Abstract
Heterotrimeric GTP-binding proteins, which consist of Gα, Gβ, and Gγ subunits, play important roles in transducing extracellular signals perceived by cell surface receptors into intracellular physiological responses. In addition to a single prototypical Gα protein (GPA1), Arabidopsis has three unique Gα-like proteins, known as XLG1, XLG2, and XLG3, that have been found to be localized in nuclei, although their functions and mode of action remain largely unknown. Through a transcriptomic analysis, we found that XLG2 and XLG3 were rapidly induced by infection with the bacterial pathogen Pseudomonas syringae, whereas the XLG1 transcript level was not affected by pathogen infection. A reverse genetic screen revealed that the xlg2 loss-of-function mutation causes enhanced susceptibility to P. syringae. Transcriptome profiling revealed that the xlg2 mutation affects pathogen-triggered induction of a small set of defense-related genes. However, xlg1 and xlg3 mutants showed no difference from wild-type plants in resistance to P. syringae. In addition, the xlg2 xlg3 double mutant and the xlg1 xlg2 xlg3 triple mutant were not significantly different from the xlg2 single mutant in the disease resistance phenotype, suggesting that the roles of XLG1 and XLG3 in defense, if any, are less significant than for XLG2. Constitutive overexpression of XLG2 leads to the accumulation of abnormal transcripts from multiple defense-related genes. Through co-immunoprecipitation assays, XLG2 was found to interact with AGB1, the sole Gβ subunit in Arabidopsis, which has previously been found to be a positive regulator in resistance to necrotrophic fungal pathogens. However, no significant difference was found between three xlg single mutants, the xlg2 xlg3 double mutant, the xlg triple mutant, and wild-type plants in resistance to the necrotrophic fungal pathogens Botrytis cinerea or Alternaria brassicicola. These results suggest that XLG2 and AGB1 are components of a G-protein complex different from the prototypical heterotrimeric G-protein and may have distinct functions in modulating defense responses.
Keywords: Defense responses, disease resistance, plant–microbe interactions, Arabidopsis, G-protein
INTRODUCTION
Heterotrimeric guanine nucleotide-binding proteins (G-proteins) transduce a wide array of extracellular signals into appropriate physiological responses (Neer, 1995; Neves et al., 2002). The signaling pathway is initiated by binding of an extracellular ligand to a cell surface receptor that belongs to the superfamily of seven transmembrane G-protein-coupled receptors (GPCRs). The recognition of the ligand by the receptor leads to activation of the G-protein, which is made up of three subunits: Gα, Gβ, and Gγ. Gα and Gβγ subunits then dissociate and activate their respective downstream effectors, which include phospholipases, nitric oxide synthase, adenyl cyclase, ion channels, MAPKs, Ca+2 channels, and other proteins (Neer, 1995; Neves et al., 2002; Cabrera-Vera et al., 2003). Although mammals have nearly 1000 GPCRs and 20 Gα, 5 Gβ, and 12 Gγ isoforms, plants have a much smaller number of G-protein components (Assmann, 2004; Temple and Jones, 2007). For instance, the Arabidopsis genome only encodes one canonical Gα (GPA1), one Gβ (AGB1), and two known Gγ subunits (AGG1 and AGG2) (Ma et al., 1990; Weiss et al., 1994; Mason and Botella, 2000, 2001), and the number of predicted GPCRs is also much smaller (Moriyama et al., 2006; Gookin et al., 2008). Despite the paucity of G-protein components, mutational analyses have revealed that G-protein signaling functions in a variety of biological processes in plants, including the auxin response, ABA-mediated inhibition of stomatal opening, cell division and expansion, selected light responses, seed germination, sugar sensing, and drought tolerance (reviewed by Jones and Assmann, 2004; Perfus-Barbeoch et al., 2004). Several studies have also indicated that different G-protein subunits play distinct roles in disease resistance (Suharsono et al., 2002; Llorente et al., 2005; Trusov et al., 2006, 2007; Zhang et al., 2008).
Unlike animals, plants do not have specified cells to defend themselves against pathogen attack. Instead, every living plant cell is generally equipped with the components necessary for detecting invading pathogens and mounting an appropriate defense response. A plant cell contains receptors that recognize conserved microbe-/pathogen-associated molecular patterns (MAMPs/PAMPs) (Gomez-Gomez and Boller, 2000; Nurnberger et al., 2004; Ausubel, 2005; Kaku et al., 2006; Zipfel et al., 2006; Wan et al., 2008). The MAMP-triggered innate immune response provides the first layer of induced defense against an invading pathogen. This non-race-specific basal resistance, together with constitutive physical and chemical barriers, successfully prevents most infections from becoming established. To overcome basal resistance, pathogens have evolved a repertoire of virulence effector proteins that are delivered into hosts to suppress the basal defense response (Abramovitch and Martin, 2004; da Cunha et al., 2007). In turn, plants have evolved Resistance (R) proteins, each of which recognizes the action of specific virulent effector(s) as a signal of invasion to trigger the hypersensitive response (HR) (Jones and Dangl, 2006). HR is a strong physiological response that often leads to cell suicide and elimination of the pathogen.
Growing evidence indicates that the basal defense response largely overlaps with the R-protein-mediated HR, and that R-proteins may function to hyper-activate the basal resistance mechanism (Tao et al., 2003; Navarro et al., 2004; Eulgem, 2005; Burch-Smith et al., 2007; Dangl, 2007; Shen et al., 2007). Recognition of a MAMP by a cell surface receptor leads to activation of WRKY transcription factors through a MAP kinase cascade (Asai et al., 2002). Recent studies have revealed that many R-proteins function by directly modulating activities of transcription factors (Deslandes et al., 2002; Holt et al., 2002; Deslandes et al., 2003; Shen et al., 2007; Shen and Schulze-Lefert, 2007). These and other studies together indicate that different signaling events triggered by pathogen recognition converge in the cell nucleus to modify transcriptional factors that regulate both the basal and the R-mediated defense responses.
In addition to GPA1, the Arabidopsis genome encodes three extra-large GTP binding proteins (XLG1, XLG2, and XLG3) (Lee and Assmann, 1999; Ding et al., 2008). To date, XLG genes have been found in plant genomes but not elsewhere (Ding et al., 2008). The C-termini of XLGs are similar to prototypical Gα proteins. The N-termini (∼400 amino acids) are homologous to each other but share little sequence similarity to other known proteins in the GenBank database. Recently, the xlg2 xlg3 double mutant and the xlg1 xlg2 xlg3 triple mutant were found to display an increased primary root length when grown in the dark (Ding et al., 2008), and insertional mutation of XLG3 impaired root waving and skewing (Pandey et al., 2008). The triple mutant also exhibited altered sensitivity in root growth and/or seed germination in response to ABA, osmotic stress, ethylene, and sugars. In addition, these XLGs were found to be localized in the nucleus by transient transformation (Ding et al, 2008).
We initially identified XLG2 as one of the early pathogen-responsive (EPR) genes from a transcriptome profiling analysis. In a subsequent phenotype screen for alteration in resistance to P. syringae in knockout mutants of over 50 EPR genes, the xlg2 mutant was identified as an enhanced disease susceptibility mutant. Our study revealed that XLG2 physically interacts with AGB1 in planta. Constitutive overexpression of XLG2 was found to cause constitutive accumulation of abnormal transcripts from defense-related genes. The results indicate that XLG2 is a component of the G-protein heterotrimer and may be involved in transcriptional or post-transcriptional regulation of defense-related genes.
RESULTS
The xlg2 Mutation Results in Enhanced Susceptibility to Pseudomonas syringae
We have obtained T-DNA insertion lines for over 50 Arabidopsis genes that were rapidly induced following infection with the P. syringae pv. tomato strain DC 3000 (Pst) (Ge et al., 2007). These lines were screened using an in-planta bacterial growth assay to search for any mutation that alters resistance to Pst. Mutations in at least three of these genes led to alteration in the disease resistance phenotype. Among them, the loss-of-function mutation in the AtNUDT7 gene resulted in enhanced resistance to the infection (Ge et al., 2007). xlg2 is one of the two enhanced disease susceptibility mutants that were found in the screen to be more susceptible to Pst than wild-type (wt) plants. The xlg2 mutation results in a moderate but significant increase (approximately five-fold) in the growth rate of Pst in multiple independent in-planta growth assays, as exemplified in Figure 1A. Growth of both the avirulent strain Pst avrRpm1 as well as P. syringae pv. phaseolicola (Psp), a non-host strain on Arabidopsis, were also moderately increased in the xlg2 mutant plants compared to wt plants (Figure 1B and 1C). We did not observe any difference between the mutant and wt plants in appearance of hypersensitive cell death triggered by Pst avrRpm1 or Pst avrRpt2.
