Abstract
The induction of localized DNA damage within a discrete nuclear volume is an important tool in DNA repair studies. Both charged particle irradiation and laser microirradiation (LMI) systems allow for such a localized damage induction, but the results obtained are difficult to compare, as the delivered laser dose cannot be measured directly. Therefore, we revisited the idea of a biological dosimetry based on the microscopic evaluation of irradiation-induced Replication Protein A (RPA) foci numbers. Considering that local dose deposition is characteristic for both LMI and charged particles, we took advantage of the defined dosimetry of particle irradiation to estimate the locally applied laser dose equivalent. Within the irradiated nuclear sub-volumes, the doses were in the range of several hundreds of Gray. However, local dose estimation is limited by the saturation of the RPA foci numbers with increasing particle doses. Even high-resolution 4Pi microscopy did not abrogate saturation as it was not able to resolve single lesions within individual RPA foci. Nevertheless, 4Pi microscopy revealed multiple and distinct 53BP1- and γH2AX-stained substructures within the lesion flanking chromatin domains. Monitoring the local recruitment of the telomere repeat-binding factors TRF1 and TRF2 showed that both proteins accumulated at damage sites after UVA–LMI but not after densely ionizing charged particle irradiation. Hence, our results indicate that the local dose delivered by UVA–LMI is extremely high and cannot be accurately translated into an equivalent ionizing radiation dose, despite the sophisticated techniques used in this study.
Introduction
In the last decade, the use of laser microirradiation (LMI) as a tool to generate localized DNA damage has become increasingly popular. In the majority of studies, the recruitment of repair factors specific for DNA double-strand breaks (DSBs) are monitored, for example 53BP1 (1), members of the phosphatidyl-inositol 3 kinase-related kinases (1–3) and components of the Mre11–Rad50–Nbs1 complex (1,4). As a common marker for DSBs, the phosphorylated form of histone H2AX (γH2AX) is also frequently monitored in LMI studies (1,5,6). In addition to DSB repair, the recruitment of proteins associated with single-stranded DNA (ssDNA) such as the Replication Protein A (RPA) (1,7) or proteins involved in single-strand breaks (8), nucleotide excision (9) or base excision repair (10) have also been the focus of several studies. The different LMI systems used in the respective studies combine all features of a research microscope—e.g. fluorescence and time-lapse imaging—with a very precise irradiation spot size of ∼1 μm in diameter. This allows the targeted irradiation of defined subnuclear regions and the simultaneous observation of fast and early damage responses (5,9). The implementation of a relatively low cost 405 nm diode laser in modern confocal laser scanning microscopes further contributes to the availability of such microirradiation systems (11).
A comparable experimental performance with respect to the discrete localization of DNA damage can also be realized by irradiation with α (12,13) or accelerated charged particles (14,15) that induce DNA lesions along the particle trajectories. In combination with the setup of single charged particle microprobes, predefined numbers of particles can be directed to the target volume with sub-micrometre precision (16). Using live cell microscopy at the beamline, it is also possible to visualize in real time the recruitment of repair proteins after charged particle irradiation (17,18).
In contrast to LMI systems, the spectrum of charged particle-induced DNA damage is better defined and can be correlated with a measurable energy deposition. Nevertheless, particle irradiation does suffer some disadvantages, such as a more complex experimental setup, restricted availability and higher costs compared to LMI systems.
Meanwhile, several different types of laser systems are frequently used to generate localized DNA damage, but a comparison between the obtained data is often difficult as the results depend strongly on the wavelength (19,20) and the energy (6,20) of the laser system used. With the application of multiphoton laser systems (2,21), the duration of laser pulses also has a major impact on the damage outcome (20,22). To unify this diversity of impact factors, it would be useful to find a single parameter correlated with the amount of laser-induced DNA damage. An appropriate parameter could be the absorbed energy as it is typically used for ionizing radiation (IR). Unfortunately, the measurement of the absorbed laser energy fails due to the thin target of cell monolayers and the low absorption rate of DNA at wavelengths used in most LMI systems (23). To facilitate a comparison of different laser-obtained data, but also between laser and IR data, several attempts were made to estimate the absorbed laser energies. The approaches were based either on calculations using standard absorption rates yielding a locally applied laser dose of >800 Gy (λ = 390 nm; 10 MJ/m2) (4) or on a biological dosimetry comparing DNA damage responses after LMI and sparsely IR with results ranging from 3 to 30 Gy (1,5,10,11).
Motivated by these extreme differences in estimates of the laser dose equivalent (LDE), we revisited the idea of a previously established biological dosimetry in which the numbers of X-ray- and laser-induced RPA foci are compared to define an X-ray equivalent laser dose (1). By using the same cell line (U2OS), repair-related protein (RPA) and LMI system (pulsed nitrogen laser; λ = 337 nm), we ensured comparability with previous results (1). Beside X-rays, we used heavy charged particles as an ionizing reference radiation. In order to account for the non-homogeneous dose distribution of LMI and charged particles, the volume in which the RPA foci were counted was restricted to the irradiated part of the nucleus. Because the resulting dose–response curve for charged particle-induced RPA foci saturated, which even high-resolution 4Pi microscopy could not resolve, the local LDE could not be accurately determined. However, the locally applied LDE can be estimated to be comparable to or even higher than the dose delivered by very densely ionizing charged particles. Recruitment studies of the telomere repeat-binding factors TRF1 and TRF2 both showed local accumulation after LMI as reported previously for TRF2 (13,24). However, no significant recruitment of TRF1 and TRF2 was observed after low energy xenon ion irradiation. These results support the assumption of an even higher LDE than previously estimated, although additional and not well-characterized differences between laser- and IR-induced DNA damage spectra cannot be ruled out.
Materials and methods
Cell culture
Confluent normal human foreskin fibroblasts (AG1522; Coriell cell repository, Camden, NJ, USA; passage 13 to 16), human osteosarcoma cells (U2OS; American Type Culture Collection, Middlesex, UK) and human dermal fibroblasts (1BR3) stably expressing yellow fluorescent protein (YFP)-TRF1 or YFP-TRF2 were used. Cells were grown at 37°C, 100% humidity and 5% CO2 in Eagle's Minimum Essential Medium, Dulbecco's Modified Eagle's Medium or Alpha-Modified Eagle's Medium, respectively (all Biochrome, Berlin, Germany). The media were supplemented with Earle's balanced salt solution, 1% glutamine, 0.5% penicillin–streptomycin and 15% fetal calf serum, with 4.5 g/l glucose Na-pyruvate and 10% fetal calf serum or with 1% glutamine and 10% fetal calf serum, respectively. Cell lines were regularly checked to be mycoplasma free.
