Abstract
Background. Female carriers of X-linked Alport syndrome (XLAS) demonstrate variability in clinical phenotype that, unlike males, cannot be correlated with genotype. X-inactivation, the method by which females (XX) silence transcription from one X chromosome in order to achieve gene dosage parity with males (XY), likely modifies the carrier phenotype, but this hypothesis has not been tested directly.
Methods. Using a genetically defined mouse model of XLAS, we generated two groups of Alport female (Col4a5+/−) carriers that differed only in the X-controlling element (Xce) allele regulating X-inactivation. We followed the groups as far as 6 months of age comparing survival and surrogate outcome measures of urine protein and plasma urea nitrogen.
Results. Preferential inactivation of the mutant Col4a5 gene improved survival and surrogate outcome measures of urine protein and plasma urea nitrogen. In studies of surviving mice, we found that X-inactivation in kidney, measured by allele-specific mRNA expression assays, correlated with surrogate outcomes.
Conclusions. Our findings establish X-inactivation as a major modifier of the carrier phenotype in X-linked Alport syndrome. Thus, X-inactivation patterns may offer prognostic information and point to possible treatment strategies for symptomatic carriers.
Keywords: collagen type IV; disease models, animal; female; longevity; nephritis, hereditary
Introduction
X-linked Alport syndrome (XLAS, OMIM #301050) is a disorder of basement membranes caused by mutations in the COL4A5 gene encoding the α5(IV) chain of type IV collagen. Affected males demonstrate deafness and progressive glomerulopathy leading to end-stage kidney disease (ESKD) in young adulthood [1]. Although early reports indicated that carrier females had few serious manifestations [2], a natural history study published in 2003 demonstrated disease burden in this population [3]. By age 40, 12% of female carriers reach ESKD, while 10% experience deafness. Genotype–phenotype correlations can be drawn in males, with patients having large deletions or nonsense mutations demonstrating severe disease as compared to those with missense or splice site mutations [4,5]. However, no such genotype–phenotype correlations have been found in females with variability in disease evident even among family members [3].
X-inactivation, the process by which early somatic cells inactivate one of two X chromosomes to achieve dosage compensation, likely contributes to variability among female carriers of X-linked disorders including XLAS, but this hypothesis has not been tested directly. While X-inactivation ratios of 50:50 are expected in a normal population of cells, X-inactivation ratios can be skewed due to statistical fluctuation, X-inactivation modifier genes or mutation selection advantages [6].
The influence of X-inactivation on the Alport carrier phenotype has been difficult to define. Guo et al. described an Alport carrier who developed ESKD by age 30 and was found to have two mutations in COL4A5 expressed in >90% of kidney cells and lymphocytes [7]. Vetrie et al. did not find a correlation between X-inactivation measured in lymphocytes with Alport disease severity in a group of 43 women [8]. Nakanishi et al. found correlations between epidermal basement membrane (EBM) and glomerular basement membrane (GBM) α5(IV) expression and between EBM α5(IV) expression and urine protein-to-creatinine ratio [9]. Massella et al. did not find a correlation between EBM α5(IV) expression and disease severity [10]. In a study of five Alport carriers, Wang et al. found that cultured skin fibroblast COL4A5 mRNA levels correlated significantly with dipstick proteinuria [11]. These findings may be inconsistent among studies because X-inactivation ratios vary depending upon patient age and tissue of origin limiting the relevance of blood or skin to kidney.
To elucidate the effects of X-inactivation in female carriers of XLAS, we utilized a mouse model that recapitulates features of human Alport syndrome [12]. Alport female (Col4a5+/−) mice exhibit mosaic expression of the α5(IV) chain and develop kidney disease variably and more slowly than affected males. In mice, the X-controlling element Xce is the major locus influencing X-inactivation choice. Three Xce alleles have been identified among inbred strains: Xcea, Xceb and Xcec [13–15]. Mice homozygous for Xce alleles demonstrate random X-inactivation. Mice heterozygous for Xce alleles demonstrate preferential X-inactivation in rank order Xcea > Xceb > Xcec. By perturbing X-inactivation experimentally, we demonstrate that X-inactivation influences clinical outcomes.
