Abstract
Biotransformation of inorganic arsenic (iAs) involves methylation catalyzed by arsenic (+ 3 oxidation state) methyltransferase (As3mt) yielding mono-, di-, and trimethylated arsenicals. To investigate the evolution of molecular mechanisms that mediate arsenic biotransformation, a comparative genomic approach focusing on the invertebrate chordate Ciona intestinalis was used. Bioinformatic analyses identified an As3mt gene in the C. intestinalis genome. Constitutive As3mt RNA expression was observed in heart, branchial sac, and gastrointestinal tract. Adult animals were exposed to 0 or 1 ppm of iAs for 1 or 5 days. Steady-state As3mt RNA expression in the gastrointestinal tract was not modulated significantly by 5 days of exposure to iAs. Tissue levels of iAs and its methylated metabolites were determined by hydride generation-cryotrapping-gas chromatography-atomic absorption spectrometry. At either time point, exposure to iAs significantly increased concentrations of iAs and its methylated metabolites in tissues. After 5 days of exposure, total speciated arsenic concentrations were highest in branchial sac (3705 ng/g), followed by heart (1019 ng/g) and gastrointestinal tract (835 ng/g). At this time point, the sum of the speciated arsenical concentrations in gastrointestinal tract and heart equaled or exceeded that of iAs; in branchial sac, iAs was the predominant species present. Ciona intestinalis metabolizes iAs to its methylated metabolites, which are retained in tissues. This metabolic pattern is consistent with the presence of an As3mt ortholog in its genome and constitutive expression of the gene in prominent organs, making this basal chordate a useful model to examine the evolution of arsenic detoxification.
Keywords: arsenic, Ciona intestinalis, comparative genomics, methyltransferases, toxicogenomics
Inorganic arsenic (iAs) is a ubiquitous component of the biosphere, and organisms have evolved a variety of strategies to handle this potentially toxic metalloid. In bacteria, genes in the ars operon encode proteins that catalyze reduction of pentavalent arsenic (As) to trivalency and extrusion of trivalent arsenic from cells (Lin et al., 2007; Rosen, 2002a,b). The bacterial arsM gene encodes an As methyltransferase that facilitates extrusion of As by catalyzing formation of volatile methylarsines (Qin et al., 2006; Yaun et al., 2008). In vertebrates, including humans, ingested or inhaled iAs is converted to methylated species (Hughes, 2006; Tseng, 2007). Therefore, inorganic, methylated, and dimethylated arsenicals are found in urine of humans exposed to iAs. Arsenic (+ 3 oxidation state) methyltransferase (As3mt), the prototype As methyltransferase in mammals, was initially identified in Rattus norvegicus (Lin et al., 2002; Thomas et al., 2007). Genomic analyses in organisms ranging in complexity from the sea urchin Strongylocentrotus purpuratus to Homo sapiens have identified candidate genes that could encode As3mt-like proteins. For vertebrates, occurrence of a predicted As3mt ortholog in the genome correlates with reports of the presence of methylated arsenicals in tissues or urine after exposure to iAs (Li et al., 2005; Thomas et al., 2007). Recombinant As3mt from human, rat, and mouse catalyzes the S-adenosylmethionine (AdoMet)-dependent conversion of arsenicals to methylated metabolites (Fomenko et al., 2007; Walton et al., 2003).
Here, we have identified an orthologous As3mt gene in the sea squirt Ciona intestinalis and examined the metabolism of iAs in this primitive chordate. Ciona intestinalis is the closest living invertebrate relative of vertebrates, representing a lineage that diverged from the last common ancestor of all vertebrates at least 500 million years ago (Blair and Hedges, 2005; Dehal et al., 2002; Delsuc et al., 2006). Evolution of C. intestinalis likely occurred in an environment that contained iAs. Background levels of total iAs in seawater range from about 1 to 5 μg/l, although higher levels are often found near point discharge sources (Sidiq, 1992). Adult specimens of C. intestinalis filter seawater at a rate of 2–5.5 l/g/day (Goodbody, 1974; Petersen and Svane, 2002). At background levels of total iAs in seawater, this would result in exposures ranging from 2 to 27.5 μg/g/day. If absorption of As from filtered water were 10%, then absorbed doses would range from about 0.2 to 2.8 μg/g/day. Although there are no data on rates of As absorption by C. intestinalis, this organism absorbs oxyanionic vanadate from seawater (Bielig et al., 1961). This suggests that an oxyanionic As species (e.g., arsenate) could be absorbed from seawater. In this case, the steady-state concentration of As in tissues of C. intestinalis would be determined by fractional absorption of As from filtered seawater and the rate of clearance of As from the organism. Coordinating the formation of methylated arsenicals, some of which are more reactive than iAs, with clearance processes can minimize accumulation of toxic species and reduce the risk associated with exposure to this toxicant (Thomas, 2007; Thomas et al., 2001). Here, we report that C. intestinalis rapidly accumulates iAs present in seawater and converts it to mono-, di-, and trimethylated arsenicals. The capacity of C. intestinalis to convert iAs is associated with the expression of a gene encoding a protein with considerable sequence identity to As3mt identified in other species.