Figure 1.
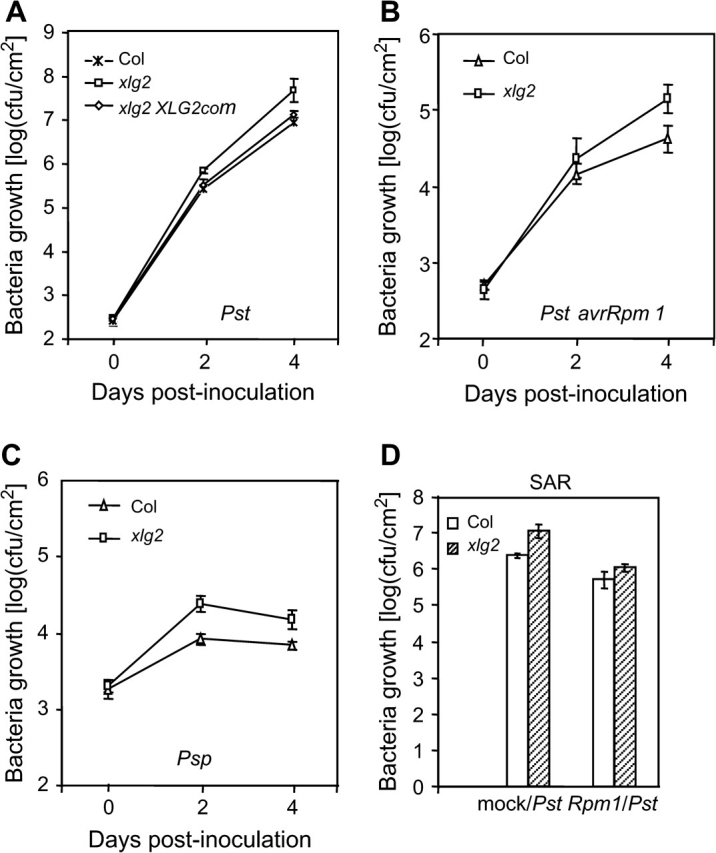
The xlg2 Mutation Leads to Enhanced Disease Susceptibility.
(A) The in-planta growth assays show a six-fold increase in bacterial growth in the mutant over the wt plants (Col). Leaves were hand-infiltrated with the virulent Pst strain and bacterial numbers in the infected leaves were counted 2 and 4 d post inoculation. The mutant's enhanced disease susceptibility phenotype was complemented by the XLG2 transgene (XLG2com) expressed in the xlg2 mutant.
(B, C) The xlg2 mutation also compromises resistance to the avirulent Pst avrRpm1 strain (B) and to the non-host strain Psp (C).
(D) The xlg2 mutant is not defective in induction of SAR. Induction of SAR by Pst avrRpm1 led to an over five-fold decrease in growth of Pst in both the mutant and wt plants. Rpm1/Pst: four to five leaves of each plant were inoculated with Pst avrRpm1 to induce SAR and, 3 d later, other upper leaves were infected with virulent Pst. Mock/Pst: no SAR induction prior to infection with Pst; plants had been mock-inoculated with water before infection with Pst. Shown are the bacterial colony forming units (cfus) in the leaves 4 d post infection with Pst. Each data point represents the average of three replicates ± standard deviation.
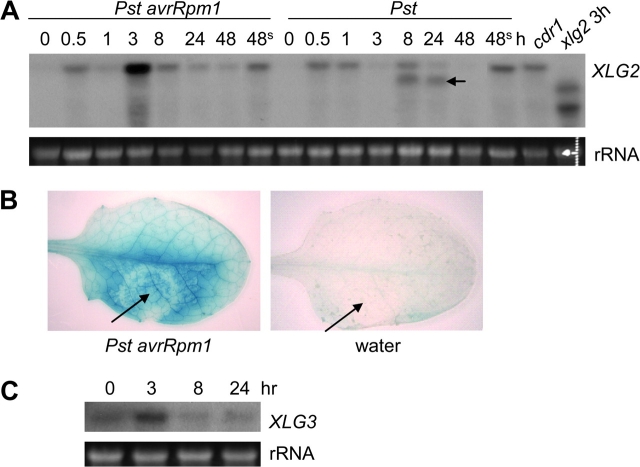
Northern blotting analysis showed that the XLG2 gene is expressed at a low level in uninfected leaves of wt plants (Figure 2A). Significant accumulation of the XLG2 transcript was detected within 30 min of inoculation with Pst avrRpm1 and reached its highest level 3 h post-inoculation (hpi). XLG2 was also quickly induced by the virulent Pst strain; however, its transcript level dropped by 3 hpi. At later time points, the Pst-infected leaves produced a smaller transcript (indicated by the arrow in Figure 2A), which is estimated to be approximately 300 bp smaller than the normal XLG2 transcript. It is unlikely that the smaller transcript was from cross-hybridization to XLG1 or XLG3 because it was not detected in the xlg2 mutant (Figure 2A). In addition, the hybridization was carried out under high stringency conditions meant to prevent such cross-hybridization. In the cdr1-D mutant, an enhanced disease resistance mutant that expresses many defense-related genes in the absence of pathogen infection (Xia et al., 2004), the XLG2 gene is constitutively expressed (Figure 2A).
Figure 2.
XLG2 Is an Early Pathogen-Responsive Gene.
(A) Expression profiles of XLG2 following infection with Pst avrRpm1 and Pst. RNA was isolated from wt leaves (except the last two lanes) taken at different time points following pathogen inoculation. The two lanes labeled with 48s are RNA samples of un-inoculated leaves taken from the plants that had previously been inoculated with either Pst avrRpm1 or Pst. cdr1: the RNA sample was isolated from uninfected cdr1-D mutant. xlg2, 3h: the RNA sample was isolated from the xlg2 mutant leaves at 3 hpi with Pst avrRpm1. The arrow points to the smaller XLG2 transcript.
(B) Induction of the XLG2pro::GUS reporter gene by Pst avrRpm1 revealed by GUS staining. The leaves were taken from XLG2pro::GUS transgenic plants 20 hpi with the pathogen (left panel) or with H2O as a negative control (right panel).
(C) XLG3 is also pathogen inducible. RNA was isolated from leaves of wt plants taken at different time points after inoculation with Pst avrRpm1.
As shown in Figure 2A, accumulation of the XLG2 transcript was detected in the uninfected leaves of the plants inoculated with Pst avrRpm1 or Pst, raising the possibility that XLG2 could be involved in systemic acquired resistance (SAR). To determine whether the xlg2 mutation affects induction of SAR, we first inoculated four to five leaves of each plant with Pst avrRpm1 by hand-infiltration with bacterial suspension at a concentration of 2 × 107 cfu ml−1 to induce SAR. Control plants were mock-inoculated (infiltrated with water). Three days later, un-inoculated leaves of these plants were infected with Pst. Growth of Pst was determined by counting bacterial numbers 4 d post-infection with Pst. In both wt and xlg2 plants pre-inoculated with Pst avrRpm1, growth of Pst was reduced by more than five-fold compared to the mock-inoculated control plants (Figure 1D). This result indicates that the xlg2 mutation does not impair induction of SAR. However, even with SAR induction, the growth rate of Pst in the xlg2 mutant was still higher than in wt plants, indicating that SAR induction did not compensate for the compromised local resistance caused by the xlg2 mutation.
Transgenic lines expressing the GUS reporter under the control of the XLG2 promoter (XLG2pro) were generated. Staining for the presence of GUS activity in Pst avrRpm1-infected and mock-infected leaves of the XLG2pro::GUS lines further revealed that pathogen infection does induce expression of the reporter gene (Figure 2B, left panel). The mock-inoculated leaves (right panel) showed a detectable but weak GUS activity, whereas the pathogen-inoculated area (left panel, as indicated by the arrow) and the surrounding area exhibited a much stronger GUS activity.
The xlg2 mutant line (SALK_062645) used in this study carries a T-DNA insertion in the second exon of the XLG2 gene (At4g34390). This T-DNA insertion line has previously been named as the xlg2-1 allele (Ding et al., 2008), and we use xlg2 to represent this mutant allele in this report. The xlg2 mutant does not produce normal XLG2 transcripts in either uninfected leaves or leaves infected with Pst avrRpm1 (Figure 2A), but rather produces two transcripts of smaller size when inoculated with Pst avrRpm1 (Figure 2A). The exact nature and role (if any) of these transcripts remain to be determined; however, no protein bands were detected from these plants by Western analysis using the anti-XLG2 antibodies (see below). To confirm that the enhanced disease susceptibility phenotype associated with the xlg2 mutant is indeed caused by disruption of XLG2 by the T-DNA insertion, we cloned the whole genomic fragment of this gene (including its 1.2-kb promoter region) from wt plants (Col-0 ecotype) and transformed it into the xlg2 mutant. The transgene (XLG2com) was found to be able to complement the mutant phenotype as revealed in the in-planta bacterial growth assay (Figure 1A). Two independent XLG2com lines were tested in the assay and both lines showed a disease resistance phenotype indistinguishable from wt plants. Shown in Figure 1A is the result from one of the XLG2com lines.