Directly before UVA–LMI, the cell samples were mounted in a Focht chamber system FCS2 (Bioptechs Inc., Butler, PA, USA) filled with conditioned medium. For the laser experiments, U2OS cells were pre-sensitized towards UVA light by incubation with 10 μM bromodeoxyuridine (BrdU) for 24 h (1,25). Pre-sensitization was omitted for 1BR3 cells.
Production of cells stably expressing YFP-TRF1 and -TRF2
Full-length human TRF1 or TRF2 complementary DNA was cloned in-frame in a FLAG-tag and YFP-containing plasmid construct derived from the retroviral vector pQCXIP (BD Biosciences, Franklin Lakes, NI, USA). All constructs were verified by sequencing. Packaged YFP-TRF1 or YFP-TRF2 viruses were used to transfect the human fibroblast cell line 1BR3 to generate puromycin-resistant cells stably expressing YFP-TRF1 and YFP-TRF2.
Irradiation and dose calculation
For UVA laser irradiation, the Leica Microdissection System LMD AS was used. The upright Leica microscope is equipped with a pulsed (5 ns; 30 Hz) Nitrogen Laser (337 nm). Measurements of the laser power were performed prior to irradiation with a UVA-sensitive photometer Orion/PD (Ophir Optotronics, Rohrsen, Germany) simulating the irradiation situation of a sample mounted in the FCS2. Irradiation was performed with a HCX PL Fluotar L 63×0.70 Corr PH 2 objective focusing the laser beam spot to a diameter of ∼2 μm. With the used laser setting, a laser irradiance of 1.5 μW was applied corresponding to a radiant exposure of 0.3 MJ/m2. For UVA–LMI of 1BR3 cells, the laser irradiance was increased to 4.4 μW but the exposure time was reduced to keep the laser exposure constant. Multiple setting of targeting marks within the field of view allowed the almost simultaneous irradiation of several nuclei. The irradiation of multiple fields took up to half an hour.
The charged particle irradiation was done at the UNILAC facility at the GSI Helmholtzzentrum für Schwerionenforschung as described previously (14,17). The ions used, their specific energies and linear energy transfers (LETs), are listed in Table I. The deposited energy for each ion species was calculated as the product of a constant LET and the summed length of all traversals per nucleus. The total traversal length was further standardized to the average value of all analyzed nuclei (18 μm) to allow for a direct comparison between ion species. The dose deposited per nucleus was calculated as the ratio of the deposited energy and the assumed average mass of a nucleus (855 ng, supplementary Figure S1, available at Mutagenesis Online).
Table I.
Calculated charged particle doses absorbed per nucleus
| Ion species | Mean energy on target | LET | Dose per nucleus after standardization |
| MeV/u | keV/μm | Gy | |
| Carbon | 5.3 | 265 | 0.9 |
| Nitrogen | 5.1 | 363 | 1.2 |
| Nickel | 4.9 | 3530 | 11.9 |
| Xenon | 2.4 | 8679 | 29.2 |
| Uranium | 1.9 | 13 438 | 45.3 |
Dose = energy/mass = (0.16 × LET × 18 μm)/(ρwater × 855 μm3)
Irradiation with X-rays was performed in a 250 keV X-ray tube (Isovolt DS1, Seiffert, Germany) at a dose rate of 3 Gy/min.
Immunofluorescence assay
For the immunocytochemical staining experiments, cells were fixed in 2% formaldehyde and permeabilized as described previously (15). Solely before Mre11 staining, the soluble protein fraction was pre-extracted (14). Mouse monoclonal anti-RPA (p34) (Lab Vision, Wedel, Germany) and the secondary antibody 488 goat anti-mouse F(ab)2 conjugate (Invitrogen, Karlsruhe, Germany) were used at a dilution of 1:200 in 0.2% bovine serum albumin in phosphate-buffered saline and 1:400, respectively. For 4Pi microscopy, the RPA antibody was combined with rabbit polyclonal anti-53BP1 (Ab-1; 1:500; Oncogene, VWR, Darmstadt, Germany) antibody, as indicated. Also for 4Pi microscopy, the anti-MRE11B rabbit polyclonal antibody (1:300; Novus-Biologicals; Abcam, Cambridge, UK) was used together with mouse monoclonal anti-phospho-(Ser139)-H2AX primary antibody (1:500; clone JBW301; Upstate Biotechnology, Hamburg, Germany) at the indicated dilutions. Secondary antibodies used were 5 μg/ml of Atto 647N goat anti-mouse-IgG (Sigma–Aldrich Corporation, St Louis, MO, USA) and Alexa 568 goat anti-rabbit IgG conjugate (Invitrogen, Karlsruhe, Germany). Cells were counterstained with 1 μM ToPro3 (Invitrogen, Karsruhe, Germany).
Microscopy
Microscopic imaging was done on a Leica TCS confocal system equipped with a DM IRBE inverted microscope (lens: HCX PlanApo 63×1.32) and an argon–krypton laser. From each sample, images containing in total 13–23 nuclei were taken as sets of 11–14 optical sections. The optical sections were recorded in increments of ∼0.25 μm across the thickness of nuclei which was usually ∼3 μm.
4Pi Microscopy was performed with a custom made 4Pi setup at the German Cancer Research Centre (DKFZ) in Heidelberg. The setup was based on a Leica confocal laser scanner TCS-SP2 equipped with two opposing HCX PL APO CS 100×1.46 Oil (Leica Microsystems, Mannheim, Germany) (26). The dyes Alexa 568 and ATTO647 in the samples were excited with HeNe-Lasers at 543 and 633 nm, respectively. The resolution in xy was 170 nm and axially 100 nm. For imaging, the samples were mounted in 97% 2,2-thiodiethanol according to the procedure described in Staudt et al. (27).
Quantitative image analysis
RPA foci were counted in average projections of acquired confocal stacks of X-ray-irradiated nuclei (up to 28 nuclei per dose), UVA laser-irradiated nuclei sensitized with BrdU (n = 53) and charged particle-irradiated nuclei (up to 25 nuclei per ion species). The analysis programme was developed in collaboration with the Department of Image Processing (R. Schäfer, D. Schubert and K. Sandau) at the University of Applied Science, Darmstadt (Germany) and programmed in IDL (V6.0; Research Systems Inc.). The software has been described elsewhere (28). Briefly, after segmentation of nuclei and extraction of signals above background, foci were detected using adaptive bimodal thresholds and fitted by 2D intensity projections of spheres with increasing radii.
