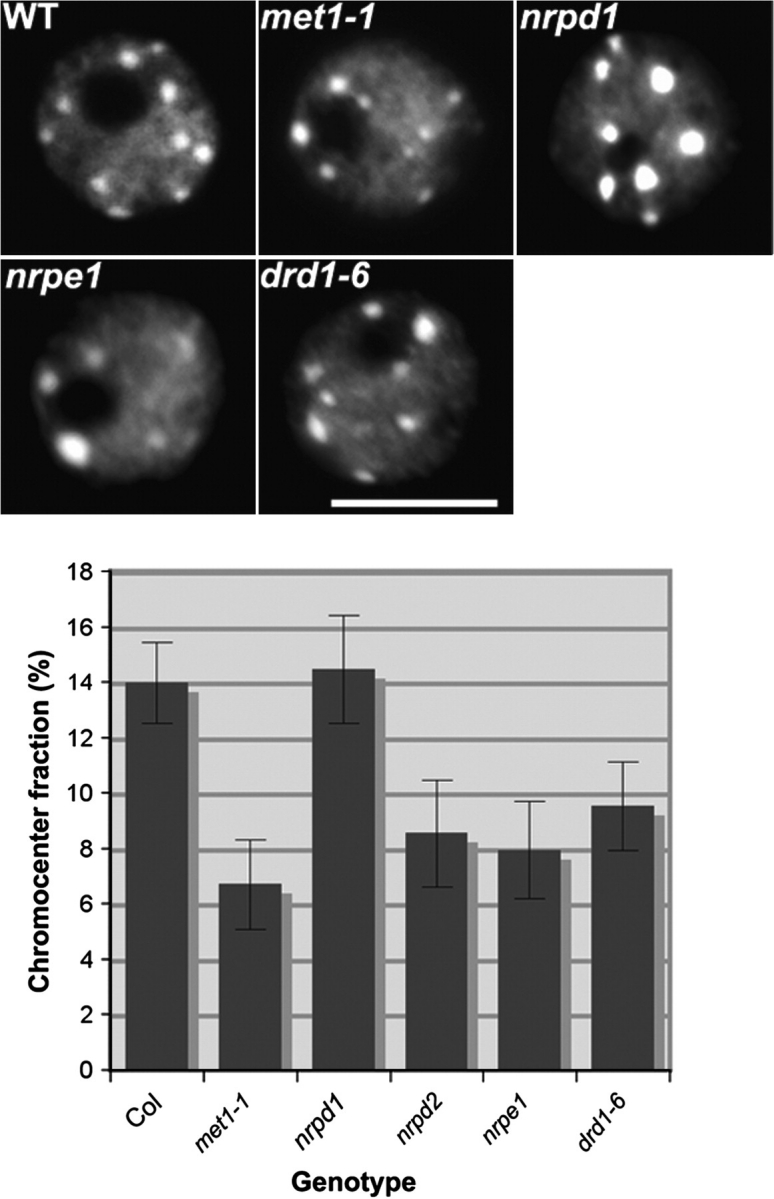
Pol V, drd1, and met1 Mutations Result in Loss of Heterochromatin at Chromocenters.
In the upper panel, representative interphase nuclei of the different mutants are displayed. DNA was counterstained by DAPI (gray). The size bar corresponds to 5 lm. The graph shows the heterochromatin content in wild-type, nrpd1, met1, pol V subunits and drd1 mutants. The mean values and standard deviation are shown in the histogram calculated from Col (n = 97), met1-1 (n = 93), nrpd1 (n = 127), nrpd2/nrpe2 (n = 90), nrpe1 (n = 171), drd1-6 (n = 116).
Decreased heterochromatin content is correlated with disruption of histone H3 lysine 9 dimethylation (H3K9me2) in interphase nuclei (Soppe et al., 2002). Immunolocalization of H3K9me2 combined with DNA-FISH shows that H3K9me2 foci co-localize with 180-bp centromeric repeats and DAPI-stained chromocenters in wild-type and nrpd1 nuclei (Figure 3). However, in met1 mutants, the H3K9me2 signals are no longer confined to the chromocenters but are dispersed into the decondensed euchromatic regions, with fainter signals detected in the chromocenters. A similar phenotype was observed in ddm1 mutants (Supplemental Figure 3 and Supplemental Table 4), in agreement with a prior report (Soppe et al., 2002). In nrpe1 and drd1 nuclei, dissociation of H3K9me2 signals from the chromocenters is also observed (Figure 3), although not as dramatically as in met1 mutants. Like the Pol IV mutant, nrpd1, other mutants disrupting the 24-nt siRNA-dependent DNA pathway, including rdr2, dcl3, and ago4 mutants, had no significant effect on H3K9me2 signals compared to wild-type (Figure 3, Supplemental Figure 3 and Supplemental Table 4).
Figure 3.