XLG3 Is Also Pathogen-Inducible
Our previous GeneChip microarray data showed that the XLG1 transcript level was not significantly changed whereas the XLG3 transcript level increased 2.3-fold (p < 0.01) after infection with Pst avrRpm1 (at 6 hpi) in wt plants. The pathogen-triggered induction of XLG3 was confirmed through RNA blot analysis (Figure 2C). However, two xlg3 T-DNA insertion lines (SALK_030162/xlg3-2 and SALK_141914/xlg3-3) and the xlg1-1 mutant (Ding et al., 2008) showed no difference in resistance to Pst from wt plants. Although the in-planta growth rates of Pst in the xlg2 xlg3-1 double mutant (Ding et al., 2008) were slightly higher than those in the xlg2 single mutant, the differences were often not statistically significant. Similarly, the xlg1-1 xlg2 xlg3-1 triple mutant (Ding et al., 2008) was found to be no different from either the xlg2 single mutant or the xlg2 xlg3-1 double mutant in the disease resistance phenotype. Furthermore, we did not find any significant difference between wt plants, the xlg1-1, xlg2, xlg3-2, xlg3-3 single mutants, the xlg2 xlg3-1 double mutant, and the triple mutant in resistance to two necrotrophic fungal pathogens, Botrytis cinerea and Alternaria brassicicola.
The xlg2 Mutation Compromises Induction of a Small Set of Pathogen-Responsive Genes
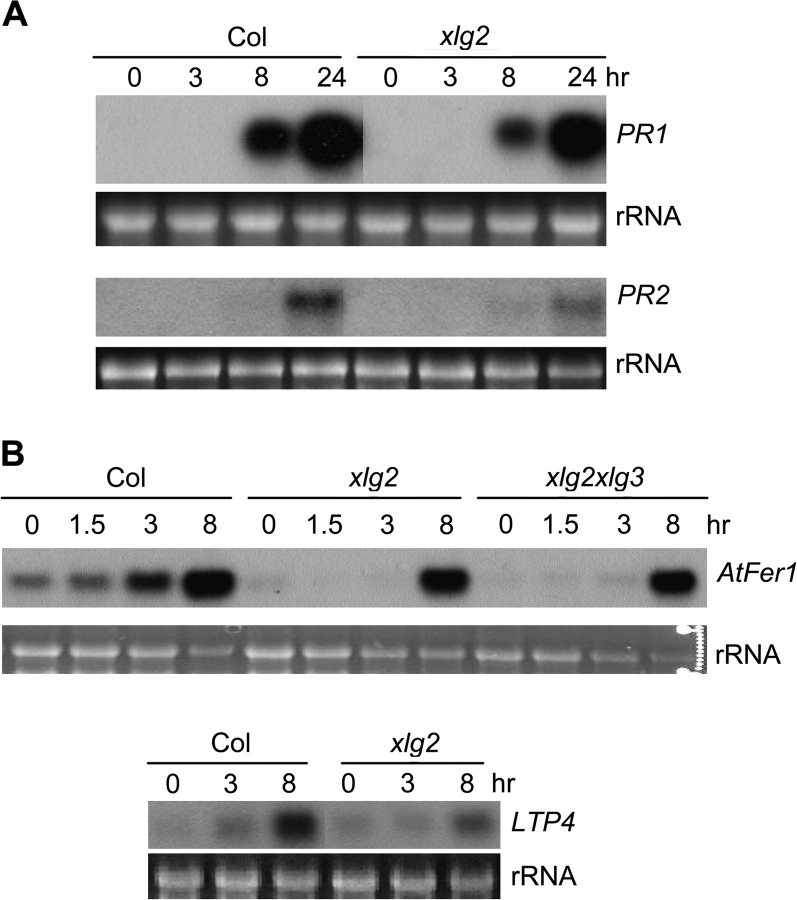
We initially used Northern blot analysis to determine whether the xlg2 mutation affects induction of pathogen-responsive genes. We compared expression profiles of several well known defense-related genes between the mutant and wt plants following inoculation with Pst avrRpm1 and/or Pst. These genes include PATHOGENESIS-RELATED GENE 1 (PR1), PR2, PR4, AvrRpt2-INDUCED GENE 1 (AIG1), and MITOGEN-ACTIVATED PROTEIN KINASE 3 (MPK3). Induction of PR1 and PR2 genes was found to be slightly weaker in the mutant following the Pst avrRpm1 infection (Figure 3A) but expression patterns of the other genes were not different between wt and the xlg2 mutant (data not shown).
Figure 3.
The xlg2 Mutation Affects Pathogen-Triggered Expression of Defense-Related Genes.
RNA samples were isolated from leaves infected with Pst avrRpm1 and uninfected leaves of wt and xlg2 mutant plants at the indicated times post infection. RNA blotting analysis was used for the detection of the transcripts. AtFer1: At5g01600; LTP4: At5g59310.
We then carried out a gene expression profiling experiment using Affymetrix's Arabidopsis gene expression chip ATH1. RNA used to hybridize the GeneChip was isolated from leaves of wt and xlg2 mutant plants 6 hpi with Pst avrRpm1. Six Affymetrix ATH1 arrays were used to hybridize the RNA samples, which included three biological replicates each for wt and the xlg2 mutant. Analyses of the transcriptome data revealed that the overall gene expression profiles in the xlg2 mutant and wt plants following pathogen infection were highly similar. Transcript levels of only a small set of genes were affected by the xlg2 mutation. From the subset of differentially expressed genes that were identified with p-values of less than 0.05, those whose expression levels differed by more than two-fold are listed in Table 1. Among them, 17 genes (including XLG2 and PR1) were expressed in the mutant at levels lower than in the wt plants, whereas another six genes were expressed at higher levels in the mutant. Expression patterns of some of the differentially expressed genes (including AtFer1 and LTP4) were further confirmed by RNA blotting analysis (Figure 3B). Eleven of the 17 genes whose expression was impaired in the xlg2 mutant were previously found to be pathogen-inducible according to the result from a separate Genechip transcriptome experiment (Table 1; see Methods).
Table 1.
A List of Genes that Were Differentially Expressed between Pst avrRpm1-Infected wt and xlg2 Mutant Leaves.
| Probe ID | Gene ID | Description of gene products | Fold difference |
|
| wt/xlg2 | Inf/uninf1 | |||
| 253257_at | At4g34390 | XLG2 | 0.03 | 8.80* |
| 251109_at | At5g01600 | FERRITIN 1 (AtFer1) | 0.15 | 1.89* |
| 247718_at | At5g59310 | non-specific lipid transfer protein (LPT4) | 0.18 | 1.56 |
| 253044_at | At4g37290 | Unknown | 0.29 | 2.19* |
| 247717_at | At5g59320 | non-specific lipid transfer protein (LPT3) | 0.38 | 0.59* |
| 264514_at | At1g09500 | cinnamyl alcohol dehydrogenase-like | 0.38 | 12.33* |
| 254098_at | At4g25100 | Fe superoxide dismutase | 0.38 | 0.74 |
| 267168_at | At2g37770 | similar to aldo/keto reductase | 0.41 | 10.11* |
| 252984_at | At4g37990 | cinnamyl alcohol dehydrogenase /ELI3-2 | 0.41 | 104.10* |
| 255127_at | At4g08300 | Nodulin MtN21 family protein | 0.43 | 1.89* |
| 266385_at | At2g14610 | PR1 | 0.44 | 76.32* |
| 263228_at | At1g30700 | Reticuline oxidase-like | 0.47 | 19.73* |
| 253103_at | At4g36110 | Auxin-induced protein | 0.47 | 2.67* |
| 265012_at | At1g24470 | similar to b-keto acyl reductase | 0.48 | 0.44* |
| 251438_s_at | At3g59930 | Defensin family protein | 0.48 | 1.35 |
| 247492_at | At5g61890 | unknown | 0.48 | 1.50* |
| 248205_at | At5g54300 | unknown | 0.50 | 1.10 |
| 246573_at | At1g31680 | copper amine oxidase-like | 2.39 | 0.59* |
| 266590_at | At2g46240 | BAG homolog | 2.39 | 1.88* |
| 246603_at | At1g31690 | copper amine oxidase-like | 2.47 | 0.55* |
| 259511_at | At1g12520 | copper/zinc superoxide dismutase | 2.63 | 1.23 |
| 265058_s_at | At1g52040 | myrosinase-binding protein | 2.71 | 2.62* |
| 266165_at | At2g28190 | copper/zinc superoxide dismutase | 3.02 | 0.73 |
The numbers in this column represent the fold difference in transcript levels of these genes between Pst avrRpm1-inoculated and mock-inoculated leaves of wt plants at 6 hpi. The * indicates that the difference is statistically significant (p < 0.01).