The actual irradiated area was defined manually in each image and afterwards subdivided into areas of 2 × 2 μm. As the foci counting was performed in stack projections, the area corresponds to a volume of 2 × 2 × 3 μm considering an average nuclear height of 3 μm. Thus, the measured foci number was related to this volume named counting cuboid (CountCub). Foci numbers related to the cubic volume (Cube; 2 × 2 × 2 μm) were then calculated based on the data obtained in CountCubs and introducing a scaling factor. Assuming a random distribution of RPA foci in depth after X-ray and laser irradiation, the respective scaling factors for both radiation types depend only on geometrical assumptions illustrated by a simplified model of a nucleus (supplementary Figure S1, available at Mutagenesis Online). For laser irradiation, the irradiated volume is mostly located in the centre of the nucleus where the thickness is given by the average value of 3 μm. Thus, the scaling factor for the number of laser-induced RPA foci per Cube is the ratio of the heights of the Cube and the nucleus that is 2:3 (67%). This is different for X-ray-induced foci being randomly distributed all over the nucleus. Here, the volume ratio of the nuclear ‘basement’ (supplementary Figure S1, available at Mutagenesis Online) and the whole nucleus has to be calculated yielding a scaling factor of 85%. For charged particle irradiation, no scaling is needed as the Cube contains the same number of foci as the CountCub.
Results
The volume used for foci counting affects the estimation of localized doses
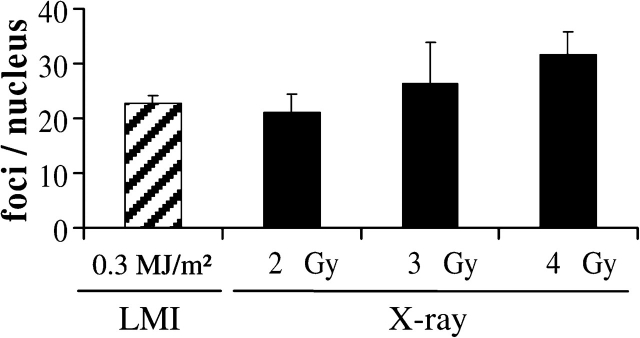
For an estimation of the absorbed X-ray equivalent dose in UVA laser experiments, we modified a previously used method of biological dosimetry (1). This method was based on the comparison of RPA foci numbers induced by UVA laser irradiation and different X-ray doses. According to the original assay (1), U2OS osteosarcoma cells were sensitized to UVA light by incorporation of BrdU for 24 h before irradiation and fixed 1 h post-irradiation. Irradiation-induced RPA foci were automatically counted in projections of the obtained confocal image stacks. The amount of RPA foci induced by the UVA laser system (337 nm Nitrogen laser) was comparable to that observed after an X-ray dose of 2–3 Gy (Figure 1), consequently defined as the X-ray equivalent laser dose. This result is in agreement with the previously reported X-ray equivalent laser dose of 3 Gy using this type of approach (1). Therefore, the laser exposure of 0.3 MJ/m2 used in this experiment was chosen as a standard setting for all subsequent UVA laser irradiations.
Fig. 1.
Amount of RPA foci per nucleus induced by UVA laser and X-ray irradiation. U2OS osteosarcoma cells were irradiated with a UVA laser and different doses of X-rays. The average amounts of RPA foci per nucleus recognized by the analyzing programme and after subtraction of the level in mock-irradiated cells (3.5 ± 0.3 RPA foci; n = 95) are depicted. Average values induced by a laser exposure of 0.3 MJ/m2 are comparable to the RPA foci induction after 2–3 Gy of X-rays. Error bars indicate the standard error of the mean (SEM).
While the number of RPA foci per nucleus was similar after laser and X-ray irradiation, we observed clear differences in terms of signal intensity and spatial distribution of the respective foci. Laser-induced foci typically appeared brighter and denser than the broadly distributed and less intense RPA foci after X-ray irradiation (Figure 2), giving the impression that UVA–LMI induced much more severe DNA damage than 3 Gy of X-rays. Therefore, we studied the suitability of the used dosimetry approach to reproduce higher and locally applied doses. For this purpose, we used accelerated charged particles that allow local deposition of a broad range of well-defined doses. To facilitate counting of charged particle-induced RPA foci along the ion trajectory, the irradiation was delivered at low angle (15°) between the cell layer and the beam direction as described earlier (14). Using this irradiation geometry, a track of clearly distinguishable foci (Figure 3; inlay) could be assigned to a single ion traversal. The number of foci can be assumed to be linearly correlated to the length of the ion traversals (28). Thus, the nuclear dose was calculated based on the LET (defined as the deposited energy per track length) and the total length of all traversals in a nucleus. This assures a linear correlation between calculated nuclear particle dose and the number of foci induced by this dose. As the total traversal length varied from nucleus to nucleus, the total length of all traversals in a nucleus was standardized to the average value for all analyzed nuclei (18 μm). As a result of the standardization, the calculated nuclear particle doses depend only on the respective LET (Table I; dose formula) and range from 0.9 Gy (carbon ions) to 45.3 Gy (uranium ions). The resulting mean numbers of RPA foci induced per 18-μm track length are depicted in Figure 3 for several ion species in comparison to the X-ray data. While the X-ray data were generally obtained in U2OS cells, for charged particle irradiation also normal human fibroblasts were used. However, neither cell line showed a significant difference in RPA foci induction as representatively shown for carbon ion irradiation (Figure 3; first two columns). According to the counted RPA foci, the X-ray equivalent doses for the used ion species were well <3 Gy. The actually deposited, standardized charged particle doses per nucleus (Table I) are also depicted in Figure 3. For carbon and nitrogen ions, they were in the range of the X-ray equivalent doses estimated from RPA foci induction. However, for charged particles with higher LET and, therefore, higher lesion density, the deposited nuclear doses (nickel: 11.9 Gy, xenon: 29.2 Gy, uranium: 45.3 Gy) and the given X-ray equivalent dose (2 Gy) diverged dramatically. Clearly, the used method of biological dosimetry is not able to reproduce nuclear doses of high-LET charged particles but underestimates their dose deposition by a factor of 15 in the case of xenon ion irradiation.
Fig. 2.

The fraction of the irradiated nuclear volume depends on the irradiation type. The depicted cell nuclei were irradiated with 2 Gy of X-rays (top), UVA laser (center; 0.3 MJ/m2) and nickel ions (bottom). The RPA foci (green) that were recognized by the IDL programme are marked with red circles. The number of foci within the nuclei (dotted line) was 23, 22 and 24, respectively. When applying the newly defined Cube of 2 × 2 × 2 μm (indicated as dashed box in the depicted 2D average projection), the average numbers of counted foci per Cube are altered. The resulting average values for the respective irradiation types are indicated to the right of each depicted nucleus. Average projections of the original image stacks are shown. The DNA is counterstained with ToPro3 (blue).
Fig. 3.