MATERIALS AND METHODS
Experimental animals.
Ciona intestinalis specimens were obtained from Marine Biology Laboratory (Woods Hole, MA) and maintained in circulating seawater tanks (average seawater temperature 15°C) at Mount Desert Island Biological Laboratory (http://www.mdibl.org/). Animals were adjusted to the new environmental conditions for at least 5 days before use.
Preparation of As-containing seawater.
Sodium arsenite (NaAsO2; Sigma, St Louis, MO) was dissolved in water at 1M concentration as stock solution. It was used at 13.3μM (equivalent to 1 ppm) by diluting the stock solution into fresh seawater containing approximately 1 ppb of iAs.
Exposure conditions.
Adult C. intestinalis specimens (average length = 5.6 ± 0.5 cm) were maintained in a 15°C seawater tank with the water constantly circulating through a standard aquarium pump. Individual animals were attached to a fiberglass mesh screen via suture of the basal tunic to enable continual submersion and equal distribution in the seawater tank. Animals were randomly divided into two groups (n = 8–11): (1) 1 ppm arsenite in seawater and (2) control unamended seawater. Two independent experiments of 1- or 5-day exposure to arsenite (or control seawater) were conducted. For the 5-day treatment, fresh seawater was replaced daily.
Tissue collection.
At exposure’s end, animals were anesthetized by immersion in a saturated magnesium chloride solution (Petersen and Svane, 2002). Animals were then placed in a petri dish and dissected under a low-magnification microscope. Heart, gastrointestinal tract, and branchial sac were collected separately, washed twice in PBS, placed in cryovials, and snap frozen in liquid N2. Tissue samples were stored at −20°C until analysis.
Genomic analyses.
The sequence of a candidate As3mt gene (ENSCING00000015865) was retrieved from the C. intestinalis genome (www.ensembl.org/Ciona_intestinalis/index.html). A systematic approach was followed to infer gene homology. This included the use of methods based on hit clustering (Wapinski et al., 2007) and using the results (hits) from sequence similarity searches among highly annotated proteins from human, rat, and mouse to derive an orthology assignment between genes. First, the widely used approach of reciprocal (bidirectional) best hit (RBH) (Wall et al., 2003) was applied. BLASTP (Altschul et al., 1997) analysis was then used to back search the full retrieved amino acid sequence against RefSeq (NCBI Reference Sequence project) protein sequences. Second, the Pfam algorithm (Bateman et al., 2004) was used for the identification of conserved domains within the predicted C. intestinalis sequence. Third, the highly annotated and curated SWISS-PROT and TrEMBL protein databases (Bairoch et al., 2004) were used to assess statistical significance of pairwise sequence similarity for the C. intestinalis As3mt sequence using probability of random shuffle (PRSS) and SSEARCH analyses (Pearson, 1991; Smith and Waterman, 1981). Finally, BLAST search analysis against the OrthoMCL database was used for the identification of orthologous groups across multiple taxa (Chen et al., 2006). Sequences from the OrthoMCL As3mt orthologous group (accession number OG2_76202) including Chlamydomonas reinhardtii (195338), C. intestinalis (ENSCINP00000027827), Cyanidioschyzon merolae_1 (CMT024C), C. merolae_2 (CME010C), Danio rerio_1 (ENSDARP00000085262), D. rerio_2 (ENSDARP00000057878), Debaryomyces hansenii (DEHA0D15466g), Dehalococcoides ethenogenes 195 (YP_182128.1), Filobasidiella neoformans (185.m02734), Gallus gallus (ENSGALP00000013184), Halobacterium sp. NRC-1 (NP_046066.1), H. sapiens (ENSP00000358896), Mus musculus (ENSMUSP0000000365), Neurospora crassa (NCU05616), Ostreococcus tauri_1 (Ot17g00760), O. tauri_2 (Ot17g00590), R. norvegicus (ENSRNOP00000051584), Rhodopirellula baltica SH 1 (NP_867437.1), Synechococcus sp. WH 8102 9 (NP_896777.1), Tetraodon nigroviridis_1 (GSTENP00022702001), T. nigroviridis_2 (GSTENP00022701001), Thalassiosira pseudonana_1 (140233), and T. pseudonana_2 (21295) were retrieved from OrthoMCL database. In addition, reference protein sequences (NCBI database) from Bos taurus (NP_001030195.1), Canis familiaris (XP_543995.2), Macaca mulatta (XP_001113391.1), Ornithorhynchus anatinus (XP_001511844.1), Pan troglodytes (XP_508007.2), and Xenopus tropicalis (AAI33181.1) were retrieved from the GenBank as well as Takifugu rubripes (ENSXETP00000020854) from Ensembl database. These sequences were aligned using the ClustalW software (Larkin et al., 2007), and phylogenetic trees were constructed with the maximum parsimony method (Eck and Dayhoff, 1966) using MEGA software version 4.0 (Tamura et al., 2007).