XLG2 Protein Turnover Is Regulated via the Proteasome-Mediated Protein Degradation Pathway
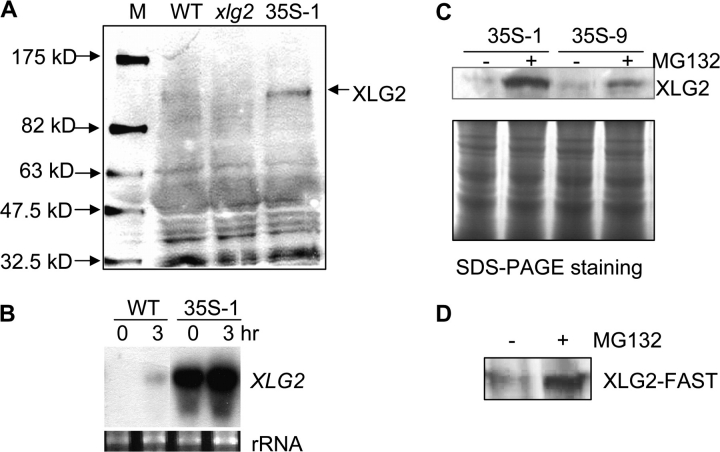
Polyclonal antibodies against a 14-amino acid peptide derived from XLG2 were raised, affinity-purified, and used to detect XLG2 in protein extracts through immunoblot analysis. The XLG2 level in crude protein extracts from uninfected leaves was below the limit of detection. In crude protein extracts from Pst avrRpm1-inoculated leaves, XLG2 protein was detectable, although faintly, by Western analysis (Figure 4A). No XLG2 protein was detected in the xlg2 mutant, as expected. We have generated XLG2 overexpression lines in which the XLG2 gene is under the control of the 35S promoter (35S). Figure 4B shows a high XLG2 transcript level in one of the 35S::XLG2 lines (35S::XLG2-1), as determined by RNA blot analysis (Figure 4B). The XLG2 protein level was also significantly higher in the overexpression line than in wt plants infected with the avirulent pathogen (Figure 4A). However, even protein extracts from the overexpression line required a long (∼5 min) exposure to X-ray film after incubation of the peroxidase-conjugated secondary antibody with its substrate in order to view the XLG2 protein band. Such a long exposure time led to the appearance of other protein bands that cross-reacted with the antibodies (Figure 4A).
Figure 4.
The XLG2 protein level is regulated by the proteasome-mediated protein degradation pathway.
(A) Detection of XLG2 protein with anti-XLG2 polyclonal antibodies through immunoblot analysis. Crude protein extracts were from leaves of wt, xlg2, and 35S::XLG2 transgenic line 35S-XLG2-1 (indicated as 35S-1 in the panel) plants 3 hpi with Pst avrRpm1. M, protein molecular weight markers.
(B) RNA blot showing XLG2 transcript levels in wt and the XLG2 overexpression line. Total RNA was isolated from uninfected (0 h) and infected (3 hpi with Pst avrRpm1) leaves of wt and 35S::XLG2-1 plants.
(C) MG132 treatment increased the XLG2 level in the two 35S::XLG2 lines (35S::XLG2-1 and 35S::XLG2-9 indicated as 35S-1 and 35S-9, respectively). MG132 (+) and water (–, as a negative control) were hand-infiltrated into leaves followed by protein extraction from the leaves 4 h after the infiltration. XLG2 protein was detected by immunoblot analysis (upper panel) using the anti-XLG2 polyclonal antibodies. The lower panel shows a Coomassie blue-stained gel with these protein samples.
(D) MG132 treatment increased the level of the XLG2–FAST fusion protein in the pathogen-infected leaves of XLG2pro::XLG2–FAST transgenic plants. Leaves were infiltrated with Pst avrRpm1 alone (–) or the pathogen together with MG132 (+). Protein was extracted 4 hpi and detected using the anti-FLAG M2 antibody.
In addition to the 35S::XLG2 lines, we generated transgenic Arabidopsis lines that carry the 35S::XLG2-GFP fusion construct. However, the GFP signal was very weak in these lines when observed by confocal microscopy, suggesting that these plants do not accumulate a sufficient level of the fusion protein. Together, these results suggest that the XLG2 protein level may be under tight control through translational and/or post-translational mechanisms. To determine whether XLG2 protein accumulation is subjected to regulation via the proteasome-mediated protein-degradation pathway, we infiltrated leaves with the synthetic proteasome inhibitor benzyloxycarbonyl-L-leucyl-L-leucyl-L-norvaline 4-methyl-coumaryl-7-amide (MG132) to block the proteasome activity. It was found that MG132 treatment significantly enhanced XLG2 accumulation in the XLG2 overexpression lines (Figure 4C). Similarly, in the XLG2pro::XLG2–FAST lines in which XLG2 is fused with the FAST epitope tag that consists of the FLAG tag and StrepII tag (Ge et al., 2005), the MG132 treatment also led to a much higher level of XLG2–FAST protein (Figure 4D).
XLG2 Interacts with AGB1 In Planta
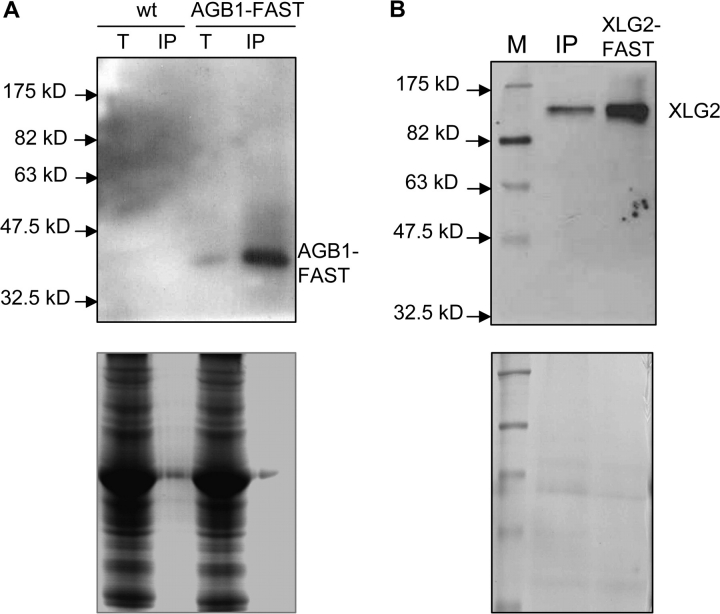
A hallmark of canonical Gα proteins is their interaction with Gβγ subunits, and, like the XLGs (Ding et al., 2008), the Arabidopsis Gβ subunit (AGB1) has been reported to be present in the nucleus (Obrdlik et al., 2000; Peskan and Oelmuller, 2000; Anderson and Botella, 2007). To determine whether XLG2 interacts with the Arabidopsis Gβ subunit of the heterotrimeric G-protein, we first used the yeast two-hybrid system. However, we were unable to detect any interaction between XLG2 and AGB1 in the yeast system. We then carried out a co-immunoprecipitation (co-IP) assay to determine whether XLG2 interacts with AGB1 in planta. We fused the epitope tag FAST to the C-terminus of AGB1 and generated Arabidopsis transgenic lines expressing the AGB1–FAST fusion protein under the control of the 35S promoter. Expression of AGB1–FAST in the transgenic lines was confirmed through immunoblot analysis of crude protein extracts from the transgenic plants using the anti-FLAG M2 monoclonal antibody (Figure 5A). For the co-IP experiment, the transgenic plants were infected with Pst avrRpm1 to induce expression of XLG2. AGB1–FAST was then immuno-precipitated from crude protein extracts of the infected leaves by its affinity to the anti-FLAG M2 antibody. AGB1–FAST, which has a predicted molecular mass of 41 kDa, was not visible from the immuno-precipitate in the Coomassie-stained SDS–PAGE gel; however, it was readily detected in the immuno-precipitate through immunoblot analysis (Figure 5A). We then used the anti-XLG2 polycolonal antibodies to detect XLG2 in the AGB1-containing immuno-precipitate. As shown in Figure 5B, XLG2 was also readily detected in the precipitate, demonstrating that XLG2 was co-precipitated with AGB1–FAST. We tried to determine if there is any interaction between AGB1 and XLG2 in uninfected leaf tissues using the same method; however, we could not detect any XLG2 in the precipitate, presumably because of an insufficient level of XLG2 in uninfected tissues, although we could not rule out the possibility that XLG2 may not interact with AGB1 in uninfected tissues.