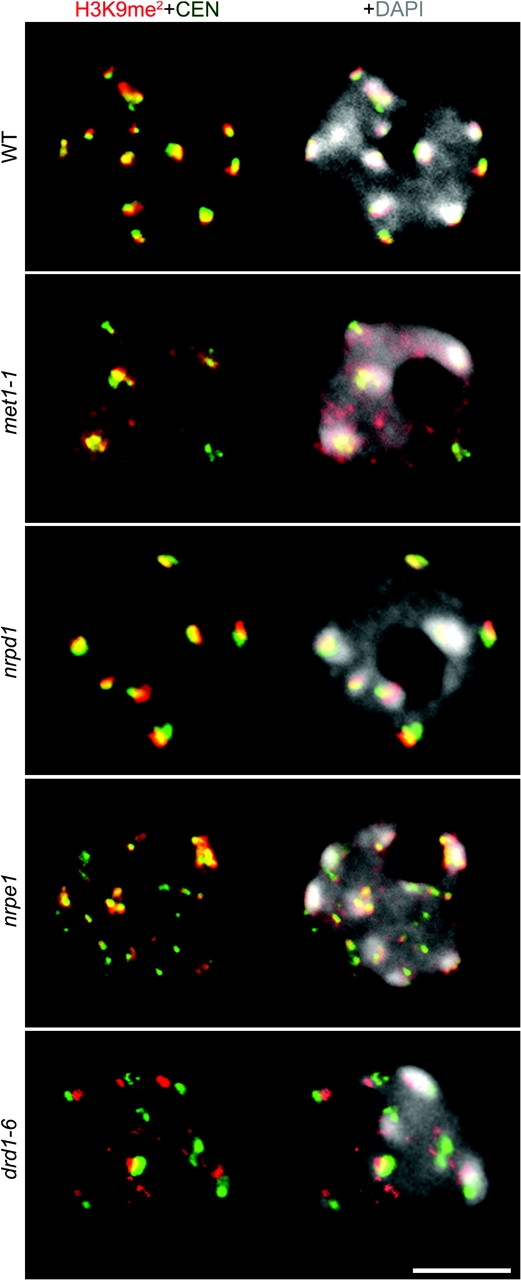
H3K9me2 Clustering at the Chromocenters Is Disrupted by nrpe1, drd1, and met1 Loss-of-Function Mutants.
The panel shows representative images of immuno-stained interphase nuclei with H3K9me2 (red) combined with DNA-FISH to detect the 180-bp centromeric repeats (green). DNA was visualized by DAPI (gray) and the size bar indicates 5 μm.
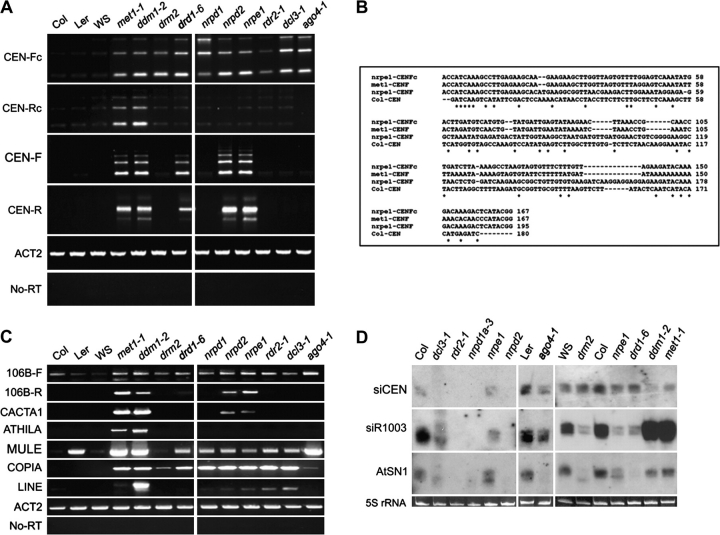
Specific Pericentromeric Transcription Units Are Suppressed by MET1, DDM1, Pol V, and DRD1, Independently of the 24-nt siRNA Biogenesis Pathway
To investigate potential links between heterochromatin disruption and pericentromeric transcription, we used RT–PCR to detect RNAs corresponding to pericentromeric 180-bp repeats. Two sets of strand-specific primers that distinguish between conserved and non-conserved 180-bp repeat families (May et al., 2005) were tested. These primers were designated CEN-Fc and CEN-Rc, and CEN-F and CEN-R, where F denotes forward-strand transcripts, R denotes reverse-strand transcripts, and c denotes conserved repeats (supplemental Figure 4 and May et al., 2005). In wild-type plants (ecotypes Col, Ler, and WS), CEN-Fc repeats are not significantly expressed. However, in met1 and ddm1 mutants, as well in all mutants that define the 24-nt siRNA-dependent DNA methylation pathway (nrpd1, nrpe1, rdr2, dcl3, ago4, drd1, and drm2), CEN-Fc transcripts are readily observed, suggesting that a combination of maintenance and de-novo cytosine methylation suppresses these transcription units. In contrast, CEN-Rc transcripts are up-regulated specifically in met1 and ddm1 mutants (Figure 4A), suggesting that maintenance cytosine methylation is primarily responsible for silencing these transcription units. Interestingly, CEN-F and CEN-R transcripts derived from non-conserved centromeric repeats were selectively de-repressed in met1, ddm1, drd1, and Pol V (nrpe1 and nrpd2/nrpe2) mutants, but not in mutants required for 24-nt siRNA-dependent DNA methylation pathway (Figure 4A). Sequencing revealed that the 180-bp, 360-bp, and 520-bp RT–PCR products correspond to transcripts that spanned one, two, or three repeats. Interestingly, the sequencing data also revealed that 180-bp repeats reactivated in pol V mutants differ in sequence from repeats de-repressed in met1 (Figure 4B).