Materials and methods
Mice
All studies were carried out under protocols approved by the Institutional Animal Care Use Committee at the University of Minnesota and adhere to the NIH Guide for the Care and Use of Laboratory Animals. Col4a5-targeted breeder mice were backcrossed onto a congenic C57BL/6 (Xceb) background. The 129.Pgk1a line, generated by backcrossing the Xcec allele onto a congenic 129SvPas background, has been maintained by brother–sister matings [15]. Group 1 mice were generated by breeding Col4a5+/− Alport females and 129SvPas (Xcea) males. Group 2 mice were generated by breeding Alport females and 129.Pgk1a (Xcec) males. Mice were genotyped as described previously [12]. Urine samples were collected after spontaneous voiding at 2, 4 and 6 months of age. At 6 months of age, mice were anaesthetized by intraperitoneal injection of pentobarbital (50 mg/kg). Blood samples were obtained by puncture of the vena cava and kidneys harvested thereafter.
Laboratory analysis
Urine protein was measured using the Bio-Rad Protein Assay (Bio-Rad Laboratories). Urine creatinine was measured by the Jaffé reaction, using the Creatinine Analyzer-2 (Beckman Coulter, Inc.). Plasma urea nitrogen was measured using the Liquid Urea Nitrogen Reagent Set (Pointe Scientific, Inc.).
Allele-specific mRNA expression assays
By database search, we identified expressed polymorphisms distinguishing B6 and 129 strains in the X-chromosome genes Srpx, Rpgr and Aff2 (Table 1). We verified these polymorphisms by sequencing PCR products amplified from genomic DNA. RNA was extracted from kidneys using Ultraspec RNA (Biotecx Laboratories, Inc.). Samples were reverse transcribed using the Superscript III Kit (Invitrogen). Products were amplified in a multiplex polymerase chain reaction using gene-specific primers (Table 1). After purification through Qiagen spin columns, products were divided and subjected to overnight restriction digestion with Sau96I (for Srpx), BsaAI (for Rpgr) or BsrDI (for Aff2). Digestion products were analysed using an Agilent 2100 Bioanalyzer (Agilent Technologies) and Xi for a given gene expressed as the sum of densitometric units for B6 bands relative to the sum for B6 and 129 bands. In preliminary experiments, we found that RT–PCR amplification of Rpgr yielded a doublet likely reflecting alternative splicing and that neither product could be digested to completion as predicted. In addition, correlation between Xi determined by Srpx and Aff2 allele-specific assays gave evidence for outlying behaviour of the Aff2-based assay (data available upon request). We did not explore the basis for this observation but suspect that heteroduplexes resistant to digestion formed under the assay conditions. Therefore, we opted to use Xi from the Srpx-based assay in our analysis.
Table 1.
Single-nucleotide polymorphisms, gene-specific primers and expected band sizes for undigested and digested PCR products used in allele-specific mRNA expression assays
| Gene | refSNP ID | PCR primer sequences | Size (bp) | Enzyme | Fragments B6 Allele | Fragments 129 Allele |
|---|---|---|---|---|---|---|
| Srpx | rs30730568 | F:5′-TCATCGGCAGGATCAGAGCAAAG-3′ | 375 | Sau96I | 154 221 | 375 |
| R:5′-GCAAAGAAAGCACATAATAAAATAGAC-3′ | ||||||
| Rpgr | rs33492631 | F:5′-AGTACAATGAAAATCCAAAAGGACAC-3′ | 602 | BsaAI | 602 | 95 507 |
| R:5′-GCTTCATTCGTCTTTCAAATATAAAAATAG-3′ | ||||||
| Aff2 | rs13483812 | F:5′-AAACAGATTCATCTACATCTGACTCC-3′ | 441 | BsrDI | 166 275 | 441 |
| R:5′-CAGGGATTTTCTCTGCAGTTTCTG-3′ |
Quantitative reverse transcription-polymerase chain reaction
We selected 12 samples of reverse-transcribed kidney RNA randomly from both Groups 1 and 2, stratifying across Xi and also three samples from both wild-type females and Alport males. Each sample was amplified in triplicate for Col4a5 and Gapdh using SYBR Green PCR Master Mix (Invitrogen). For Col4a5, we used forward primer 5′-AGAGAAGAATGCAAGTGCGTG-3′ recognizing the wild-type sequence but not the Alport mutation c.13G>T [12] and reverse primer 5′-CTTCTGGACCTGGAAATCCT-3′. For Gapdh, we used a commercial primer pair (SA Biosciences). We verified linear behaviour for the primer pairs in preliminary experiments. Threshold cycles were averaged for each sample and data transformed and analysed by standard methods [16]. Col4a5 are expressed relative to Gapdh mRNA levels and scaled to give a mean value of 1 for wild-type females. Wild-type Col4a5 mRNA was undetectable in kidneys from Alport males.