Manual annotation of the C. intestinalis As3mt gene was further validated with in silico cloning analysis (prediction of a gene product sequence using only genomic and expressed sequence tags [ESTs] sequence data) (Boguski et al., 1994). The larger transcript sequence (ENSCINT00000028073) of the predicted As3mt gene was retrieved from Ensembl project website and used as query for BLASTN analysis (Zhang et al., 2000) against the GenBank EST database (Boguski et al., 1993).
Gene expression analysis.
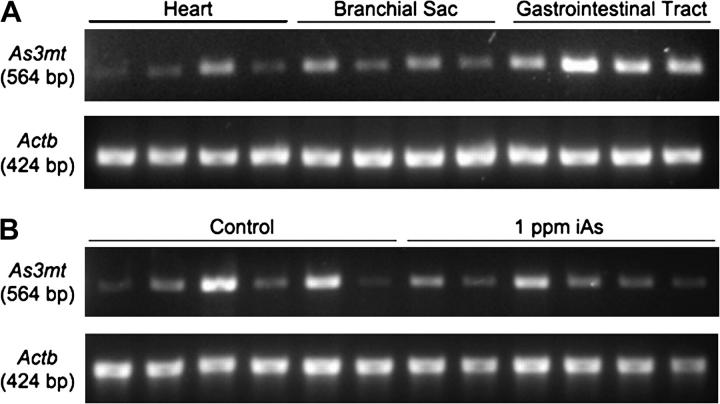
End-point reverse transcription (RT)-PCR analysis was used to examine constitutive As3mt RNA expression in heart, branchial sac, and gastrointestinal tract and the effects of 5 days of exposure to iAs in the gastrointestinal tract of C. intestinalis. Total RNA was extracted with the RNeasy kit (Qiagen, Valencia, CA). The One-Step RT-PCR Kit (Qiagen) was used to detect an As3mt messenger RNA amplicon (564 bp) with primers (forward: CCG TTC CAC TTG TGA CCT GAA and reverse ACC CAT ACC CTC ACC CCA TAA) designed from the overlapping sequence of the two As3mt candidate transcripts. Beta-actin (Actb) was targeted as a constitutively expressed control gene (forward: TTG TAC GCC AAC ACC GTT CTC TCT and reverse TAG CAG CTG AAG CCG GTT TAG GAA). Reactions were performed in a total volume of 25 μl with approximately 40 ng total RNA/μl. First, complementary DNA (cDNA) synthesis and pre-denaturation were performed in single cycles at 50°C for 30 min and 95°C for 15 min, respectively. Next, PCR amplification was performed for 35 cycles: 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s with a final extension cycle at 72°C for 6 min. Two negative controls, RT-PCR grade water, and no RT reaction samples were included in each run to distinguish possible false-positive results. RT-PCR reactions targeting Actb in which reverse transcriptase was omitted did not generate amplicons, indicating that the RNA samples were not contaminated with genomic DNA (data not shown). PCR products were size separated by agarose gel electrophoresis and visualized with ethidium bromide.
Speciated arsenical analysis in C. intestinalis tissues.
Tissues were homogenized in deionized water (20% wt/vol), and homogenates were digested as previously described (Hughes et al.,2005). Speciated arsenicals in digestates were determined by hydride generation-cryotrapping-gas chromatography-atomic absorption spectrometry with hydride production at pH 1 and pH 6, respectively (Hernández-Zavala et al., 2008). This analysis quantified iAs, methyl arsenic (MAs), dimethyl arsenic (DMAs), and trimethyl arsenic (TMAs) species in tissues. Total speciated As concentrations of tissue samples were calculated as the sum of iAs, MAs, DMAs, and TMAs concentrations. Statistical comparisons of the concentrations of arsenicals in tissues were made using the Mann-Whitney rank sum test.
RESULTS AND DISCUSSION
Automatic ortholog prediction using the Ensembl project website identified a C. intestinalis As3mt candidate gene located on scaffold_67 at location 111,078–114,780. The single As3mt candidate gene (accession number ENSCING00000015865) encodes two putative proteins. The larger protein (ENSCINP00000027827, 293 amino acids) is encoded by eight exons; the last three exons are missing for the shorter protein (ENSCINP00000027826, 199 residues). Both As3mt putative proteins possess the methyltransferase domain (Table 1). However, the ENSCINP00000027827 protein sequence harbors 94 additional amino acids at the C-terminus that are highly conserved among As3mt proteins. Thus, the larger As3mt candidate protein was used for RBH analysis (Wall et al., 2003) against RefSeq databases, which revealed higher homology with well-annotated and biochemically characterized As3mt proteins from mammals (Table 2).
TABLE 1.
Conserved Domains in As3mt Amino Acid Sequences Identified in Pfam Database
| Species | Accession number | Pfam accession | Pfam description | E value |
| Human | NP_065733.1 | PF08241 | Methyltransferase domain | 2.00E-14 |
| Rat | NP_543166.1 | PF08241 | Methyltransferase domain | 2.00E-12 |
| Mouse | NP_065602.2 | PF08241 | Methyltransferase domain | 2.00E-14 |
| Ciona intestinalis | ENSCINP00000027827 | PF08241 | Methyltransferase domain | 6.00E-11 |
TABLE 2.