Figure 4.
Specific Centromere Sequences Are De-Repressed in pol V and drd1 Mutants.
(A) Strand-specific centromere transcription was evaluated by RT–PCR using primers specific for different subsets of 180-bp repeat families: forward conserved (CEN-Fc), reverse conserved (CEN-Rc), forward non-conserved (CEN-F), and reverse non-conserved (CEN-R) 180-bp repeats.
(B) Different centromere sequences are up-regulated in nrpe1. The figure displays aligned centromere transcript consensus sequences obtained by cloning RT–PCR products from the following mutants: nrpe1 CEN-Fc 180-bp (n = 12), nrpe1 CEN-F (n = 15), and met1 CEN-F 180-bp (n = 10).
(C) Transposable element transcription detected by RT–PCR.
(D) Northern blot analysis of centromere-derived siRNAs (siCEN). Ethidium bromide-stained 5S rRNA resolved by agarose gene electrophoresis serves as a loading control.
Next, we examined several classes of transposable elements and repeats enriched in pericentromeric heterochromatin to compare the genetic requirements for their suppression (Figure 4C). Strand-specific RT–PCR of 106B-dispersed repeats (May et al., 2005) indicates that forward-strand transcription is detectable, even in wild-type plants, but is up-regulated in met1 and ddm1 mutants as well as in mutants that define the 24-nt siRNA-dependent DNA methylation pathway (Figure 4C). However, transcripts corresponding to the reverse-strand of the 106B repeats were abundant in met1, ddm1, and pol V mutants (nrpe1 and nrpd2/nrpe2) and were detectable in the drd1 mutant, but were not observed in the other mutants tested. Similarly, CACTA1-like transposons are de-repressed in met1 and ddm1 plants, in agreement with prior studies (Mathieu et al., 2005; Kato et al., 2003), and are also de-repressed in the Pol V catalytic subunit mutants (nrpe1 and nrpd2/nrpe2) but not in Pol IV, rdr2, dcl3, ago4, or drm2 mutants (Figure 4C). In contrast, transcripts of Mutator-like DNA transposons (MULE), Copia4 transposons, and LINE transcripts are detected in the majority of the mutants analyzed (Figure 4C). Athila retroelement transcription was only detected in met1 and ddm1 mutants. Collectively, these results indicate that Pol V collaborates with MET1 and DDM1-dependent cytosine methylation in the suppression of specific classes of pericentromeric repeats, but can function independently of the 24-nt siRNA biogenesis or RNA-directed DNA methylation pathway.
Transcription or Silencing of Pericentromeric Regions Is Not Correlated with siRNA Accumulation
Although mutations disrupting the 24-nt siRNA biogenesis pathway do not phenocopy Pol V mutants with respect to chromocenter organization or the de-repression of CEN-F, CEN-R, 106B-R, or CACTA1 elements (Figure 4A and 4C), siRNAs generated by other pathways might be involved. Using small RNA Northern blotting, we examined the abundance of siRNAs corresponding to 180-bp centromere repeats in mutants that have differential effects on pericentromeric repeat silencing (Figure 4D). No strict correlation between CEN siRNA abundance and silencing was observed. For instance, CEN siRNAs are undetectable in nrpd1, nrpd2/nrpe1, and rdr2 mutants, in which CEN-F, CEN-R, 106B-R, or CACTA1 elements remain silenced, yet siRNAs are also reduced in met1 and ddm1 mutants, in which the elements are de-repressed (Figure 4D). Likewise, in Pol V mutants (nrpe1), siRNA levels persist at near wild-type levels, yet CEN-F, CEN-R, 106B-R, and CACTA1 elements are de-repressed. A smear of differently sized siRNAs is present in dcl3 mutants, likely attributed to the redundant action of the three remaining functional DCL enzymes present in A. thaliana (Gasciolli et al., 2005). Collectively, the RT–PCR and siRNA data suggest a lack of correlation between transcription or silencing and siRNA abundance, at least with respect to siRNAs that correspond to 180-bp centromere repeats.