Statistics
Comparison of premature death rate was performed using a chi-square test for the 2 × 2 table of group by premature death; we estimated the proportion of variation explained by group with maximum-rescaled R2 from logistic regression. Urine protein excretion and plasma urea nitrogen were both analysed on the log scale to adjust right skew towards normality, and presented results are transformed back to the original scale, as geometric means with 95% confidence intervals (CI) that represent the range of uncertainty in the presented mean due to random sampling. Similarly, Col4a5 mRNA levels were analysed on the log2 scale and transformed back by a scaled power of 2 where the scale factor was estimated based on the mean of the wild-type females for presenting means, 95% confidence intervals and graphs. Longitudinal urine protein excretion measurements at 2, 4 and 6 months were modelled with a random intercept for each mouse, accounting for correlation between measurements within the same mouse and adjusting for unbalanced number of observations across mice who died prematurely; this model was used to estimate an overall linear trend across time and the main effect for group, which was also used to test for a difference in mean urine protein excretion across groups [17]. Plasma urea nitrogen was only measured in mice surviving to 6 months, and its mean difference across groups was tested using an independent two-sample t-test.
Histograms of the Srpx-based Xi distribution for each group were presented, and a test of mean difference between groups was performed using a two-sample t-test. Each group’s mean Xi was compared to the value of 0.5 by one-sample t-test. Spearman correlations were used to test association between Xi and 6-month urine protein excretion and plasma urea nitrogen. The association between Xi and trends in urine protein excretion over time was examined in mice that did not die prematurely. Longitudinal measurements of urine protein excretion at 2, 4 and 6 months were modelled as a function of age, Xi and an age*Xi interaction. All analyses were performed using SAS Version 9.1, and graphics were made using R [18].
Results
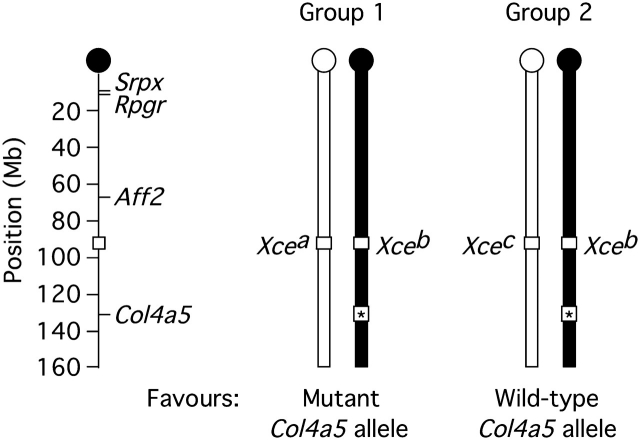
We generated two groups of Alport females sharing a [C57BL/6 (B6) × 129SvPas (129)] F1 genetic background but diverging at the Xce locus (Figure 1). In Group 1 (Xcea/b), inactivation of the X chromosome carrying the wild-type Col4a5 allele was favoured. In Group 2 (Xceb/c), inactivation of the X chromosome carrying the mutant Col4a5-null allele was favoured. Hypothesizing more severe manifestations of Alport kidney disease in Group 1, we designed our comparison between Groups 1 and 2 with power to detect a significant difference in urine protein excretion—a marker of kidney disease estimated by the ratio of protein to creatinine in spot samples—at 6 months of age. Unexpectedly, some mice died before 6 months: 37% (14/38) in Group 1 and 8% (4/52) in Group 2, with a significantly higher rate in Group 1 (chi-square P = 0.0006). Xce genotype (group) explained 20% of the variance in survival (maximum-rescaled R2).
Fig. 1.
Groups 1 and 2. Representation of paired 129 (white) and B6 (black) X chromosomes illustrating map positions and genotypes. Strain-specific polymorphisms in the Srpx, Rpgr and Aff2 genes were used in X-inactivation assays.