Amount of RPA foci per nucleus induced by charged particle and X-ray irradiation. The amount of RPA foci per nucleus induced by different ion species compared with the X-ray data from Figure 1 allocates the nuclear charged particle doses at ∼2 Gy of X-rays or lower. The indicated calculated nuclear particle doses and the corresponding foci numbers per nucleus are standardized to a total length of all traversals per nucleus of 18 μm. Error bars indicate the SEM. Inlay: U2OS cell nucleus irradiated with carbon ions under a low angle. The nucleus was hit by a single carbon ion as visualized by immunocytochemical staining of RPA (green). The RPA foci form typical streaks. The DNA is counterstained with ToPro3 (blue).
One reason for the failure of this assay to reproduce high nuclear doses of charged particle irradiation can be assigned to the different spatial dose distribution of X-rays and charged particles. In contrast to X-rays, particles do not deposit their energy homogeneously but localize along the particle trajectory and hence in a small sub-volume of the nucleus (14,29). However, in the hitherto applied assay, the RPA foci were counted throughout the whole nucleus.
Estimation of a local LDE by foci counting in reduced subnuclear volumes
In order to accommodate for the localized dose deposition of charged particles, we reduced the volume in which the RPA foci were counted. The dimensions of the newly defined cubic volume (Cube; 2 × 2 × 2 μm) were chosen considering the track structure of charged particles and the range of induced δ-electrons (29,30). Figure 2 is a representative example illustrating the impact of the volume in which the X-ray-, UVA laser- and charged particle-induced RPA foci are counted. Within the depicted nuclei, similar foci numbers (23, 22 and 24 foci) were counted after different types of irradiation, X-rays, UVA laser and nickel ions, respectively. Accordingly, X-ray equivalent doses of 2 Gy would be defined for the laser and nickel ion irradiations, even though the nickel irradiation applies an actual nuclear dose of 20.7 Gy at the given total track length of 31 μm within the depicted nucleus (Figure 2; lower panel). In contrast, the foci numbers counted within the Cube were 0.3 (X-ray), 1.9 (laser) and 1.5 (nickel) foci per Cube for these particular nuclei, showing that the local LDE could be even higher than for nickel ion irradiation. Note that foci quantification is typically done in average projections comprising the whole nucleus in depth (here up to 3 μm). The resulting foci numbers were subsequently scaled according to the 2 μm depth of the Cube (for details see Materials and methods section).
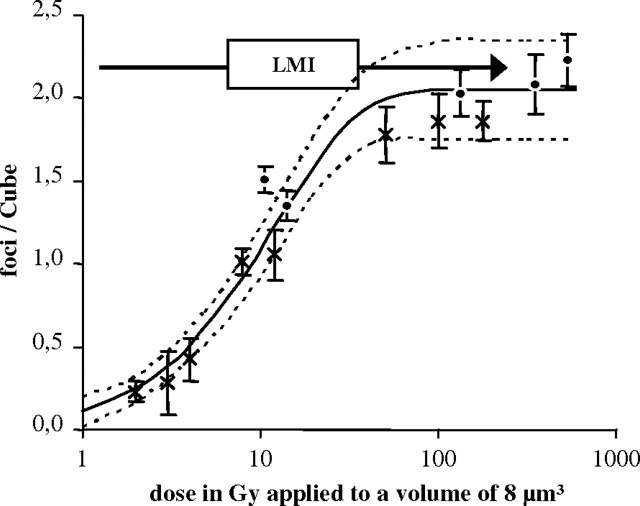
The mean numbers of RPA foci counted per Cube are depicted in Figure 4 for different X-ray and local charged particle doses. The particle doses absorbed within the Cube were calculated based on the dose definition formula (Table II). The absorbed energy is the product of LET and the edge length of the Cube (2 μm) and the mass (8 ng) is given by the respective volume and the density of water. A combined dose–response curve was fitted into the charged particle and X-ray data points. The monoexponential curve levels off at ∼2.0 foci per Cube with a 95% confidence interval of ±0.3. The fact that foci numbers above the fitted saturation level could still be counted indicates that the observed saturation is not due to a limited optical resolution of RPA foci. The mean RPA foci number induced by laser irradiation was 2.2 ± 0.2 foci per Cube (Figure 4; arrow) showing no significant difference to the fitted saturation level. Thus, the damage potential of laser irradiation seems to be in the range of high-LET charged particles corresponding to a locally applied dose of hundreds of Gray, confirming our hypothesis of a more severe damage induction after UVA–LMI than after a few Gray of X-rays. Unfortunately, the saturation behaviour of the curve does not allow for an accurate definition of local LDEs.
Fig. 4.
Dose–response curve for irradiation-induced RPA foci using a cubic volume (8 μm3) for foci quantification. Resulting dose–response curve over a logarithmical x-axis after application of the Cube volume. The monoexponential curve (solid line) is fitted in the charged particle and X-ray data points (R2 = 0.96). The allocation of the laser data points to the fit curve is indicated for a laser exposure of 0.3 MJ/m2 (2.2 ± 0.2 foci per Cube; arrow). Error bars indicate the standard error of the mean, whereas the dotted lines represent the 95% confidence interval of the fit curve.
Table II.
Calculated charged particle doses absorbed per Cube
| Ion species | LET | Absorbed energy | Absorbed dose |
| keV/μm | 10−15 J | Gy | |
| Carbon | 265 | 85 | 10.7 |
| Nitrogen | 363 | 116 | 14.0 |
| Nickel | 3530 | 1073 | 134.1 |
| Xenon | 8679 | 2781 | 347.7 |
| Uranium | 13 438 | 4306 | 583.2 |
Dose = energy/mass
High-resolution 4Pi microscopy of RPA foci does not circumvent limitations in the laser dose estimation
Because charged particle irradiation produced far fewer RPA foci compared with the expected number of induced DSBs (28), we set out to explore if the limited resolution of the confocal microscope could account for the saturation of the RPA foci dose–effect curve. We attempted to resolve better the single lesions within the individual RPA foci by utilizing high-resolution 4Pi microscopy. Resolving possible substructures would increase the RPA foci number and thus, the dynamic range of the charged particle-induced foci dose-response, potentially allowing a more accurate local LDE definition.
The used 4Pi microscope setup provides a lateral resolution of 170 nm and an axial resolution of 100 nm by applying constructive interference illumination produced by two opposing high aperture lenses. Thus, the axial resolution is ∼5-fold higher than in conventional confocal systems. However, even with the improved resolution of 170 × 170 × 100 nm, a substructure of the RPA foci could not be observed (Figure 5A). Therefore, more detailed imaging did not allow further specification of a local LDE, although the size of the RPA foci was far larger than this resolution limit.
Fig. 5.