Figure 5.
Co-Immunoprecipitation Assay Reveals that XLG2 Interacts with AGB1 In Planta. Proteins were extracted from leaves of the 35S::AGB1–FAST transgenic line (AGB1–FAST) and non-transgenic wt plants (wt) infected with Pst avrRpm1. T, total protein extract (wt); IP, immunoprecipitated protein.
(A) Detection of AGB1–FAST in the total protein extracts and the immunoprecipitates with the anti-FLAG M2 antibody in immunoblot analysis (upper panel). The lower panel shows Coomassie blue staining of the SDS–PAGE gel containing the same protein samples.
(B) Detection of XLG2 in the proteins co-immunoprecipitated by anti-FLAG M2 monoclonal antibody against AGB1–FAST. XLG2 presence in the immunoprecipitate (IP) was detected with anti-XLG2 polyclonal antibodies (upper panel). XLG2 protein in the precipitate was readily detectable. The lane labeled ‘XLG2–FAST’ contains XLG2–FAST protein, which was pulled down from 35S::XLG2–FAST transgenic plants using the anti-FLAG M2 antibody affinity gel, as a positive control for XLG2 detection in the immunoblot analysis. The lower panel shows Coomassie blue staining of the SDS–PAGE gel containing the same protein samples as the immunoblot.
Constitutive Overexpression of XLG2 Leads to Constitutive Accumulation of Abnormal Transcripts from Defense-Related Genes
Two XLG2 overexpression lines (35S::XLG2-1 and 35S::XLG2-9) were subjected to disease resistance phenotype analysis. None of the XLG2 overexpression lines showed any visible morphological difference from a wt plant. Nor did we find any difference between the overexpression lines and the wt plants in resistance to Pst or to the fungal pathogens B. cinerea and A. brassicicola. We also compared expression patterns of six well known pathogen-responsive genes between the overexpression lines and wt plants through RNA blot analysis. We did not find any significant difference in expression patterns of three of the genes (PR1, PR4, and AIG1) that were probed in the analysis (Figure 6). However, both XLG2 overexpression lines showed constitutive accumulation of transcripts from three other defense-related genes (AtMPK3/At3g45640, RbohC/At5g51060, and PAD3/At3g28630; Figure 6). Surprisingly, all three of these genes generated abnormally small transcripts in the overexpression lines. In the case of the RbohC gene, the smaller transcript (estimated to be 0.8–1 kb smaller than the normal transcript) appeared in both pathogen-infected wt tissues and the overexpression line, whereas the smaller transcripts from MPK3 (estimated to be 300–500 bp smaller than the normal transcript) and PAD3 were only detected in the overexpression lines. To investigate whether these abnormal transcripts result from alternative splicing, we attempted to amplify the alternative transcripts of these genes through reverse transcription PCR (RT–PCR) from RNA samples isolated from the XLG2 overexpression lines. Although multiple sets of primer combinations were used in the PCR analysis (Supplementary Data), we only detected RT–PCR products that have predicted normal sizes. The failure to detect alternatively spliced transcripts suggests that these abnormal transcripts may be derived from alternative transcription initiation or termination. It is also possible that these abnormal transcripts may be derived from an RNA processing mechanism that specifically removes either a 5’ or 3’ portion of the transcripts.
Figure 6.
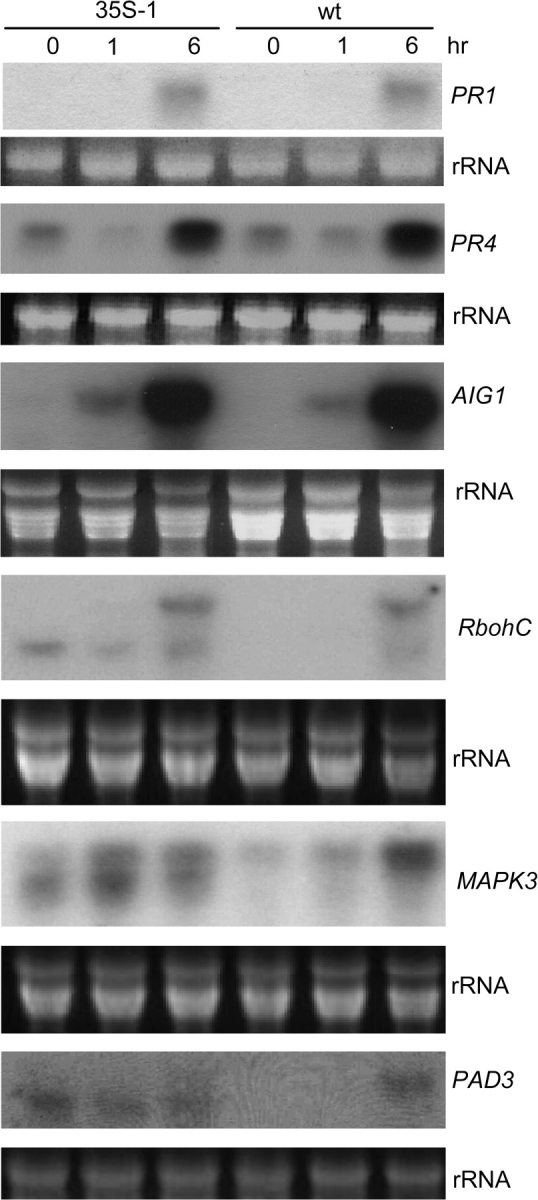
Constitutive Overexpression of XLG2 Leads to Constitutive Accumulation of Abnormal Transcripts of a Few Defense-Related Genes.
Transcript profiles of PR1, PR4, and AIG1 are similar between wt and the overexpression line 35-XLG2-1 (indicated as 35S-1); however, abnormal transcripts from three other genes were detected from the XLG2 overexpression line. RNA was isolated from uninfected leaves (0 h) and leaves infected with Pst avrRpm1, and evaluated with probes for defense-related genes in RNA blot analysis. The detection of PAD3 was carried out using Roche's DIG High Prime DNA labeling and Detection kit and the probes for all other genes were labeled with 32P.
DISCUSSION
Basal resistance, analogous to horizontal resistance, is largely a quantitative trait controlled by multiple genes (Young, 1996). Several important genes involved in basal as well as R-mediated resistance, including NPR1, EDS1, PAD4, and RAR1, have been identified through genetic screens (Cao et al., 1997; Falk et al., 1999; Jirage et al., 1999; Shirasu et al., 1999). That being said, mutations in many of as yet unknown genes that regulate basal immunity may result in only a minor change in the disease resistance phenotype, making it difficult to identify them through a classical forward genetic approach. The loss-of-function mutation of XLG2, which encodes one of the three unique Gα-like XLG proteins, was found to cause moderately enhanced susceptibility to P. syringae in a reverse genetic screen aiming to identify genes that function in basal immunity. The xlg2 mutation also compromises resistance to an avirulent strain of Pst and to the non-host strain Psp. Together, the results indicate that XLG2 has a positive regulatory role in the plant defense response against the bacterial pathogen. In addition to XLG2, transcription of XLG3 is induced by pathogen infection. However, the xlg2 xlg3 double mutant and the xlg1 xlg2 xlg3 triple mutant were not found to be significantly different from the xlg2 mutant in the tested disease resistance phenotypes.
Heterotrimeric G-proteins are involved in transducing extracellular signals from cell surface receptors into intracellular effectors (Neer, 1995; Neves et al., 2002). Although plants have only a small number of genes encoding the G-protein subunits, they have been implicated in a wide variety of biological processes (for reviews, see Jones and Assmann, 2004; Perfus-Barbeoch et al., 2004). Until now, very few downstream effectors of G-proteins have been identified from plants, and the molecular mechanisms by which G-proteins activate downstream signaling pathways in plant biological processes remain elusive. It has been proposed that a G-protein may activate a signaling event in response to stress conditions that lead to activation of NADPH oxidase(s) and production of reactive oxygen species (Suharsono et al., 2002; Joo et al., 2005).