Pericentromere-Derived Transcripts Accumulate in the Nucleus
To determine if we could visualize 180-bp centromere repeat transcripts and determine their spatial distribution, we used RNA-FISH with probes specific for pericentromeric CEN-F or CEN-R transcripts (Figure 5). Interestingly, CEN-F and CEN-R transcripts were found to have different spatial distributions within the nucleus. CEN-R signals are dispersed throughout the nucleoplasm and nucleolus and this pattern is not affected by any of the mutants analyzed (data not shown). However, CEN-F transcript localization patterns differed in the mutants relative to wild-type. In wild-type nuclei (ecotype Col-0), we detected weak punctuate CEN-F signals in the nucleoplasm and a conspicuous strong signal associated with the nucleolus, which appears as a prominent dark region in DAPI-stained nuclei due to the relative paucity of nucleolar DNA in this nuclear territory (Figure 5, top row nuclei in red; see also Supplemental Table 4). By contrast, multiple strong punctate CEN-F signals, located at the edges of the chromocenters, are observed in the met1, nrpe1, and drd1 mutants (Figure 5 and supplemental quantitative data in Supplemental Table 5), which correlates with the de-repression of CEN-F transcripts determined by RT–PCR (see Figure 4). The Pol IV mutant, nrpd1, displays a CEN-F RNA phenotype similar to wild-type nuclei, consistent with the fact that CEN-F transcripts are not de-repressed in nrpd1, except that the nucleolar signal is markedly reduced. RNase treatment prior to in-situ hybridization abolished the signals detected using CEN-F or CEN-R probes whereas DNase I treatment had no effect, indicating that only RNA is detected (Figure 5, bottom-most nuclei).
Figure 5.
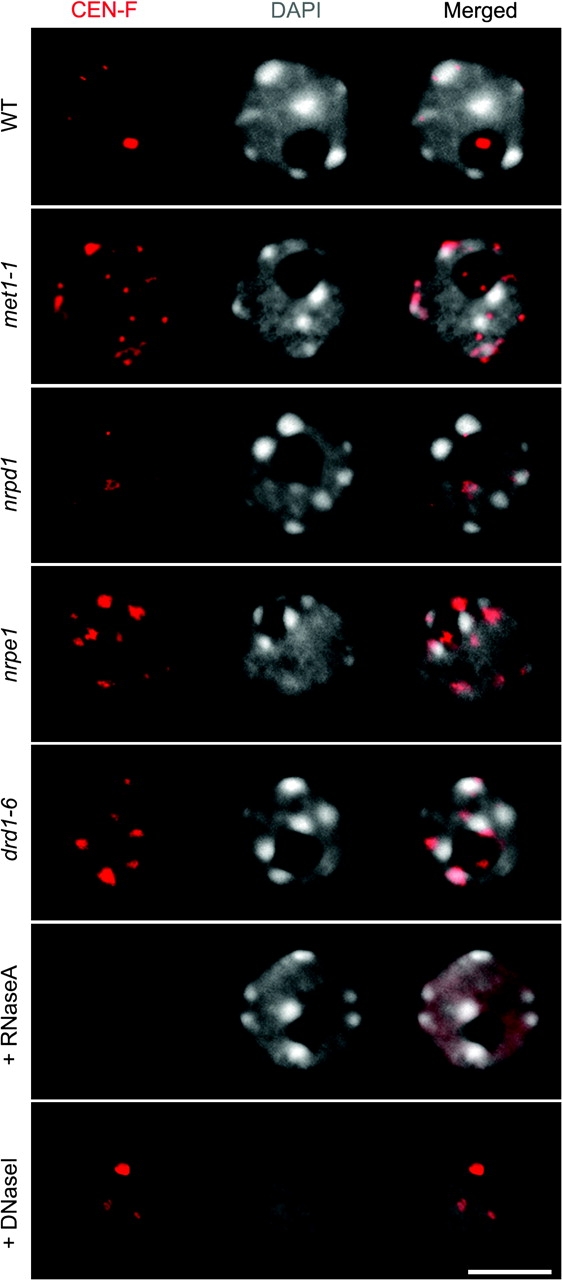
Pericentromere-Repeat RNAs Localized by RNA-FISH during Interphase.
The probe specifically detects forward transcripts (CEN-F, red). RNase A treatment prior to in-situ hybridization eliminates the FISH signals, indicating that the method predominantly detects RNA (bottom nuclei). DNA was DAPI-counterstained (gray). The size bar corresponds to 5 μm.