RBH Analysis of Ciona intestinalis As3mt Gene in GenBank
The predicted sequence of the C. intestinalis As3mt protein contains features consistent with its identity as an AdoMet-dependent As methyltransferase (Thomas et al., 2007). As shown in Figure 1, it contains the four methyltransferase sequence motifs (I, I′, II, and III) that are present in human, rat, and mouse As3mt (Kagan and Clarke, 1994). For each predicted protein, the invariant amino acid residues in the motifs are conserved. In addition, the five highly conserved Cys residues, which have been identified in all As3mt sequences, are also present in C. intestinalis As3mt (Fig. 1). The function of As3mt as an As methyltransferase is dependent on the presence of Cys156 in rat and Cys157 in mouse. Replacement of these residues with Ser leads to a loss of catalytic function (Fomenko et al., 2007; Li et al., 2005); replacement of both Cys156 and Cys205 with Ser in human AS3MT nullifies its catalytic function (Thomas et al., 2007). The persistence of these Cys residues in species that diverged over a period of about 500 million years suggests that their presence determines the protein’s catalytic activity.
FIG. 1.
Sequence alignments for human, rat, mouse, and Ciona intestinalis As3mt showing the presence of motifs I, I′, II, and III in the methyltransferase domain and the positions of conserved cysteine residues. Sequences of human, rat, and mouse As3mt from NCBI GenBank (Homo sapiens [NP_065733], Rattus norvegicus [NP_543166], Mus musculus [NP_065602], and C intestinalis [ENSCINP00000027827]) were aligned using M-Coffee software, a meta-method for assembling multiple sequence alignments (Wallace et al., 2006).
The statistical significance of pairwise sequence similarity for the C. intestinalis As3mt protein using PRSS and SSEARCH analyses (Pearson, 1991; Smith and Waterman, 1981) is consistent with its high homology to human, rat, and mouse proteins (Table 3). Both construction of orthologous groups across multiple eukaryotic taxa (OrthoMCL algorithm) and protein phylogenetic analysis (Fig. 2) reveal a clear evolutionary relation among homologous As3mt proteins. The phylogenetic tree indicates that As3mt might have diverged from the base of the chordate tree in another lower eukaryote. In sum, these analyses validate the accuracy of the manual annotation of the As3mt gene in C. intestinalis.
TABLE 3.
Statistical Significance of Pairwise Sequence Similarity for Ciona intestinalis As3mt Gene Using PRSS and SSEARCH Analyses
FIG. 2.
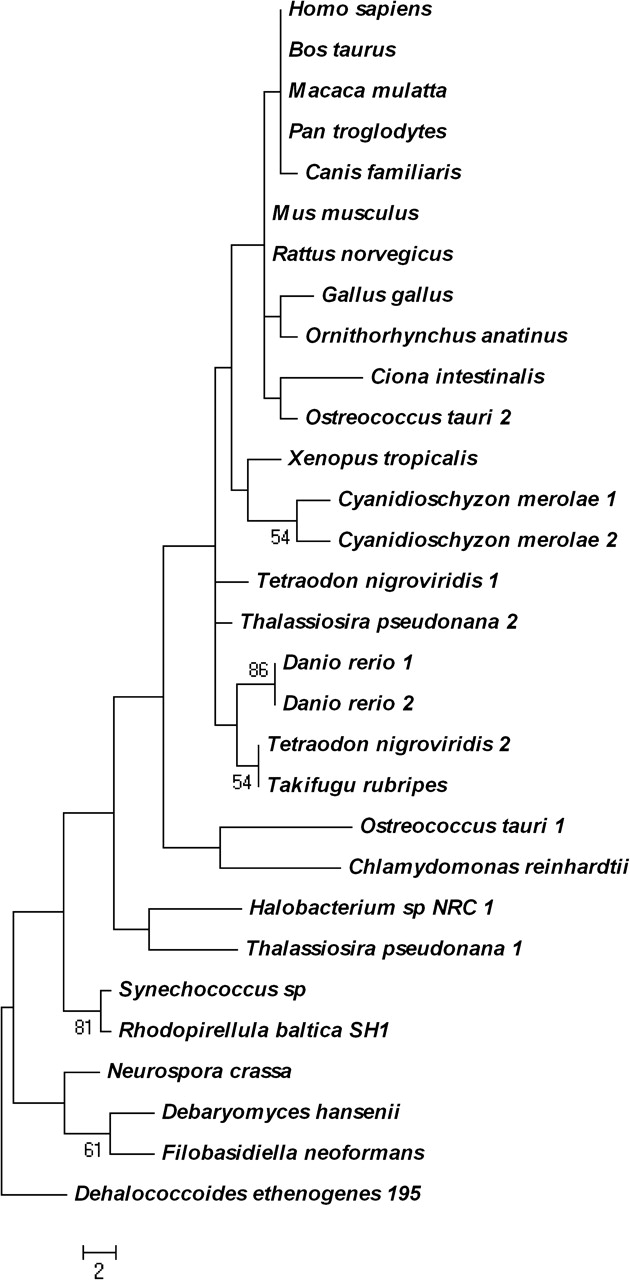
Phylogeny of As3mt protein sequences in Eukarya, Archaea, and Bacteria. The phylogenetic tree was constructed with the maximum parsimony method using the PAUP* 4.0 b10 program. The statistical significance of branch order (numbers on branches) was estimated by the generation of 1000 replications of bootstrap resampling of the originally aligned amino acid sequences. The deduced amino acid sequence of arsenite S-adenosyl methyltransferase (YP_182128) from the bacterium Dehalococcoides ethenogenes was used as a root sequence. Scale bar indicates the number of amino acid changes among the aligned sequences.