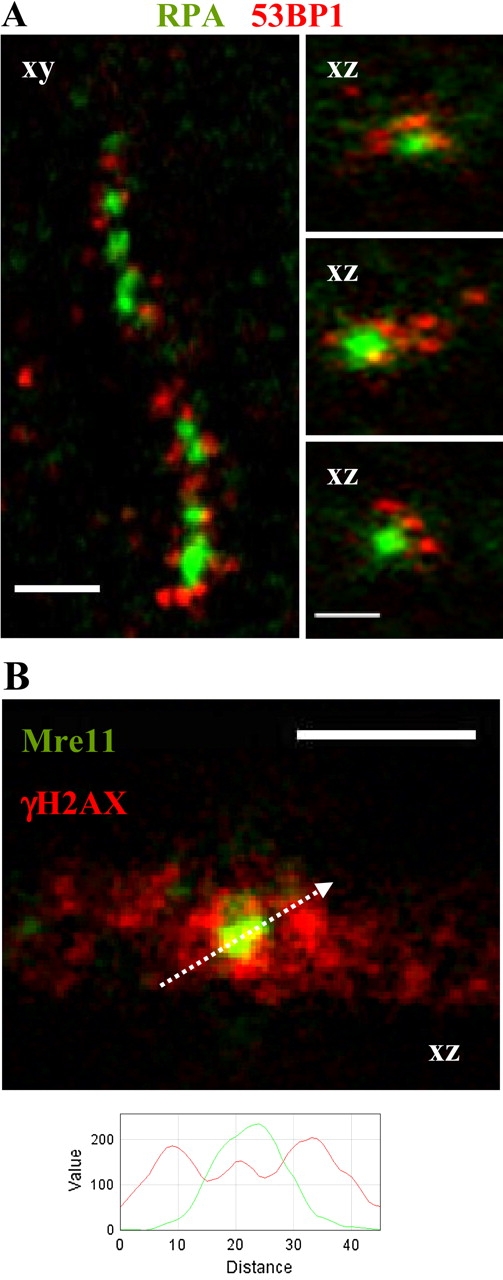
High-resolution 4Pi microscopy of a charged particle-induced RPA streak. (A) The high-resolution image of a xenon ion track shows a streak pattern of charged particle-induced RPA (green) and 53BP1 foci (red). Three RPA foci from the depicted streak are also given in the xz perspective showing no further substructure (resolution: 170 × 170 × 100 nm). All three RPA foci are surrounded by several smaller 53BP1 foci, but both proteins do not co-localize. (B) A similar behaviour is observed for Mre11 foci (green) and γH2AX (red). The depicted Mre11 focus is surrounded by an intense γH2AX signal that is reduced within the Mre11 focus (see profile). Scale bars: 2 μm.
However, high-resolution microscopy revealed a ∼400 nm-sized substructure within individual 53BP1 foci, thus demonstrating the capability of the system to resolve even smaller structures than the observed RPA foci. Interestingly, several of the resolved multiple 53BP1 spots surrounded a single RPA focus without co-localizing (Figure 5A). Comparable behaviour regarding the substructure and the lack of co-localization was observed using 4Pi microscopy for the DSB markers Mre11 after pre-extraction and the DSB-flanking H2AX phosphorylation at this high resolution (Figure 5B).
Recruitment of TRF1 and TRF2 after LMI but not after charged particle irradiation
Further support for a high local LDE came from the observation of the recruitment of telomere repeat-binding factors TRF1 and TRF2. These proteins are known for their interaction with telomeric regions and with repair-related proteins (reviewed in ref. 31). Recently, the DNA damage-induced phosphorylation of TRF2 (32) as well as its recruitment to laser-damaged sites were demonstrated in sensitized (13,24) and non-sensitized cells (13). However, TRF2 recruitment to damage sites was not observed following IR exposures such as 5 Gy γ-rays or α-particles. Even localized irradiation with multiple helium ions at the Columbia University microbeam facility (RARAF) failed to accumulate TRF2 (13). Here, using high LET (8679 keV/μm) xenon ions, no detectable amounts of TRF1 or TRF2 were recruited to the sites of ion traversals (Figure 6B) in human fibroblasts (1BR3) stably expressing YFP-tagged TRF1 or TRF2, respectively. Note that a single xenon ion deposits a local dose of ∼350 Gy (Table II) to the Cube (8 μm3) corresponding to an estimated induction of ∼120 DSBs (33).
Fig. 6.
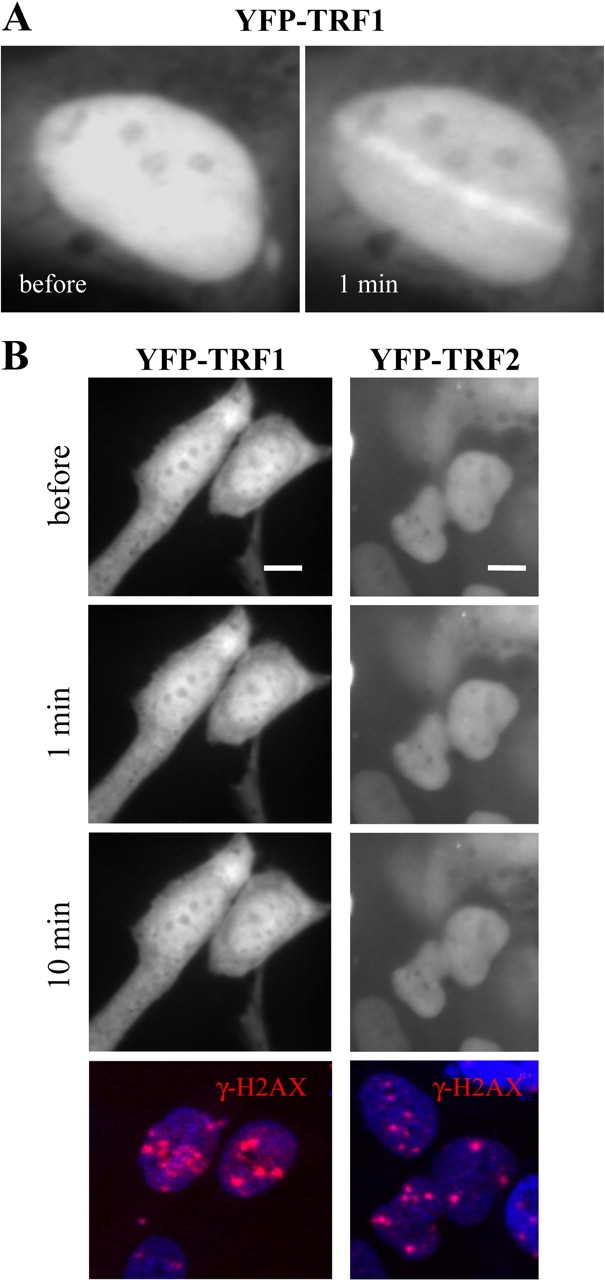
TRF1 and TRF2 recruitment after LMI but not charged particle irradiation. 1BR3 cells stably expressing YFP-tagged TRF1 or TRF2. (A) LMI induces the fast recruitment of YFP-TRF1. The irradiated nucleus is shown directly before and 1 min after the irradiation. (B) The same cell line irradiated with xenon ions does not show TRF1 recruitment within 10 min after irradiation (left panel). Hitting of the depicted nuclei was confirmed by subsequent immunostaining against γH2AX (red; lower panel). Similar results were obtained with YFP-tagged TRF2 also stably expressed in 1BR3 cells (right panel). Scale bar: 10 μm.