The increased RNA-FISH signals at the periphery of the chromocenters may correspond to pericentromeric CEN-F transcripts at the sites of transcription, in agreement with the RT–PCR analyses (Figure 4A). The reduced nucleolar localization of CEN RNA signals in mutants involved in siRNA formation, such as nrpd1 and rdr2, strongly suggest that these nucleolar signals reflect the process of siRNA biogenesis, consistent with evidence that 24-nt siRNAs and siRNA pathway proteins localize to a nucleolar compartment bearing markers of Cajal Bodies (Li et al., 2006; Pontes et al., 2006).
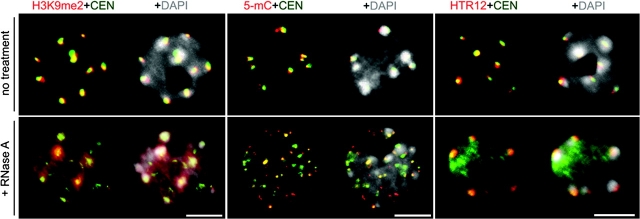
RNA Is Required for the Topology of Pericentromeric Heterochromatic Marks
In mouse, pericentromeric heterochromatin is RNase-sensitive, as assayed by histones bearing heterochromatin marks, such as H3K9me2 or the presence of non-histone proteins such as Heterochromatin Protein 1; HP1 (Maison et al. 2002; Muchardt et al., 2002). These observations indicate that RNA is necessary for the large-scale organization of heterochromatin. To test the hypothesis that RNA is also essential for assembly of A. thaliana pericentromeric heterochromatin, immunolocalization of H3K9me2 and 5-methylcytosine (5-mC), combined with DNA-FISH localization of 180-bp centromere repeats, was performed in RNase A-treated and untreated nuclei. As shown in Figure 6 (top row), H3K9me2, 5-mC, and 180-bp centromere repeats co-localize within discrete nuclear foci that correspond to the DAPI-enriched heterochromatic chromocenters in non-treated interphase nuclei. In contrast, RNase A treatment causes H3K9me2 to become distributed throughout the nucleoplasm (Figure 6, bottom-left panel), and causes 5-mC and centromere repeats to disperse into smaller foci (Figure 6, middle panel). Interestingly, DAPI-stained chromocenters persist in RNase-treated nuclei and HTR12, the A. thaliana centromeric histone H3 (Talbert et al., 2002), largely maintains its co-localization with these chromocenters and a fraction of the 180-bp centromeric repeats.
Figure 6.
Effects of RNase A Treatment on the Interphase Organization of Centromere Repeats (Green), H3K9me2 (Red), and 5-mC (Red) Detected by DNA-FISH and Immunostaining.
Representative images of untreated and nuclease treated (+RNase) nuclei are shown. The right-most nuclei depict immunostaining of HTR12 (red) and 180-bp pericentromeric repeats (green) in non-treated and RNase A-treated nuclei. DNA was visualized by DAPI (gray). Size bars indicate 5 μm.
DISCUSSION
Our study shows that Pol V has functions in heterochromatin organization and gene silencing that are independent of Pol IV-dependent 24-nt siRNA production and DRM2-dependent de-novo cytosine methylation. In this role, Pol V and DRD1, a chromatin remodeler required for Pol V transcription (Wierzbicki et al., 2008), apparently act in partnership with MET1 and DDM1, whose activities are required to maintain the majority of the methylation in the Arabidopsis genome. The correspondence between heterochromatin disruption and the de-repression of specific classes of 180-bp pericentromeric repeats, which accumulate within the nucleus in met1, ddm1, nrpe1, and drd1 mutants, suggests that suppression of a subset of transcription units is important for pericentromeric repeat clustering within chromocenters. However, the transcription machinery that modulates the transcription of centromere repeats is not yet defined in plants. It is not yet clear how met1, ddm1, nrpe1, and drd1 might work together within a discrete pathway to suppress these transcription units. Importantly, nrpe1 and drd1 mutants do not cause losses of methylation at 180-bp centromere repeats, unlike met1 and ddm1 mutants (Kanno et al., 2008, 2005), suggesting that NRPE1 and DRD1 act independently or downstream of MET1- and DDM1-dependent DNA methylation.
An apparent paradox revealed by our study is that despite the correlation between transcriptional silencing and large-scale pericentromeric heterochromatin, formation of the resulting heterochromatic structures is RNase A-sensitive. This observation suggests that RNA is an important structural component of pericentromeric heterochromatin in A. thaliana. To this end, Pol V or Pol V-generated transcripts might serve as structural RNAs that are important for long-range heterochromatin interactions, possibly forming a structural network or complex that retains or recruits heterochromatin-forming complexes. Similar roles for non-coding RNAs have been proposed in the control of sex chromosome dosage in mammals and Drosophila (Wutz, 2003), in the formation of the yeast telomerase complex (Zappulla and Cech, 2004), and in the organization of human pericentromeric heterochromatin (Maison et al., 2002). Importantly, RNase A treatment does not affect the organization and architecture of the core centromere domain, as indicated by the persistence of an HTR12 centromeric histone signal, which marks a domain that is structurally distinct from the flanking pericentromere (Choo, 2001).