In silico cloning (Boguski et al., 1994) of As3mt in C. intestinalis confirms the presence of its transcripts in tissues from juvenile and mature adult organisms and in the egg and dividing embryo (Table 4). Moreover, RT-PCR analysis from replicate animals demonstrated constitutive expression of As3mt in heart, branchial sac, and gastrointestinal tract (Fig. 3A). The level of steady-state As3mt RNA expression was variable among replicate animals for all three tissues, and expression in the gastrointestinal tract did not appear to be modulated by 5 days of exposure to iAs (Fig. 3B). As3mt is also constitutively expressed in many tissues of the adult rat (Lin et al., 2002). Although exposure to iAs does not appear to increase As3mt gene transcription or protein levels in mice (Drobna et al., 2009; Xing, unpublished data), single nucleotide polymorphisms in the human As3mt gene, sequence variation in 5′ flanking regions, and the number of tandem repeats in the 5′ untranslated region affect protein expression and activity (Wood et al., 2006). Hence, regulation of As3mt activity and expression is probably a complex interplay of transcriptional and translational controls.
TABLE 4.
In silico Cloning of the As3mt Gene in Ciona intestinalis
| Accession number | Description | E value |
| BW379540.1 | cDNA library, adult digestive gland | 0 |
| BW418659.1 | cDNA library, mature adult whole animal | 0 |
| BW462498.1 | cDNA library, juvenile whole animal | 0 |
| BW063681.1 | cDNA library, young adult | 0 |
| BW107407.1 | cDNA library, cleaving embryo | 0 |
| AV863336.1 | cDNA library, egg | 0 |
| BW056695.1 | cDNA library, blood cells | 0 |
FIG. 3.
End-point RT-PCR analysis of constitutive As3mt RNA expression in heart, branchial sac, and gastrointestinal tract of Ciona Intestinalis using four replicate animals (A). Steady-state As3mt RNA expression in the gastrointestinal tract was compared in six animals exposed to 1 ppm arsenite in seawater for 5 days to six animals maintained under similar conditions in control unamended seawater to examine the extent to which As3mt expression is modulated by exposure to iAs (B). The RNA isolation protocol, primer sequences, and reaction conditions are provided in the “Materials and Methods” section.
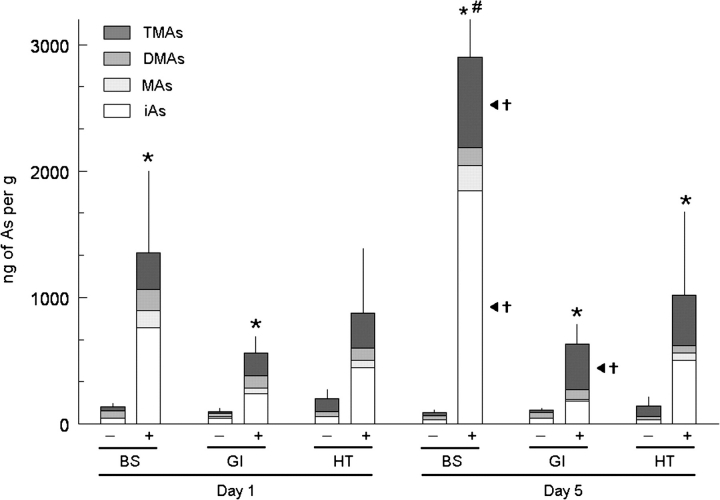
In these experiments, organisms were exposed to iAs (added as arsenite) for up to 5 days. The background concentration of iAs in seawater used in these studies was approximately 0.001 ppm; in amended seawater, iAs concentration was approximately 1 ppm. Tissues from control animals maintained in unamended seawater contained iAs and its methylated metabolites (Fig. 4). The presence of methylated arsenicals in tissues under basal exposure conditions indicates that these organisms constitutively methylate iAs. At day 1 of exposure, the summed concentrations of speciated arsenicals in branchial sac or gastrointestinal tract from animals in amended seawater versus unamended seawater were significantly different (p < 0.001). However, the summed concentrations of speciated arsenicals in heart were not significantly different in these two treatment groups on day 1 of exposure. At day 5 of exposure, the summed concentrations of speciated arsenicals in all tissues from animals in amended seawater or unamended seawater were significantly different (p ≤ 0.008). The summed concentrations of speciated arsenicals in tissues of control animals maintained in unamended seawater did not significantly change (p > 0.05) between days 1 and 5 of exposure. Ciona intestinalis exposed to iAs in amended seawater quickly accumulated iAs and its methylated arsenicals in tissues. After 1 day of exposure, iAs and methylated arsenicals were present in all tissues; branchial sac contained the highest summed concentrations of speciated arsenicals. Although the summed concentrations of speciated arsenicals increased in all tissues between days 1 and 5 of exposure, this increase was statistically significant (p = 0.002) only in branchial sac. In this tissue, the concentrations of iAs (p = 0.001) and TMAs (p = 0.001) were significantly different on days 1 and 5 of exposure. In the gastrointestinal tract, the concentration of TMAs was significantly different (p = 0.003) on days 1 and 5 of exposure.