All three regular subunits of the Arabidopsis G-protein have been reported to have roles in disease resistance. The Arabidopsis gpa1 mutant exhibited enhanced resistance to several necrotrophic fungal pathogens, including Plectosphaerella cucumerina, A. brassicicola, and Fusarium oxysporum (Llorente et al., 2005; Trusov et al., 2006). In contrast, mutations in AGB1 and AGG1 lead to enhanced susceptibility to necrotrophic fungal pathogens (Llorente et al., 2005; Trusov et al., 2006, 2007). These results suggest that the Gα subunit is a negative regulator while Gβ and Gγ are positive regulators in disease resistance. However, the rice Gα loss-of-function mutation (the d1 mutation) compromises resistance against an avirulent strain of the biotrophic fungal pathogen Magnaphorthe grisea, which causes rice blast (Suharsono et al., 2002). The d1 mutant is impaired in the hypersensitive response, H2O2 production, and defense gene induction triggered by the pathogen (Suharsono et al., 2002). In Arabidopsis, neither the Gα mutation nor the Gβ mutation alters resistance to virulent or avirulent strains of P. syringae, a biotrophic bacterial pathogen (Trusov et al., 2006). These studies suggest that the role of G-protein in plant disease resistance depends on the type of pathogen (necrotropic versus biotrophic and bacterial versus fungal) and that the different G-protein subunits may have different functions. The molecular mechanism by which the various subunits of G-proteins affect disease resistance also remains obscure. The agb1 mutant was found to be impaired in induction of PDF1.2 by either methyl jasmonate (MeJA) or A. brassicicola (Trusov et al., 2006). Our study revealed that XLG2 interacts with AGB1 and acts a positive regulator in resistance to P. syringae that largely triggers the SA response pathway. However, we did not observe any obvious differences between wt plants and any of the xlg mutants (including their double and triple mutants) in resistance to B. cinerea or A. brassicicola, which trigger the JA-responsive pathway, suggesting that XLG2 and AGB1 have distinct, and possibly antagonistic, functions in disease resistance through their interaction.
Because of significant sequence divergence between the XLGs and the canonical Gα, it was proposed by Temple and Jones (2007) that the XLGs are not likely to interact with the Gβγ subunit in a traditional fashion, if at all. In the yeast two-hybrid system, we could not detect interaction of XLG2 with AGB1; however, the co-immunoprecipitation assay demonstrated that XLG2 does interact with AGB1 in planta. These results indicate that interaction of XLG2 with AGB1 may be regulated through certain conformational changes that could be mediated through an intramolecular event (such as protein modification) or an intermolecular event (such as interaction with another protein).
The N-termini of the three XLGs contain a putative nuclear localization signal, and GFP fused with XLGs is localized in the nuclei in Vicia faba leaves as determined by a transient expression assay (Ding et al., 2008). The heterotrimeric forms of canonical G-proteins are generally associated with the plasma membrane, and localization of GPA1, AGB1, AGG1, and AGG2 at the plasma membrane has been confirmed (Adjobo-Hermans et al., 2006; Zeng et al., 2007; Wang et al., 2008). Accordingly, the discovery that XLGs localize in the nucleus suggests that XLGs might have a different mode of action from canonical Gα. However, the subunits of the canonical G-protein in plants have also been found in multiple compartments. For instance, the Gβ subunit has also been reported to be targeted to the nucleus when expressed in both Arabidopsis and tobacco cells (Obrdlik et al., 2000; Peskan and Oelmuller, 2000; Anderson and Botella, 2007). In another report, AGB1 was found to be largely associated with the endoplasmic reticulum (ER) and to play a role in the unfolded protein response (Wang et al., 2007). It is tempting to speculate that XLG2 may mediate information transfer from cell surface to nucleus in response to an extracellular signal such as pathogen recognition. In animals, Gβ5 undergoes nucleocytoplasmic shuttling, which is mediated by binding of regulators of G-protein signaling (RGS) to Gβ5 (Rojkova et al., 2003).
The gene expression microarray data show that overall defense transcriptomes in pathogen-infected leaves are very similar between the xlg2 mutant and wt plants. About 24 genes showed more than a two-fold difference in their expression levels between the mutant and wt plants. Most of these differentially expressed genes are pathogen-inducible. Although the result suggests that the xlg2 mutation alters defense gene induction, this small set of differentially expressed genes does not offer a clear insight into which particular defense signaling pathway is affected by the xlg2 mutation. Interestingly, constitutive overexpression of XLG2 leads to constitutive expression of three defense-related genes that were examined by the Northern blot analysis (MPK3, RbohC, and PAD3). Surprisingly, the accumulated transcripts of these three genes had abnormal sizes in the overexpression lines. These abnormal transcripts may be derived from alternative transcription initiation or termination or could be a product of RNA processing. The irregularity in transcriptional and/or post-transcriptional processing of these genes through the overexpression of XLG2, together with XLG2 localization in the nucleus, raise the possibility that XLG2 is a component of transcriptional and/or post-transcriptional regulation of defense-related genes.
METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis plants used in this study are the Columbia (Col-0) ecotype. The xlg2-1 (SALK_062645) and xlg3-2 and xlg3-3 T-DNA insertion mutants (SALK_030162 and SALK_141914) were obtained from the Arabidopsis Biological Resource Center (Columbus, OH). The xlg1-1 single mutant, xlg2-1 xlg3-1 double mutant, and xlg1-1 xlg2-1 xlg3-1 triple mutant were previously described (Ding et al., 2008). Arabidopsis plants used in the experiments were grown in a growth room with the following conditions: an 8-h light/16-h dark light cycle at a light intensity of 125 mol m−2 s−1 provided by cool-white fluorescent bulbs, 50% humidity and 21°C.
PCR Verification of T-DNA Insertions at XLG2 and XLG3
The T-DNA insertion in SALK_62645 was verified by PCR analysis using a T-DNA left-border primer (LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3’) and a XLG2-specific primer (XLGp1, 5′-AACTGGCAGAGAGAACACAGC-3’). The primers LBb1 and XLG3p1r (5′-TGGCCTGCAAATGAAGCCTAA-3’) were used to amplify the insertion-flanking sequence in SALK_030162 while the primer pair LBb1 and XLG3p3r (5′-GGGGTGCTTAACCATTGATTCG-3’) were used to amplify the insertion flanking sequence in SALK_141914.
RNA Blot Analysis
Total RNA was isolated from leaf tissues using the Trizol extraction buffer (Invitrogen, Carlsbad, CA) and the subsequent electrophoresis, probe labeling, and Northern blotting analysis was conducted according to standard procedures (Sambrook et al., 1989). A new RNA blot was used for each probe. DNA fragments used for making probes in RNA blot analysis were amplified from the genomic DNA using the following primer pairs: 5′-ATGAACACCGGCGGTGGCCAATAC-3’ and 5′-GCTCGGCTTTAAATCCCTATGAATAATG-3’ for AtMPK3, 5′-AATGAAAGTTTCAGTATCAC-3’ and 5′-GCAGTTAACTTGTTGTTGGC-3’ for WAK1, 5′-ATGTCTAGAGTGAGTTTTGAAG-3’ and 5′-TCCGGTCTAACTTAGCCGGTC-3’ for RbohC, 5′-TTAGTCAACGTTTATGCGATGGGTC-3’ and 5′-TGAAGAACTTGAAAGAAGGCTAGAA-3’ for PAD3, 5′-GGAGAGAGCGAGTAGGAAATAAG-3’ and 5′-CATAGGAGACGTTGTATTCCACAT-3’ for AtFer1/At5g01600, 5′-CCCAAAAGAGAAGAGCAAACAC-3’ and 5′-CTTACGTGGCGCAGTTGGTG-3’ for LTP4/At5g59310. The detection of PAD3 was carried out using Roche's DIG High Prime DNA labeling and Detection kit (Indianapolis, IN) and the probes for all other genes were labeled with 32P.
Microarray Gene Profiling and Data Analyses
Leaves of wt and the xlg2 mutant plants were inoculated with Pst avrRpm1 and collected for RNA isolation 6 h post-inoculation. RNA was isolated using the Trizol extraction method followed by RNA purification using the RNeasy MiniElute Cleanup kit (QIAGEN, Valencia, CA). Six Affymetrix ATH1 arrays were used to hybridize the RNA samples, including three biological replicates each for both wt and the xlg2 mutant. cRNA synthesis, labeling, hybridization and scanning were carried out at Iowa State University GeneChip Facility. The original microarray data (.cel files) were background corrected, quantile normalized, and summarized for each probe set using the affy package (Irizarry et al., 2003) in BioConductor with default settings. The summary scores were then analyzed probe set by probe set using the MAANOVA package (Wu et al., 2003) in BioConductor. A shrinkage-based t-test was conducted to compare the two groups (Cui et al., 2005). An empirical p-value was obtained for each probe set based on permutation of the observed data (Yang and Churchill, 2007). Differentially expressed genes were identified with the constraint of p-values of less than 0.05. From this subset of genes, the genes that showed a fold-change of greater than two-fold are listed in Table 1. The fold changes of these genes between pathogen-inoculated and mock-inoculated plants shown in Table 1 are from the result of a separate Genechip experiment aimed to identify pathogen-responsive genes. In that experiment, leaves of wild-type plants (Col) were inoculated with Pst avrRpm1 or water (control) and collected at 6 hpi for RNA extraction. Three biological replicates were included in the analysis. The methods for chip hybridization and data analysis are the same as above except that the false-discovery rate (FDR) was used as the multiple comparison correction. The genes that showed a significant difference in their transcript levels between the pathogen-infected sample and the control sample (with FDR-corrected p-values of less than 0.01) are indicated with * in Table 1.