In addition to potentially generating structural RNAs, Pol V, DRD1, MET1, and DDM1 could play a direct role in silencing specific pericentromeric repeats, such as AtSN1 and solo LTR retrotransposon elements that are subject to siRNA-directed DNA methylation (Wierzbicki et al., 2008). However, silencing of these latter elements requires the canonical 24-nt siRNA-directed DNA methylation pathway that includes Pol IV, RDR2, DCL3, AGO4, and DRM2. Pol V generates non-coding transcripts at AtSN1 and solo LTR loci, leading to the hypothesis that Pol V transcripts serve as scaffolds for the binding of siRNA-AGO4 silencing complexes (Wierzbicki et al., 2008). Importantly, Pol IV, RDR2, DCL3, AGO4, and DRM2 are not required for silencing CEN-F, CEN-R, 106B, and CACTA elements, whose de-repression specifically in Pol V (nrpe1), drd1, met1, and ddm1 mutants correlates with long-range heterochromatin disruption. Interestingly, in an nrpe1 mutant background, solo LTR transcription is increased relative to WT levels (Huettel et al., 2006)—an increase that correlates with a higher occupancy of Pol II at the locus (Wierzbicki et al., 2008). Additionally, transcriptional analysis of two other intergenic regions located in the pericentromeric regions showed their transcription to be dependent on Pol V (Wierzbicki et al., 2008). These previous findings, together with our new data concerning expression patterns of CEN-F, CEN-R, 106B, and CACTA elements, indicate an extensive alteration in pericentromeric region transcription in the absence of Pol V.
Collectively, the available evidence suggests that Pol V may play a role in gene silencing via one or more pathways distinct from the 24-nt siRNA-directed pathway; however, the possibility that other small RNA pathways may be involved cannot be excluded. Knowing more about how pericentromere repeats are transcribed, the nature of these non-coding RNAs, and the identification of any interacting proteins likely reveals new insights into the epigenetic regulation of pericentromeric regions.
METHODS
Plant Material and Growth Conditions
Jim Carrington provided Arabidopsis thaliana mutants rdr2-1, dcl3-1, and hen1-1. drd1-6 was provided by Marjori Matzke; drm2-1 and ago4-1 were provided by Steve Jacobsen. nrpd1 (nrpd1a-3), nrpd2 (nrpd2a-2 nrpd2b-1), and nrpe1 (nrpd1b-11) were described previously (Kanno et al., 2005; Onodera et al., 2005). drm2-1 is in the WS genetic background; ago4-1 is in the Landsberg (Ler) genetic background. Other mutants are in the Columbia (Col-0) genetic background. Plants were grown either in sterile conditions on Murashigi-Skoog medium or on soil under long-day conditions (16 h light/8 h dark) at 22°C.
Isolation and RNA Blot Analysis of Small RNA
RNA was extracted from immature inflorescence using the mirVana® miRNA Isolation Kit (Ambion). Small RNA (smRNA) blot hybridization was performed as described (Onodera et al., 2005). Probes for smRNA blot hybridizations were derived from custom-made DNA oligonucleotides designed according to the mirVana® miRNA probe construction kit (Ambion). A T7-driven transcription reaction was used to generate labeled RNA probes with α-32P CTP, which were subsequently used for RNA:RNA hybridization. The probes used to detect 5S genes smRNAs (siR1003) and AtSN1 were described previously (Pontes et al., 2006; Xi et al., 2006) and the following oligonucleotide was used to analyze 180-bp centromeric repeats smRNAs: 5′-TGTATGATTGAGTATAAGAACTTAAAACGCAACCGCATCTTAAAAGCCTAAGTAGTATTTC-3′.
Transcript Analysis by Reverse Transcription PCR (RT–PCR)
To detect strand-specific pericentromeric transcripts, total RNA extracted as described above was treated with RQ1-DNase (Promega) and further purified using Trizol (Sigma). 100 ng was used in RT–PCR reactions using conditions and primers described previously (May et al., 2005). RT–PCR conditions and primers used to specifically amplify different elements are described in Supplemental Table 6. Negative controls were performed without reverse transcriptase to detect contaminant DNA and the constitutively expressed ACTIN2 gene was used as internal control.