FIG. 4.
Distribution of arsenicals in tissues of Ciona intestinalis following exposure to unamended seawater (−) or seawater with 1 ppm of arsenic (as arsenite) (+) for 1 day (day 1) or 5 days (day 5). Results shown for branchial sac (BS), gastrointestinal tract (GI), and heart (HT). Detected species are iAs, MAs, DMAs, and TMAs. Means and SDs shown for summed concentrations of speciated arsenicals in tissues (n = 3–8). *A significant difference (p ≤ 0.008) between treatment groups in the summed concentrations of arsenicals in a tissue on day 1 or day 5. #A significant difference (p = 0.002) in the summed concentrations of arsenicals in the branchial sac between days 1 and 5. †A significant difference (p ≤ 0.003) in the concentration of a speciated arsenical in a tissue between days 1 and 5.
Notably, the pattern of metabolites in C. intestinalis exposed to iAs resembles that found in chronically exposed rats. In rats exposed to arsenate in drinking water for 2 weeks, the As-containing methylated metabolites DMA and TMA were found in tissues and excreta (Adair et al., 2007). Recombinant rat As3mt has been shown to efficiently catalyze the mono-, di-, and trimethylation reactions (Waters et al., 2004a). Catalysis of As methylation reactions by rat As3mt is dependent on physiological reductants (thioredoxin, glutaredoxin, and dihydrolipoic acid), suggesting that these or similar dithiol reductants must be available in C. intestinalis. In addition, glutathione (GSH) is an important modulator of the activity of rat As3mt (Waters et al., 2004b). Addition of GSH to in vitro reaction mixtures containing rat As3mt, AdoMet, and a coupled thioredoxin/thioredoxin reductase/NADPH system suppressed formation of TMAs from either arsenite or dimethylarsinous acid. A recent analysis of the genome of C. intestinalis annotated a number of genes involved in the biosynthesis and utilization of GSH (Nava et al., 2009). These investigators identified genes encoding the two enzymes required for GSH synthesis (catalytic and regulatory subunits of glutamate cysteine ligase and glutathione synthetase), which were expressed in branchial sac, heart, and gastrointestinal tract of adult C. intestinalis. Further investigation is required to determine whether GSH may modulate As methylation in C. intestinalis.
Other factors could also affect the pattern of As-containing metabolites found in tissues. Translocation of iAs and its methylated metabolites among tissues during the course of exposure could obscure differences among tissues in the capacity to form metabolites. Trivalent inorganic and methylated arsenicals enter eukaryotic cells via aquaporins and hexose permease transporters and exit cells via ATP-binding cassette (ABC) transporters (Thomas, 2007). Hence, intracellular concentrations of individual methylated arsenicals will be determined by the balance of influx and efflux processes. Variation among tissues in expression of these membrane-spanning proteins may determine levels of individual metabolites in different tissues. Genes encoding aquaporin-like proteins and ABC transporters have been identified in C. intestinalis (Annilo et al., 2006; Okamura et al., 2005). Notably, the branchial sac, which consistently showed the highest levels of As accumulation, also avidly accumulates vanadium (V) added as oxyanionic vanadate (Cheney et al., 1997; Michibata, 1993). The relation, if any, between accumulation of V and of As in tissues is unclear. For V, there is evidence of specific binding proteins in tissues that sequester this element (Trivedi et al., 2003). Analogous proteins in C. intestinalis could be involved in the binding of iAs or its methylated metabolites. For example, nicatransferrin, a monolobal transferrin from C. intestinalis (Tinoco et al., 2008), could bind arsenicals in a manner analogous to As binding by mammalian transferrin (Zhang et al., 1998).
Functional similarities between the metabolism of As in C. intestinalis and in more complex organisms support the hypothesis that enzymatically catalyzed methylation of As is a fundamental component of the organismic response to this metalloid, which arose early in the evolution of the chordate lineage. Similarities among developmental and physiological processes in this primitive chordate and higher organisms may make C. intestinalis a useful model organism for studies of molecular processes involved in the accumulation, methylation, binding, and excretion of As and for studies of modes of action of iAs and its methylated metabolites as toxins or carcinogens.
FUNDING
National Institute for Environmental Health Sciences Center for Comparative Toxicology (P30 ES03828) at Mount Desert Island Biological Laboratory; DK 25636 to J.L.B.; Yale Liver Center (P30 34989).
Acknowledgments
The authors thank Ann Benefiel and Debbie Piper for editorial assistance. The authors declare no conflict of interest.