Although the high number of DSBs following charged particle irradiation was insufficient to induce TRF1 or TRF2 recruitment, both proteins were readily recruited to laser-irradiated sites within seconds as reported previously for TRF2 (13,24) and shown here for TRF1 after UVA laser irradiation of non-sensitized cells (Figure 6A). This might indicate even higher amounts of induced DSBs. For this experiment, the standard laser power (1.5 μW) was increased 3-fold to compensate for the expected reduced damage induction in non-sensitized cells according to Williams et al. (13), who monitored the recruitment of TRF2 in sensitized and non-sensitized cells using 800 nm two-photon LMI.
Discussion
The aim of this study was to compare damage induction after UVA–LMI and IR in order to estimate the IR equivalent local laser dose deposited by a UVA-LMI system. We first used a commonly applied assay described by Bekker-Jensen et al. (1) based on counting radiation-induced RPA foci. The nuclear LDE deposited in an irradiated mammalian nucleus by a 337-nm laser with a radiant exposure of 0.3 MJ/m2 and prior incorporation of BrdU was found to be equivalent to 2–3 Gy (Figure 1). This is in agreement with previously reported nuclear LDE values of 3 Gy comparing RPA foci numbers (1), 6–9 Gy comparing γH2AX foci numbers (10) or 5.25 Gy comparing γH2AX signal intensities (5).
Nevertheless, we felt these low nuclear LDE values might be misleading as the possible impact of the different spatial dose deposition of LMI and the used sparsely ionizing reference radiation was not taken into account. For example, high locally applied charged particle doses have been shown to produce more efficient DNA fragmentation compared to X-rays (34). Accordingly, an immunofluorescent comet assay monitoring fragment size distributions yielded a higher nuclear LDE of 20 Gy (35).
Since previously estimated nuclear LDEs based on radiation-induced foci formation were obtained disregarding the spatial dose distribution, we set out to test the validity of the RPA foci assay for well-known locally applied doses. For this purpose, we used densely ionizing charged particle irradiation allowing localized energy deposition similar to LMI and estimated its X-ray equivalent dose according to the previously described assay (1). Interestingly, despite depositing particle doses of up to 45 Gy per nucleus (Table I), the assay yielded X-ray equivalent charged particle doses of only ∼2 Gy (Figure 3). Clearly, the difference of the irradiated volumes leads to a discrepancy between estimated and actually applied doses in the case of local irradiation. The assay used is based on the counting of radiation-induced RPA foci within the whole nucleus, but low energy charged particles deposit their energies only within a small confined sub-volume (14,29). Consequently, nuclear particle doses are systematically underestimated using this approach. Since energy is also locally applied in the case of LMI, the LDE will also be underestimated. To account for the differences in the irradiated nuclear volumes after X-rays, charged particles and LMI, we reduced the volume for foci quantification and defined a cubic volume of 2 × 2 × 2 μm (Cube) to compensate for the radial symmetrical dose deposition of charged particles. The numbers of foci per Cube were quantified based on foci counting in average projections as commonly done. Since projections encompass the whole nucleus in depth (up to 3 μm) but the Cube's height is only 2 μm, scaling factors were introduced as described in detail in the Materials and methods section.
Following the application of the Cube, the charged particle-induced RPA foci dose–response curve reached a plateau for the high locally applied doses of xenon and uranium ions (Figure 4). Since the RPA foci induction after UVA–LMI was located in this saturation region, the local LDE could only be estimated to be roughly in the range of a few hundred Gray. This estimation is in agreement with calculated laser doses based on standard absorption rates (4). In contrast, previous attempts of a local laser dose estimation using sparsely IR as the only reference, but applying volumes for foci counting 7-fold larger than the Cube, yielded lower values of 10 Gy (1) or 30 Gy (11). However, in these studies, the defined volumes for foci counting were designed to encompass the whole laser streak, thereby comprising significant proportions of unirradiated volume. The obtained local LDE values were, therefore, comparably low. In the present study, the used UVA–LMI could be attributed to dose values exceeding 100 Gy, but the accuracy of the estimation is still limited by the observed saturation of the RPA foci numbers with increasing dose deposition. As for single measurements, foci numbers often exceeded the mean values given here; a detection limit of the used image software can be excluded. Thus, the saturation is most probably based on biological effects. A possible depletion of the RPA protein, a key mechanism in the replication/repair switch (36), may only explain the observed saturation of the X-ray dose–response curve, as the total number of induced RPA foci is much higher than after charged particle irradiation and UVA–LMI. We investigated further whether the observed plateau in the number of charged particle-induced RPA foci was a result of the resolution limit of the confocal microscope that might be insufficient to resolve single lesions within RPA foci. In order to circumvent the limiting resolution of the confocal microscope, perhaps masking additional RPA foci, we performed high-resolution 4Pi microscopy aiming to reveal the substructure of individual foci. Although the RPA foci induced after xenon ion irradiation were generally larger than the resolution limit of the 4Pi microscope (170 nm lateral, 100 nm axial), a substructure could not be resolved (Figure 5A). Hence, even high-resolution microscopy was not able to improve the estimation of the locally applied LDE.
While RPA foci could not be resolved further by 4Pi microscopy, interestingly, the damage marker 53BP1 showed a substructure of multiple smaller domains surrounding the RPA focus but remarkably without co-localizing. As 53BP1 specifically binds to a histone modification (37) and RPA binds to ssDNA (38) most probably unwound from nucleosomes, this might explain the nearby but not overlapping foci. However, even high-resolution microscopy did not reveal separate RPA-marked ssDNA filaments. This observation supports the idea that the RPA foci do not mark the processing of separate individual DNA lesions after densely IR as suggested previously (28). Instead, multiple DNA damages originally induced in near vicinity within a chromatin domain might be processed subsequently at a common place resulting in a central RPA focus surrounded by several smaller 53BP1 stained regions (Figure 5A). The finding that damage-induced histone modifications are flanking but not co-localizing with the actual sites of DSB repair was also observed after Mre11 and γH2AX staining. 4Pi microscopy revealed that the focus of Mre11 remaining after in situ extraction is surrounded by a bright γH2AX staining, whereas in its centre, the γH2AX signal appears less intense (Figure 5B). Notably, the Mre11 protein involved in DSB end processing has exonuclease/endonuclease activity providing a potential basis for ssDNA formation, a prerequisite to RPA binding. The substructure observed for the phosphorylated form of histone H2AX is in agreement with previously described distributions of γH2AX into microdomains using 4Pi microscopy (39).