Nuclease Sensitivity Assays
For RNAse sensitivity analysis, unfixed leaf nuclei were released into isolation buffer (25 mM HEPES, 1 mM MgCl2, 1 mM DTT, 10 mM KCl, 25 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg ml−1 pepstin, pH 7.4) (Abranches et al., 1998; Jasencakova et al., 2000), washed in PBS and permeabilized in the nuclei isolation buffer containing 0.05% Triton-X100 for 2 min. An incubation of 10 min at RT was performed with RNAse A (100 μg ml−1, Roche) or without RNase A plus 100 U ml−1 RNase inhibitor (RNA Guard, Pharmacia) as a control. After washing in nuclei isolation buffer and PBS, cells were fixed for 20 min in 4% paraformaldehyde/PBS, centrifuged, and applied to the slides for immunolocalization and DNA FISH.
Probe Labeling
5S and 180-bp pericentromeric gene probes were labeled with biotin-16-dUTP or digoxigenin-11-dUTP (Roche) by PCR as previously described (Pontes et al., 2003). BAC F28D6 was obtained from the Arabidopsis Biological Resource Center (ABRC, Accession No. AF147262) and labeled with biotin-Nick Translation Mix (Roche). For the detection of strand specific pericentromere transcripts, digoxigenin-11-UTP and biotin-16-UTP (Roche) labeled sense and antisense probes were prepared by in-vitro transcription of the 180-bp repeat and the custom-made oligonucleotide specific to centromeric repeats used for smRNA blot hybridization using T7 and SP6 polymerase (Ambion).
Immunofluorescence
Root meristem nuclei were isolated as described previously (Jasencakova et al., 2000). Upon 4% paraformaldehyde post-fixation, the nuclei were incubated overnight at 4°C with primary antibodies for anti-H3K9me2 (1:200, AbCam) and anti-HTR12 (1:200, kindly provided by Steve Henikoff). Secondary antibody anti-rabbit Alexa 594 (Invitrogen) was diluted at 1:500 in PBS and incubated for 2 h at 37°C. DNA was counterstained with 1 μg ml−1 DAPI in Vectashield mounting medium (Vector Laboratories). For immuno-FISH experiments, immunolocalization was performed first; cells were post-fixed in 4% paraformaldehyde and the standard DNA in-situ hybridization protocol was then followed.
RNA and DNA Fluorescence In-Situ Hybridization
Meristem nuclei were processed for DNA fluorescence in-situ hybridization (FISH) as described previously (Pontes et al., 2003), using as hybridization stringency conditions 50% formamide and 2 SSC. Post-hybridization washes were performed in 50% formamide and 0.1 SSC at 42°C. RNA in-situ hybridization was carried out at 50°C overnight using a probe solution containing 500 ng RNA probe, 5 μg yeast tRNA (Roche), 50% dextran sulphate, 100 mM PIPES, pH 8.0, 10 mM EDTA, and 3 M NaCl as described previously (Pontes et al., 2006). Slides were washed sequentially in 2 SSC, 50% formamide at 50°C followed by 1 SSC, 50% formamide. Digoxigenin-labeled probes were detected using mouse anti-digoxigenin antibody (1:250, Roche) followed by rabbit anti-mouse antibody conjugated to Alexa 488 (1:200, Molecular Probes). Biotin labeled probes were detected using goat anti-biotin conjugated with avidin (1:200, Vector Laboratories) followed by streptavidin-Alexa 546 (1:200, Molecular Probes). Nuclear DNA was counterstained with DAPI in Vectashield anti-fade medium (Vector Laboratories).
5-Methylcytosine Detection
Root tips were harvested, fixed in ethanol/acetic acid (3:1) and nuclei isolated as described (Pontes et al., 2003). Slide preparations were baked at 60°C, denatured in 70% formamide, 2 SSC, 50 mM sodium phosphate at 80°C for 3 min, washed in ice-cold PBS, blocked with 1% BSA, and incubated with the 5-methylcytosine specific antibody (1:100, AbD-Serotec). Methylated DNA was detected by incubation in rabbit anti-mouse antibody Alexa 546 (1:200, Molecular Probes) secondary antibody.
Measurement of the Heterochromatin Fraction
Nuclei were isolated from ethanol/acetic acid (3:1)-fixed material and stained with DAPI. Digital grayscale raw images were analyzed with the freeware software NIH-ImageJ. The chromocenter/heterochromatin ratio was correlated from the area and DAPI-fluorescence intensity of chromocenters relative to the whole nucleus (Soppe et al., 2002).
Microscopy and Imaging
The preparations were inspected with a Nikon Eclipse E800i epifluorescence microscope equipped with a Photometrics Coolsnap ES Mono digital camera. Images were acquired by the Phylum software and pseudocolored and merged in Adobe Photoshop.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
O.P. was supported by a fellowship SFRH/BPD/17508/2004 from the Fundação para a Ciência e Tecnologia, Portugal, and the Edward Mallinckrodt Foundation Award. P.C.N. was supported by SFRH/PBD/30386/2006 from the Fundação para a Ciência e Tecnologia, Portugal. Pikaard lab work was supported by NIH grants GM077590.