GUS Activity Staining
GUS activity staining of leaves from the XLG2pro::GUS transgenic plants was carried out as previously described (Xia et al., 1997). Leaves were stained overnight (approximately 15 h), cleared in 100% ethanol overnight, and kept in 70% ethanol before images were taken.
Pathogen Treatment and MG132 Treatment
P. syringae pv. tomato (Pst), Pst avrRpm1, Pst avrRpt2, P. syringae pv. phaseolicola (Psp), and P. syringae pv. maculicola (Psm) were grown as previously described (Cameron et al., 1994). For in-planta growth assays, bacterial suspensions at a concentration of 5 × 104 cfu ml−1 (for Pst), 1 × 105 cfu ml−1 (for Pst avrRpm1), or 2 × 105 cfu ml−1 (Psp) were hand-infiltrated into Arabidopsis leaves. To prepare tissues used for studying pathogen-induced expression of defense-related genes or proteins, leaves were infiltrated with bacterial suspensions at a concentration of 2 × 107 cfu ml−1. Mock inoculations were infiltrated with water.
For MG132 treatment, a 50-μM solution of MG132 (Sigma-Aldrich, St Louis, MO) was hand-infiltrated into leaves. In the pathogen and MG132 co-treatment experiment, MG132 was added to pathogen suspensions to a concentration of 50 μM and the mixture was then immediately infiltrated into the leaves.
Construction of the XLG2 Complementation Vector (XLG2com), XLG2pro::GUS, XLG2 pro::XLG2–FAST, 35S::AGB1–FAST, and 35S::XLG2
The XLG2 genomic fragment, including the 1.3-kb promoter region, was PCR-amplified from Col plants by using the primer pair XLG2p5Bam (5′-GGATTCGCATGAAGGAAGTGGC-3’) and XLG2p3Sal (5′-GTCGACCGATAATTTATTGCTACTCCG-3’), subcloned into pCR–BluntII–TOPO vector, verified by sequencing, excised by cutting with BamHI and SalI, and ligated into the binary vector pBI101.3 to generate XLG2comp. To construct XLG2pro::GUS, a pair of primers, (XLG2p5Sal, 5′-GTCGACGAGCCAGCAGCATCTTC-3’) and (XLG2p3Bam, 5′-GGATCCCAATCAAGCACACATACAAAC-3’), was used to amplify the 1.3-kb promoter sequence. The PCR fragment was subcloned into pCR–BluntII–TOPO, excised by cutting with SalI/BamHI, and inserted upstream of the GUS reporter gene in the binary vector pBI101.3. For construction of XLG2pro::XLG2–FAST, a ∼4.5-kb XLG2 genomic fragment, including its ∼1.2-kb promoter region, was PCR amplified with the primer pair (5′-TCCCCCGGGAAGTGGCGCGTGGAGTTC-3’ and 5′-GCTCTAGAAGAGGACGAGCTGGCCTC-3’), cloned into pCR–BluntII–TOPO, verified by sequencing, released by cutting with SmaI and XbaI, and subcloned into the SmaI/XbaI-digested vector pAKK–BAR–FAST, a plant binary vector constructed by modifying the epitope-tagging vector previously described (Ge et al., 2005). The clone results in XLG2 fused in frame with the FLAG and StrepII tags. To construct 35S::AGB1–FAST, the AGB1 cDNA clone, including the entire ORF, was amplified with primers (5′-ATGGTACCATGTCTGTCTCCGAGCTC-3’ and 5′-CTTCTAGAAATCACTCTCCTGTGTCCTC-3’) by RT–PCR and cloned into pCR–BluntII–TOPO. The KpnI/XbaI fragment was digested out and subcloned into 35S::pAKK–BAR–FAST (a binary vector based on pAKK–BAR–FAST with the 35S promoter upstream of the FAST tag). The resulting clone was named 35S:AGB1–FAST. Construction of 35S::XLG2 was carried out by amplifying the XLG2 genomic fragment (without its promoter region) with the primer pair XLG2p5BamC (5′-GGATCCTTGATTGGGTAAGAAGATGG-3’) and XLG2p3Sal. The PCR product was cloned into pCR–BluntII–TOPO and then inserted downstream of the 35S promoter in the binary vector pCHF3 to generate 35S::XLG2.
Arabidopsis Transformation
Arabidopsis was transformed via Agrobacterium tumefaciens-mediated transformation as previously described (Clough and Bent, 1998).
Immunoblot Assays and Antibodies
To isolate protein extract, Arabidopsis tissues were homogenized in CelLytic™ P Plant Cell Lysis/Extraction Reagent (Sigma-Aldrich) with 1×protease inhibitor cocktail (Sigma-Aldrich). The extract was centrifuged at 12 000 g at 4°C for 15 min. The supernatant was collected and the protein concentration was determined by Bradford assay (Bio-Rad, Hercules, CA). Protein samples were boiled in SDS-loading buffer, separated on SDS–PAGE gels and transferred onto nitrocellulose membranes (Osmonics, Gloucester, MA). For production of anti-XLG2 polyclonal antibodies, a peptide of XLG2 (KRLDVPEEVKSPADC) was synthesized and used for raising polyclonal antibodies in rabbits (carried out by Sigma-Genosys). The antibodies were affinity-purified with the peptide by Sigma-Genosys and used for immunoblot analyses at a 1:1000 dilution.
Co-Immunoprecipitation Assay
Leaves from 35S::AGB1–FAST transgenic plants and from non-transgenic wt plants were hand-infiltrated with Pst avrRpm1 and collected at different time points (20 min, 30 min, 1 h, 3 h, and 6 h post infection). A mixture of infected leaves was used for the total protein extraction. Protein extraction and co-immunoprecipitation with the anti-FLAG M2 antibody gel (Sigma-Aldrich) were carried out according to the procedure previously described (Feng et al., 2004). Briefly, total protein extract was clarified by centrifuging at 12 000 g at 4°C for 15 min. Samples of 5–10 ml of supernatant were incubated with 50–100 μl of anti-FLAG affinity gel (Sigma) at 4°C for 4–5 h with gently shaking. The samples with resin were transferred into Pierce spin columns (cat. # 69705, Pierce, Rockford, IL) and spun at 300 g briefly to collect resin. After washing with the extraction buffer four times, protein bound to the affinity gel was eluted with 3 FLAG peptide (Sigma-Aldrich). The eluted immunoprecipitate was kept at –80°C before use in immunoblot analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by the National Institutes of Health (grant no. GM076420 to Y.-J.X.) and by grants from the USDA (2003-35304-13924) and the NSF (MCB-0209694) to S.M.A.
Supplementary Material
Acknowledgments
We thank Barbara Kunkel for all Pst strains, Jianmin Zhou for the Psp strain and Anita Snyder for editing the manuscript. No conflict of interest declared.