A comparison of the laser exposure used in this study with values in the literature (Table III) shows that the chosen setting for the UVA laser system (radiant exposure of 0.3 MJ/m2) is comparable with settings of similar UVA laser types. Thus, it can be excluded that the high local LDE found in the present study is a result of an inappropriately high laser power. It also reflects the validity of the presented estimation and applies to other UVA–LMI systems.
Table III.
Typically used UVA laser exposuresa
| Wavelength in nm | Radiant exposure in MJ/m2 | References |
| 337 | 0.1 | Referred in (20) |
| 0.3 | Present study | |
| 0.4 | (20) | |
| 0.5 | (1,20,40) | |
| 1 | (41) | |
| 390 | 10, 30 | (4) |
In the case of pre-sensitization by BrdU incorporation.
Further support for a high locally applied LDE arises from the differences in the recruitment of the telomere repeat-binding factors TRF1 and TRF2 after charged particle irradiation and LMI. The TRF2 protein has been shown to accumulate after LMI in both pre-sensitized (13,24) and non-sensitized cells (13), and recently, a function in strand invasion during non-telomeric homologous recombination repair of DSBs was proposed (42). Remarkably, no recruitment of TRF2 could be shown after IR even after local (20 μm2) application of 416 Gy α-particles using a microprobe setup for targeted irradiation (13). In our study, we used xenon ion irradiation applying an even more confined local dose of ∼350 Gy in a volume of 8 μm3. We also saw no accumulation of TRF1 or TRF2 (Figure 6B) at sites of particle traversals. In contrast, LMI rapidly induced recruitment of TRF2 (13,24) and TRF1 (Figure 6A). Different scenarios can be imagined to explain these contrasting results. The density of DSBs could be higher after two-photon LMI (13) or UVA–LMI (used here) than with heavy charged particles resulting in a detectable amount of accumulated proteins. This leads to the conclusion that the LDE of our laser system is much >350 Gy under the conditions used (increased laser power and omitted sensitization). Despite the reported influence of sensitization treatments on the yielded damage spectra (20), we obtained comparable LDE estimations using either sensitization and standard laser settings or omitting sensitization and increasing the laser power. The ability of higher laser power to compensate for a reduced damaging capability has been shown previously for TRF2 recruitment following two-photon LMI (13).
Alternatively, possible differences in the induced damage spectra could also be a reason for the differential recruitment after LMI and charged particle irradiation, as discussed by Williams et al. (13). According to the authors, the additional induction of UV-related damages after LMI might be responsible for the recruitment of TRF2. Regardless of the damage type, UVA- and two-photon LMI clearly generate conditions that permit TRF1 and TRF2 recruitment to laser-induced DNA damage sites. In this particular context, it is interesting that the mechanism of DSB induction classically described for UVA bulb irradiation does not explain the proposed high amount of DSBs induced by LMI (43). Therefore, further damage induction related to non-linear absorption effects needs to be considered (20,40). This might also be an explanation for the recruitment of TRF1 (see Figure 6A) and TRF2 (13) showing that sensitization is not mandatory for damage induction if the laser power is increased. The occurrence of damage induced by non-linear laser absorption would make a direct comparison of repair data obtained after LMI and ionizing irradiation even more difficult.
In summary, in the present study, by considering the locally applied dose deposition, we estimated the local UVA–LDE to be in the range of a few hundred Gray. Due to the saturation of the foci dose–response curve, a more accurate specification of the local LDE could not be achieved, despite the application of very sophisticated techniques such as low-angle ion irradiation and 4Pi microscopy. However, 4Pi microscopy does facilitate visualization of changes in the substructure of DSB markers such as γH2AX and 53BP1, which might allow further characterization of high local doses. Clearly, a more detailed knowledge of the laser-induced damage spectrum and the mechanism of damage generation is needed. Nevertheless, to at least improve understanding of comparability of data from different LMI systems, it would be useful to give information about LMI parameters as the totally applied laser energy, the spot size of the laser beam, the wavelength, pulse duration and the energy per laser pulse, as well as the use or lack of pre-sensitization treatments.
Supplementary data
Supplementary Figure S1 is available at Mutagenesis Online.
Funding
Bundesministerium für Bildung und Forschung (03NUK001A); National Institutes of Health (CA50519 and CA134991); NASA (NNA05CM04G).
Supplementary Material
Acknowledgments
We are grateful to Dr G. Kraft for continuous support and encouragement. We thank G. Becker and A.L. Leifke for competent cell culturing and assistance, W. Becher and G. Lenz for technical support during irradiation and F. Tobias for helpful suggestions on the manuscript.
Conflict of interest statement: None declared.