Urine protein excretion was measured at 2, 4 and 6 months, and group means were compared by longitudinal analysis after controlling for trends. Group 1 [mean = 20 mg/mg creatinine, 95% confidence interval (CI): 17–24, n = 38] was significantly higher than Group 2 (mean = 16 mg/mg creatinine, 95% CI: 14–18, n = 50) (P = 0.045). In a comparison of plasma urea nitrogen at 6 months—a marker of kidney function measurable in surviving mice—Group 1 (mean 68 mg/dl, 95% CI: 51–89, n = 23) was significantly higher than Group 2 (mean 44 mg/dl, 95% CI: 37–52, n = 47) (P = 0.0061). Together, these findings confirmed more severe Alport kidney disease in Group 1.
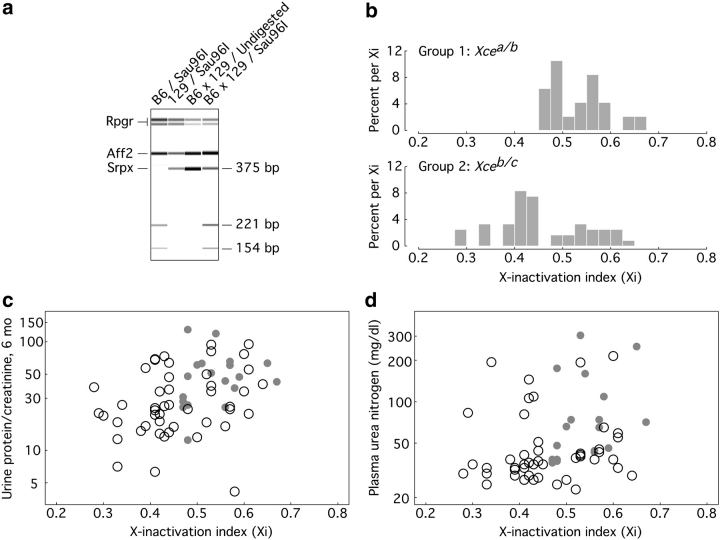
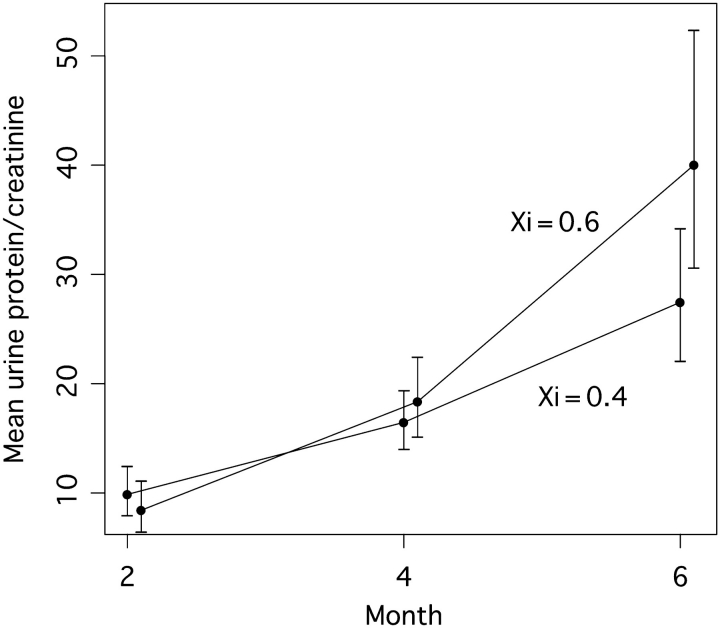
Our comparison between Groups 1 and 2 established firmly and for the first time that Xce resides within a region modifying X-linked disease. To more closely investigate the relationships between X-inactivation and disease manifestations, we determined an X-inactivation index (Xi) from allele-specific mRNA expression assays of kidneys from surviving mice and correlated the value to surrogate markers. Xi was measured as B6 allele-specific expression of the Srpx gene relative to total Srpx expression (Figure 2a), with higher values of Xi thereby indicating preferential activation of the X chromosome bearing the null allele. As expected, Xi was significantly higher in Group 1 (0.53 ± 0.06, n = 19) than in Group 2 (0.46 ± 0.09, n = 48) (P = 0.0014) (Figure 2b), with the Group 1 mean higher than 0.5 (P = 0.023) and the Group 2 mean lower than 0.5 (P = 0.0025). Xi was positively correlated with both 6-month urine protein excretion (Spearman r = 0.39; P = 0.0013; Figure 2c) and plasma urea nitrogen (Spearman r = 0.39; P = 0.0013; Figure 2d). In longitudinal analysis of urine protein excretion, we found a significant effect of age (P = 0.03) and a significant age-by-Xi interaction (P = 0.031) indicating that the rate of disease progression in carrier females was positively associated with Xi. Estimated example trend lines for mice with Xi = 0.4 compared to 0.6 (Figure 3) indicate no significant difference at 2 months but a statistically significant difference by 6 months (P = 0.039).