References
- Adair BM, Moore T, Conklin SD, Creed JT, Wolf DC, Thomas DJ. Tissue distribution and urinary excretion of dimethylated arsenic and its metabolites in dimethylarsinic acid- or arsenate-treated rats. Toxicol. Appl. Pharmacol. 2007;222:235–242. doi: 10.1016/j.taap.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annilo T, Chen ZQ, Shulenin S, Costantino J, Thomas L, Lou H, Stefanov S, Dean M. Evolution of the vertebrate ABC gene family: analysis of gene birth and death. Genomics. 2006;88:1–11. doi: 10.1016/j.ygeno.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Bairoch A, Boeckmann B, Ferro S, Gasteiger E. Swiss-Prot: juggling between evolution and stability. Brief Bioinform. 2004;5:39–55. doi: 10.1093/bib/5.1.39. [DOI] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielig HJ, Pfleger K, Rummel W, Seifen E. Uptake and distribution of vanadium by the tunicate Ciona intestinalis L. Hoppe Seylers Z. Physiol. Chem. 1961;326:249–258. doi: 10.1515/bchm2.1961.326.1.249. [DOI] [PubMed] [Google Scholar]
- Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol. Biol. Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST—database for “expressed sequence tags.”. Nat. Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Tolstoshev CM, Bassett DE., Jr Gene discovery in dbEST. Science. 1994;265:1993–1994. doi: 10.1126/science.8091218. [DOI] [PubMed] [Google Scholar]
- Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheney MA, Berg JR, Swinehart JH. The uptake of vanadium (V) and other metals by the isolated branchial sacs of the ascidians Ascidia ceratodes, Ciona intestinalis, and Styela montereyensis. Comp. Biochem. Physiol. 1997;116C:49–153. [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- Drobna Z, Naranmandura H, Kubachka KM, Edwards BM, Herbin-Davis K, Styblo M, Le XC, Creed JT, Maeda N, Hughes MF, et al. Disruption of the arsenic (+3 oxidation state) methyltransferase gene in the mouse alters the phenotype for methylation of arsenic and affects distribution and retention of orally administered arsenate. Chem. Res. Toxicol. 2009;22:1713–1720. doi: 10.1021/tx900179r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eck RV, Dayhoff MO. Atlas of Protein Sequence and Structure. Silver Spring, MD: National Biomedical Research Foundation; 1966. [Google Scholar]
- Fomenko DE, Xing W, Adair BM, Thomas DJ, Gladyshev VN. High-throughput identification of catalytic redox-active cysteine residues. Science. 2007;315:387–389. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]
- Goodbody I. The physiology of ascidians. In: Russell FS, Yonge M, editors. Advances in Marine Biology. Vol. 12. London, UK: Academic Press; 1974. pp. 1–149. [Google Scholar]
- Hernández-Zavala A, Matousek T, Drobna Z, Paul DS, Walton F, Adair BM, Jiri D, Thomas DJ, Stýblo M. Speciation analysis of arsenic in biological matrices by automated hydride generation-cryotrapping-atomic absorption spectrometry with multiple microflame quartz tube atomizer (multiatomizer) J. Anal. At. Spectrom. 2008;23:342–351. doi: 10.1039/b706144g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MF. Biomarkers of exposure: a case study with inorganic arsenic. Environ. Health Perspect. 2006;114:1790–1796. doi: 10.1289/ehp.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MF, Devesa V, Adair BM, Stýblo M, Kenyon EM, Thomas DJ. Tissue dosimetry, metabolism and excretion of pentavalent and trivalent monomethylated arsenic in mice after oral administration. Toxicol. Appl. Pharmacol. 2005;208:186–197. doi: 10.1016/j.taap.2005.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan RM, Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch. Biochem. Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li J, Waters SB, Drobna Z, Devesa V, Stýblo M, Thomas DJ. Arsenic (+3 oxidation state) methyltransferase and the inorganic arsenic methylation phenotype. Toxicol. Appl. Pharmacol. 2005;204:164–169. doi: 10.1016/j.taap.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Lin S, Shi Q, Nix FB, Stýblo M, Beck MA, Herbin-Davis KM, Hall LL, Simeonsson JB, Thomas DJ. A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol. J. Biol. Chem. 2002;277:10795–10803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- Lin YF, Yang J, Rosen BP. ArsD: an As(III) metallochaperone for the ArsAB As(III)-translocating ATPase. J. Bioenerg. Biomembr. 2007;39:453–458. doi: 10.1007/s10863-007-9113-y. [DOI] [PubMed] [Google Scholar]
- Michibata H. The mechanism of accumulation of high levels of vanadium by ascidians from seawater: biophysical approaches to a remarkable phenomenon. Adv. Biophys. 1993;29:105–133. doi: 10.1016/0065-227x(93)90007-r. [DOI] [PubMed] [Google Scholar]