References
- Abramovitch RB, Martin GB. Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 2004;7:356–364. doi: 10.1016/j.pbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Adjobo-Hermans MJW, Goedhart J, Gadella TWJ., Jr Plant G protein heterotrimers require dual lipidation motifs of Gα and Gγ and do not dissociate upon activation. J. Cell Sci. 2006;119:5087–5097. doi: 10.1242/jcs.03284. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Botella JR. Expression analysis and subcellular localization of the Arabidopsis thaliana G-protein β-subunit AGB1. Plant Cell Reports. 2007;26:1469–1480. doi: 10.1007/s00299-007-0356-1. [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- Assmann SM. Plant G proteins, phytohormones, and plasticity: three questions and a speculation. Science's Stke [Electronic Resource]: Signal Transduction Knowledge Environment. 2004;2004 doi: 10.1126/stke.2642004re20. re20. [DOI] [PubMed] [Google Scholar]
- Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A novel role for the TIR domain in association with pathogen-derived elicitors. Plos. Biology. 2007;5:e68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Vera TM, Vanhauwe J, Thomas TO, Medkova M, Preininger A, Mazzoni MR, Hamm HE. Insights into G protein structure, function, and regulation. Endocr. Rev. 2003;24:765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- Cameron RK, Dixon RA, Lamb CJ. Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J. 1994;5:715–725. [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- da Cunha L, Sreerekha M-V, Mackey D. Defense suppression by virulence effectors of bacterial phytopathogens. Curr. Opin. Plant Biol. 2007;10:349–357. doi: 10.1016/j.pbi.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Dangl JL. PLANT SCIENCE: Nibbling at the plant cell nucleus. Science. 2007;315:1088–1089. doi: 10.1126/science.1138991. [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl Acad. Sci. U S A. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl Acad. Sci. U S A. 2002;99:2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Pandey S, Assmann SM. Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J. 2008;53:248–263. doi: 10.1111/j.1365-313X.2007.03335.x. [DOI] [PubMed] [Google Scholar]
- Eulgem T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 2005;10:71–78. doi: 10.1016/j.tplants.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JD, Daniels MJ, Parker JE. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl Acad. Sci. U S A. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Shen Y, Sullivan JA, Rubio V, Xiong Y, Sun TP, Deng XW. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell. 2004;16:1870–1882. doi: 10.1105/tpc.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Dietrich C, Matsuno M, Li G, Berg H, Xia Y. An Arabidopsis aspartic protease functions as an anti-cell-death component in reproduction and embryogenesis. EMBO Rep. 2005;6:282–288. doi: 10.1038/sj.embor.7400357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Li G-J, Wang S-B, Zhu H, Zhu T, Wang X, Xia Y. AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol. 2007;145:204–215. doi: 10.1104/pp.107.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- Gookin T, Kim J, Assmann S. Whole proteome identification of plant candidate G-protein coupled receptors in Arabidopsis, rice, and poplar: computational prediction and in-vivo protein coupling. Genome Biol. 2008;9:R120. doi: 10.1186/gb-2008-9-7-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt BF, Boyes DC, Ellerström M, Siefers N, Wiig A, Kauffman S, Grant MR, Dangl JL. An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Developmental Cell. 2002;2:807–817. doi: 10.1016/s1534-5807(02)00174-0. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl Acad. Sci. U S A. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Assmann SM. Plants: the latest model system for G-protein research. EMBO Rep. 2004;5:572–578. doi: 10.1038/sj.embor.7400174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Joo JH, Wang S, Chen JG, Jones AM, Fedoroff NV. Different signaling and cell death roles of heterotrimeric G protein α and β subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell. 2005;17:957–970. doi: 10.1105/tpc.104.029603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl Acad. Sci. U S A. 2006;103:11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Assmann SM. Arabidopsis thaliana ‘extra-large GTP-binding protein’ (AtXLG1): a new class of G-protein. Plant Mol. Biol. 1999;40:55–64. doi: 10.1023/a:1026483823176. [DOI] [PubMed] [Google Scholar]
- Llorente F, Alonso-Blanco C, Sanchez-Rodriguez C, Jorda L, Molina A. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 2005;43:165–180. doi: 10.1111/j.1365-313X.2005.02440.x. [DOI] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM. Molecular cloning and characterization of GPA1, a G protein α subunit gene from Arabidopsis thaliana. Proc. Natl Acad. Sci. U S A. 1990;87:3821–3825. doi: 10.1073/pnas.87.10.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR. Completing the heterotrimer: isolation and characterization of an Arabidopsis thaliana G protein γ-subunit cDNA. Proc. Natl Acad. Sci. U S A. 2000;97:14784–14788. doi: 10.1073/pnas.97.26.14784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason MG, Botella JR. Isolation of a novel G-protein γ-subunit from Arabidopsis thaliana and its interaction with Gbeta. Biochim. Biophys. Acta. 2001;1520:147–153. doi: 10.1016/s0167-4781(01)00262-7. [DOI] [PubMed] [Google Scholar]
- Moriyama EN, Strope PK, Opiyo SO, Chen Z, Jones AM. Mining the Arabidopsis thaliana genome for highly-divergent seven transmembrane receptors. Genome Biol. 2006;7:R96. doi: 10.1186/gb-2006-7-10-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 2004;135:1113–1128. doi: 10.1104/pp.103.036749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer EJ. Heterotrimeric G proteins: organizers of transmembrane signals. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 2002;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- Nurnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- Obrdlik P, Neuhaus G, Merkle T. Plant heterotrimeric G protein β subunit is associated with membranes via protein interactions involving coiled-coil formation. FEBS Lett. 2000;476:208–212. doi: 10.1016/s0014-5793(00)01706-3. [DOI] [PubMed] [Google Scholar]
- Pandey S, Monshausen GB, Ding L, Assmann SM. Regulation of root-wave response by extra large and conventional G proteins in Arabidopsis thaliana. Plant J. 2008;55:311–322. doi: 10.1111/j.1365-313X.2008.03506.x. [DOI] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Jones AM, Assmann SM. Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr. Opin. Plant Biol. 2004;7:719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Peskan T, Oelmuller R. Heterotrimeric G-protein β-subunit is localized in the plasma membrane and nuclei of tobacco leaves. Plant Mol. Biol. 2000;42:915–922. doi: 10.1023/a:1006477631166. [DOI] [PubMed] [Google Scholar]
- Rojkova AM, Woodard GE, Huang TC, Combs CA, Zhang JH, Simonds WF. Gγ subunit-selective G protein β5 mutant defines regulators of G protein signaling protein binding requirement for nuclear localization. J. Biol. Chem. 2003;278:12507–12512. doi: 10.1074/jbc.M207302200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. New York: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Shen QH, Schulze-Lefert P. Rumble in the nuclear jungle: compartmentalization, trafficking, and nuclear action of plant immune receptors. EMBO J. 2007;26:4293–4301. doi: 10.1038/sj.emboj.7601854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- Shirasu K, Lahaye T, Tan M-W, Zhou F, Azevedo C, Schulze-Lefert P. A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell. 1999;99:355–366. doi: 10.1016/s0092-8674(00)81522-6. [DOI] [PubMed] [Google Scholar]
- Suharsono U, Fujisawa Y, Kawasaki T, Iwasaki Y, Satoh H, Shimamoto K. The heterotrimeric G protein α subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc. Natl Acad. Sci. U S A. 2002;99:13307–13312. doi: 10.1073/pnas.192244099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang H-S, Han B, Zhu T, Zou G, Katagiri F. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell. 2003;15:317–330. doi: 10.1105/tpc.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple BR, Jones AM. The plant heterotrimeric G-protein complex. Ann. Rev. Plant Biol. 2007;58:249–266. doi: 10.1146/annurev.arplant.58.032806.103827. [DOI] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, Botella JR. Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiol. 2006;140:210–220. doi: 10.1104/pp.105.069625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, Anderson D, Chen J-G, Jones AM, Botella JR. Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. Plant Cell. 2007;19:1235–1250. doi: 10.1105/tpc.107.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang X-C, Neece D, Ramonell KM, Clough S, Kim S-Y, Stacey MG, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Assmann SM, Fedoroff NV. Characterization of the Arabidopsis heterotrimeric G protein. J. Biol. Chem. 2008 doi: 10.1074/jbc.M801376200. M801376200. [DOI] [PubMed] [Google Scholar]
- Wang S, Narendra S, Fedoroff N. Heterotrimeric G protein signaling in the Arabidopsis unfolded protein response. Proc. Natl Acad. Sci. U S A. 2007;104:3817–3822. doi: 10.1073/pnas.0611735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss CA, Garnaat CW, Mukai K, Hu Y, Ma H. Isolation of cDNAs encoding guanine nucleotide-binding protein β-subunit homologues from maize (ZGB1) and Arabidopsis (AGB1) Proc. Natl Acad. Sci. U S A. 1994;91:9554–9558. doi: 10.1073/pnas.91.20.9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Kerr MK, Cui X, Churchill GA. MAANOVA, a software package for the analysis of spotted cDNA microarray experiments. In: Parmigiani G, Garret ES, Irizarry RA, Zeger SL, editors. The Analysis of Gene Expression Data: An Overview of Methods and Software. New York: Springer, NY; 2003. [Google Scholar]
- Xia Y, Nikolau BJ, Schnable PS. Developmental and hormonal regulation of the Arabidopsis CER2 gene that codes for a nuclear-localized protein required for the normal accumulation of cuticular waxes. Plant Physiol. 1997;115:925–937. doi: 10.1104/pp.115.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Suzuki H, Borevitz J, Blount J, Guo Z, Patel K, Dixon RA, Lamb C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 2004;23:980–988. doi: 10.1038/sj.emboj.7600086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Churchill G. Estimating p-values in small microarray experiments. Bioinformatics. 2007;23:38–43. doi: 10.1093/bioinformatics/btl548. [DOI] [PubMed] [Google Scholar]
- Young ND. QTL mapping and quantitative disease resistance in plants. Annu. Rev. Phytopathol. 1996;34:479–501. doi: 10.1146/annurev.phyto.34.1.479. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Wang X, Running MP. Dual lipid modification of Arabidopsis Gγ-subunits is required for efficient plasma membrane targeting. Plant Physiol. 2007;143:1119–1131. doi: 10.1104/pp.106.093583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, He SY, Assmann SM. The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. Plant J. 2008;56:984–996. doi: 10.1111/j.1365-313X.2008.03657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.