Supplementary Material
Acknowledgments
O.P. performed all microscopy and RT–PCR analysis; P.C.N. generated the siRNA blot; P.V. performed the centromere repeat cloning and sequencing. O.P. and C.S.P. wrote the paper. We thank Jeremy Haag and Thomas Ream for suggestions to improve the manuscript. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of NIH or the Edward Mallinckrodt Foundation. No conflict of interest declared.
References
- Abranches R, Beven AF, Aragón-Alcaide L, Shaw PJ. Transcription sites are not correlated with chromosome territories in wheat nuclei. J. Cell Biol. 1998;1431:5–12. doi: 10.1083/jcb.143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelgren H, Kniola B, Ekwall K. Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 2003;116:4035–4042. doi: 10.1242/jcs.00707. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Attwood JT, Yung RL, Richardson BC. DNA methylation and the regulation of gene transcription. Cell Mol. Life Sci. 2002;59:241–257. doi: 10.1007/s00018-002-8420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J. Chromatin-based silencing mechanisms. Curr. Opin. Plant Biol. 2004;7:521–526. doi: 10.1016/j.pbi.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;5662:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Choo KH. Centromere DNA dynamics: latent centromeres and neocentromere formation. Am. J. Hum. Genet. 1997;61:1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KH. Domain organization at the centromere and neocentromere. Dev. Cell. 2001;2:165–177. doi: 10.1016/s1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl Acad. Sci. U S A. 2002;22:14584–14589. doi: 10.1073/pnas.212325299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Gaubatz JW, Cutler RG. Mouse satellite DNA is transcribed in senescent cardiac muscle. J. Biol. Chem. 1990;29:17753–17758. [PubMed] [Google Scholar]
- Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl Acad. Sci. U S A. 2003a;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SE, Kettler G, Preuss D. Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Res. 2003b;13:195–205. doi: 10.1101/gr.593403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Haupt W, Fischer TC, Winderl S, Fransz P, Torres-Ruiz RA. The centromere1 (CEN1) region of Arabidopsis thaliana: architecture and functional impact of chromatin. Plant J. 2001;27:285–296. doi: 10.1046/j.1365-313x.2001.01087.x. [DOI] [PubMed] [Google Scholar]
- Heitz E. Das Heterochromatin der Moose. Jahrb Wiss Botanik. 1928;69:762–818. [Google Scholar]
- Henikoff S. Near the edge of a chromosome's ‘black hole’. Trends Genet. 2002;18:165–167. doi: 10.1016/s0168-9525(01)02622-1. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Murata M, Ogura Y, Schwarzacher T, Motoyoshi F. Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. Plant Cell. 1999;11:31–42. doi: 10.1105/tpc.11.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;12:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z, Meister A, Walter J, Turner BM, Schubert I. Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell. 2000;12:2087–2100. doi: 10.1105/tpc.12.11.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T, Jeddeloh JA, Richards EJ. Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 1995;23:130–137. doi: 10.1093/nar/23.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Bucher E, Daxinger L, Huettel B, Böhmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJ. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Atypical RNA polymerase subunits required for RNA directed DNA methylation. Nat. Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T. Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 2003;13:421–426. doi: 10.1016/s0960-9822(03)00106-4. [DOI] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater J, Estelle MA, Somerville CR. A highly repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 1986;204:417–423. [Google Scholar]
- Mathieu O, Probst AV, Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 2005;24:2783–2791. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May BP, Lippman ZB, Fang Y, Spector DL, Martienssen RA. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 2005;1:e79. doi: 10.1371/journal.pgen.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Okamoto Y, Ohzeki J, Masumoto H. Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes. J. Cell Sci. 2003;116:4021–4034. doi: 10.1242/jcs.00697. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SI. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;11:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Pontes O, Lawrence RJ, Neves N, Silva M, Lee JH, Chen ZJ, Viegas W, Pikaard CS. Natural variation in nucleolar dominance reveals the relationship between nucleolus organizer chromatin topology and rRNA gene transcription in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2003;100:11418–11423. doi: 10.1073/pnas.1932522100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;7114:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Round EK, Flowers SK, Richards EJ. Arabidopsis thaliana centromere regions: genetic map positions and repetitive DNA structure. Genome Res. 1997;7:1045–1053. doi: 10.1101/gr.7.11.1045. [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz PF. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;23:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;5:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson H, Schmidt R, Brandes A, Heslop-Harrison JS, Dean C. A novel repetitive sequence associated with the centromeric regions of Arabidopsis thaliana chromosomes. Mol. Gen. Genet. 1996;1–2:247–252. doi: 10.1007/s004380050319. [DOI] [PubMed] [Google Scholar]
- Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl Acad. Sci. U S A. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA Polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;14:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A. RNAs templating chromatin structure for dosage compensation in animals. Bioessays. 2003;25:434–442. doi: 10.1002/bies.10274. [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:642–652. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl Acad. Sci. U S A. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.