- Nava GM, Lee DY, Ospina JH, Cai S-Y, Gaskins HR. Genomic analyses reveal a conserved glutathione homeostasis pathway in the invertebrate chordate Ciona intestinalis. Physiol. Genomics. 2009;39:183–194. doi: 10.1152/physiolgenomics.00025.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y, Nishino A, Murata Y, Nakajo K, Iwasaki H, Ohtsuka Y, Tanaka-Kunishima M, Takahashi N, Hara Y, Yoshida T, et al. Comprehensive analysis of the ascidian genome reveals novel insights into the molecular evolution of ion channel genes. Physiol. Genomics. 2005;11:269–282. doi: 10.1152/physiolgenomics.00229.2004. [DOI] [PubMed] [Google Scholar]
- Pearson WR. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics. 1991;11:635–650. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- Petersen JK, Svane I. Filtration rate in seven Scandinavian ascidians: implications of the morphology of the gill sac. Mar. Biol. 2002;140:397–402. [Google Scholar]
- Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen BP. Biochemistry of arsenic detoxification. FEBS Lett. 2002a;529:86–92. doi: 10.1016/s0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- Rosen BP. Transport and detoxification systems for transition metals, heavy metals and metalloids in eukaryotic and prokaryotic microbes. Comp. Biochem. Physiol. A. Mol. Integr Physiol. 2002b;133:689–693. doi: 10.1016/s1095-6433(02)00201-5. [DOI] [PubMed] [Google Scholar]
- Sidiq M. Toxic Metal Chemistry in Marine Environments. New York: Dekker; 1992. Arsenic in marine environments; pp. 61–105. [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. 2007. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thomas DJ. Molecular processes in cellular arsenic metabolism. Toxicol. Appl. Pharmacol. 2007;222:365–373. doi: 10.1016/j.taap.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Li J, Waters SB, Xing W, Adair BM, Drobna Z, Devesa V, Stýblo M. Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals. Exp. Biol. Med. 2007;232:3–13. [PMC free article] [PubMed] [Google Scholar]
- Thomas DJ, Styblo M, Lin S. The cellular metabolism and systemic toxicity of arsenic. Toxicol. Appl. Pharmacol. 2001;176:127–144. doi: 10.1006/taap.2001.9258. [DOI] [PubMed] [Google Scholar]
- Tinoco AD, Peterson CW, Lucchese B, Doyle RP, Valentine AM. On the evolutionary significance and metal-binding characteristics of a monolobal transferrin from Ciona intestinalis. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3268–3273. doi: 10.1073/pnas.0705037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi S, Ueki T, Yamaguchi N, Michibata H. Novel vanadium-binding proteins (vanabins) identified in cDNA libraries and the genome of the ascidian Ciona intestinalis. Biochim. Biophys. Acta. 2003;1630(2–3):64–70. doi: 10.1016/j.bbaexp.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Tseng CH. Arsenic methylation, urinary arsenic metabolites and human diseases: current perspective. J. Environ. Sci. Health C. Environ. Carcinog. Ecotoxicol. Rev. 2007;25:1–22. doi: 10.1080/10590500701201695. [DOI] [PubMed] [Google Scholar]
- Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- Wallace IM, O'Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucl. Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton FS, Waters SB, Jolley SL, LeCluyse EL, Thomas DJ, Stýblo M. Selenium compounds modulate the activity of recombinant rat AsIII-methyltransferase and the methylation of arsenite by rat and human hepatocytes. Chem. Res. Toxicol. 2003;16:261–265. doi: 10.1021/tx025649r. [DOI] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics. 2007;23:549–558. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- Waters SB, Devesa V, Del Razo LM, Stýblo M, Thomas DJ. Endogenous reductants support the catalytic function of recombinant rat cyt19, an arsenic methyltransferase. Chem. Res. Toxicol. 2004a;17:404–409. doi: 10.1021/tx0342161. [DOI] [PubMed] [Google Scholar]
- Waters SB, Devesa V, Fricke MW, Creed JT, Stýblo M, Thomas DJ. Glutathione modulates recombinant rat arsenic (+3 oxidation state) methyltransferase-catalyzed formation of trimethylarsine oxide and trimethylarsine. Chem. Res. Toxicol. 2004b;17:1621–1629. doi: 10.1021/tx0497853. [DOI] [PubMed] [Google Scholar]
- Wood TC, Salavagionne OE, Mukherjee B, Wang L, Klumpp AF, Thomae BA, Eckloff BW, Schaid DJ, Wieben ED, Weinshilboum RM. Human arsenic methyltransferase (AS3MT) pharmacogenetics: gene resequencing and functional genomics studies. J. Biol. Chem. 2006;281:7364–7373. doi: 10.1074/jbc.M512227200. [DOI] [PubMed] [Google Scholar]
- Yaun C, Lu X, Qin J, Rosen BP, Le XC. Volatile arsenic species released from E. coli expressing AsIII S-adenosylmethionine methyltransferase gene. Environ. Sci. Technol. 2008;42:3201–3206. doi: 10.1021/es702910g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Cornelis R, DeKimpe J, Mees L, Lamiere N. Study of arsenic-protein binding in serum of patients on continuous ambulatory peritoneal dialysis. Clin. Chem. 1998;44:141–147. [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J. Comp. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]