References
- 1.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mari PO, Florea BI, Persengiev SP, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl Acad. Sci. USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uematsu N, Weterings E, Yano K, et al. Autophosphorylation of DNA-PKcs regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 5.Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mortusevicz O, Amé JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mortusevicz O, Rothbauer U, Cardoso MC, Leonhardt H. Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res. 2006;34:3523–3532. doi: 10.1093/nar/gkl492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haince JF, McDonald D, Rodrigue A, Déry U, Masson JY, Hendzel MJ, Poirier GG. PARP-1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 11.Hong Z, Jiang J, Lan L, Nakajima S, Kanno S, Koseki H, Yasui A. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res. 2008;36:2939–2947. doi: 10.1093/nar/gkn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aten JA, Stap J, Krawczyk PM, Van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- 13.Williams ES, Stap J, Essers J, Ponnaiya B, Luijsterburg MS, Krawczyk PM, Ullrich RL, Aten JA, Bailey SM. DNA double-strand breaks are not sufficient to initiate recruitment of TRF2. Nat. Genet. 2007;39:698–699. doi: 10.1038/ng0607-696. [DOI] [PubMed] [Google Scholar]
- 14.Jakob B, Scholz M, Taucher-Scholz G. Biological imaging of heavy charged-particle tracks. Radiat. Res. 2003;159:676–684. doi: 10.1667/0033-7587(2003)159[0676:biohct]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Jakob B, Scholz M, Taucher-Scholz G. Characterization of CDKN1A (p21) binding to sites of heavy-ion-induced damage: colocalization with proteins involved in DNA repair. Int. J. Radiat. Biol. 2002;78:75–88. doi: 10.1080/09553000110090007. [DOI] [PubMed] [Google Scholar]
- 16.Heiss M, Fischer BE, Jakob B, Fournier C, Becker G, Taucher-Scholz G. Targeted irradiation of mammalian cells using a heavy-ion microprobe. Radiat. Res. 2006;165:231–239. doi: 10.1667/rr3495.1. [DOI] [PubMed] [Google Scholar]
- 17.Jakob B, Rudolph JH, Gueven N, Lavin MF, Taucher-Scholz G. Live cell imaging of heavy-ion-induced radiation responses by beamline microscopy. Radiat. Res. 2005;163:681–690. doi: 10.1667/rr3374. [DOI] [PubMed] [Google Scholar]
- 18.Jakob B, Splinter J, Durante M, Taucher-Scholz G. Live cell microscopy analysis of radiation-induced DNA double-strand break motion. Proc. Natl Acad. Sci. USA. 2009;106:3172–3177. doi: 10.1073/pnas.0810987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohanty SK, Rapp A, Monajembashi S, Gupta PK, Greulich KO. Comet assay measurements of DNA damage in cells by laser microbeams and trapping beams with wavelengths spanning a range of 308 nm to 1064 nm. Radiat. Res. 2002;157:378–385. doi: 10.1667/0033-7587(2002)157[0378:camodd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 20.Kong X, Mohanty SK, Stephens J, Heale JT, Gomez-Godinez V, Shi LZ, Kim JS, Yokomori K, Berns MW. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009;37:e68. doi: 10.1093/nar/gkp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meldrum RA, Botchway SW, Wharton CW, Hirst GJ. Nanoscale spatial induction of ultraviolet photoproducts in cellular DNA by three-photon near-infrared absorption. EMBO Rep. 2003;4:1144–1149. doi: 10.1038/sj.embor.7400028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berns MW, Wang Z, Dunn A, Wallace V, Venugopalan V. Gene inactivation by multiphoton-targeted photochemistry. Proc. Natl Acad. Sci. USA. 2000;97:9504–9507. doi: 10.1073/pnas.97.17.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutherland BM, Griffin KP. Absorption spectrum of DNA for wavelengths greater than 300 nm. Radiat. Res. 1981;86:399–409. [PubMed] [Google Scholar]
- 24.Bradshaw PS, Stavropoulos DJ, Meyn MS. Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat. Genet. 2005;37:193–197. doi: 10.1038/ng1506. [DOI] [PubMed] [Google Scholar]
- 25.Limoli CL, Ward JF. A new method for introducing double-strand breaks into cellular DNA. Radiat. Res. 1993;134:160–169. [PubMed] [Google Scholar]
- 26.Lang M, Engelhardt J, Hell SW. 4Pi microscopy with linear fluorescence excitation. Opt. Lett. 2007;32:259–261. doi: 10.1364/ol.32.000259. [DOI] [PubMed] [Google Scholar]
- 27.Staudt T, Lang M, Medda R, Engelhardt J, Hell SW. 2,2’-Thiodiethanol: a new water soluble mounting medium for high resolution optical microscopy. Microsc. Res. Tech. 2007;70:1–9. doi: 10.1002/jemt.20396. [DOI] [PubMed] [Google Scholar]
- 28.Jakob B, Splinter J, Taucher-Scholz G. Positional stability of damaged chromatin domains along radiation tracks in mammalian cells. Radiat. Res. 2009;171:405–418. doi: 10.1667/RR1520.1. [DOI] [PubMed] [Google Scholar]
- 29.Scholz M, Kellerer AM, Kraft-Weyrather W, Kraft G. Computation of cell survival in heavy ion beams for therapy. Radiat. Environ. Biophys. 1997;36:59–66. doi: 10.1007/s004110050055. [DOI] [PubMed] [Google Scholar]
- 30.Krämer M, Kraft G. Calculations of heavy-ion track structure. Radiat. Environ. Biophys. 1994;33:91–109. doi: 10.1007/BF01219334. [DOI] [PubMed] [Google Scholar]
- 31.de Boeck G, Forsyth RG, Praet M, Hogendoom PCW. Telomere-associated proteins: cross-talk between telomere maintenance and telomere-lengthening mechanisms. J. Pathol. 2008;217:327–344. doi: 10.1002/path.2500. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka H, Mendonca MS, Bradshaw PS, Hoelz DJ, Malkas LH, Meyn MS, Gilley D. DNA damage-induced phosphorylation of the human telomere-associated protein TRF2. Proc. Natl Acad. Sci. USA. 2005;102:15539–15544. doi: 10.1073/pnas.0507915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prise KM, Ahnström G, Belli M, et al. A review of DSB induction data for varying quality radiations. Int. J. Radiat. Biol. 1998;74:173–184. doi: 10.1080/095530098141564. [DOI] [PubMed] [Google Scholar]
- 34.Löbrich M, Cooper PK, Rydberg D. Non-random distribution of DNA double-strand breaks induced by particle irradiation. Int. J. Radiat. Biol. 1996;70:493–503. doi: 10.1080/095530096144680. [DOI] [PubMed] [Google Scholar]
- 35.Grigaravicius P, Rapp A, Greulich KO. A direct view by immunofluorescent comet assay (IFCA) of DNA damage induced by nicking and cutting enzymes, ionizing 137Cs radiation, UV-A laser microbeam irradiation and the radiometric drug bleomycin. Mutagenesis. 2009;24:191–197. doi: 10.1093/mutage/gen071. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Zhou XY, Wang H, Huq MS, Iliakis G. Roles of Replication Protein A and DNA-dependent Protein Kinase in the regulation of DNA replication following DNA damage. J. Biol. Chem. 1999;274:22060–22064. doi: 10.1074/jbc.274.31.22060. [DOI] [PubMed] [Google Scholar]
- 37.Huyen Y, Zgheib O, DiTullio RA, Jr., et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 38.Kim C, Snyder RO, Wold MS. Binding properties of Replication Protein A from human and yeast cells. Mol. Cell. Biol. 1992;12:3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bewersdorf J, Bennett BT, Knight KL. H2AX chromatin structures and their response to DNA damage revealed by 4Pi microscopy. Proc. Natl Acad. Sci. USA. 2006;103:18137–18142. doi: 10.1073/pnas.0608709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogel A, Lorenz K, Horneffer V, Hüttmann G, von Smolinski D, Gebert A. Mechanisms of laser-induced dissection and transport of histologic specimens. Biophys. J. 2007;93:4481–4500. doi: 10.1529/biophysj.106.102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tashiro S, Walter J, Shinohara A, Kamada N, Cremer T. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol. 2000;150:283–291. doi: 10.1083/jcb.150.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao Z, Seluanov A, Jiang Y, Gorbunova V. TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc. Natl Acad. Sci. USA. 2007;104:13068–13073. doi: 10.1073/pnas.0702410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peak JG, Peak MJ. Ultraviolet light induces double-strand breaks in DNA of cultured human P3 cells as measured by neutral filter elution. J. Photochem. Photobiol. 1990;52:387–393. doi: 10.1111/j.1751-1097.1990.tb04194.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.