Fig. 2.
(a) Representative Srpx-based assays for Xi. Mouse kidneys from the indicated strains were analysed by multiplex RT-PCR and subsequent digestion as described in Animals and Methods and Table 1. Sau96I digests the B6, but not the 129 Srpx RT-PCR product, generating fragments of 154 and 221 bp from the original 375-bp product. (b) Distribution by group of Xi, measured as allele-specific Srpx expression. (c) Correlation between Xi and 6-month urine protein-to-creatinine ratio. Group 1 mice are indicated by filled grey circles and Group 2 by open circles. (d) Correlation between Xi and 6-month plasma urea nitrogen. Group 1 mice are indicated by filled grey circles and Group 2 by open circles.
Fig. 3.
Predicted mean level and slope change in urine protein-to-creatinine ratio based on longitudinal analysis including Xi as a moderator of slope (i.e. including an Xi-by-age interaction). Specific examples of trajectories are given for Xi = 0.6 and Xi = 0.4. Vertical bars represent 95% CI of predicted mean urine protein-to-creatinine ratio at each time point. At 6 months, the 95% CIs do not overlap with the other mean values and this difference is statistically significant (P = 0.039).
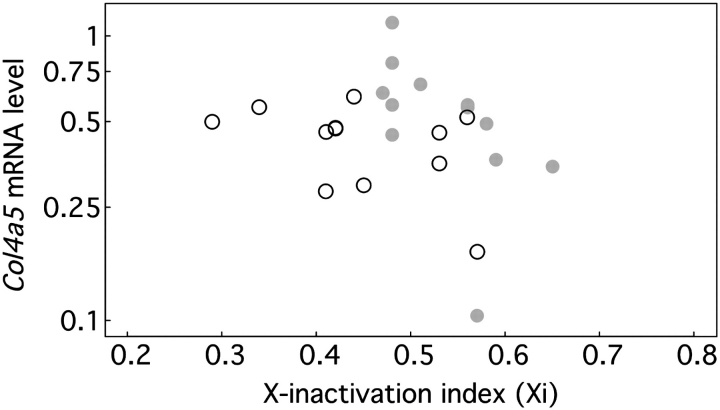
The basis for relationships between X-inactivation and clinical manifestations in our model is unknown, although the simplest explanation would be that favourable X-inactivation increases Col4a5 expression. For a randomly chosen subsample, we measured Col4a5 mRNA levels confirming that they were lower in Alport (mean 0.45, 95% CI: 0.37–0.55, n = 24) than wild-type (mean 1.00, 95% CI: 0.68–1.48, n = 3) females (P = 0.011). While there was a negative trend between Xi and Col4a5 mRNA levels (Figure 4), the relationship was not statistically significant (Spearman r = −0.23; P = 0.279, n = 24). Furthermore, Col4a5 mRNA levels did not correlate with urine protein excretion or plasma urea nitrogen (data not shown) implicating additional factors in the relationships between Xi and outcomes.
Fig. 4.
Correlation between Xi and Col4a5 mRNA levels. Group 1 mice are indicated by filled grey circles and Group 2 by open circles.
Discussion
After controlling for disease mutation and genetic background, we have shown clearly that clinical manifestations in female carriers of XLAS are modified by X-inactivation. Our findings establish X-inactivation as a risk factor for disease progression and shed light on an abiding conundrum in Alport syndrome relevant to other X-linked disorders, namely, that in large human studies, genotype–phenotype correlations can be drawn for males but not females [3,5]. In future attempts at genotype–phenotype correlations, including those relating to risk of hearing loss [3,19], it may be useful to adjust for X-inactivation. In this connection, we note that there are differences between mechanisms of X-chromosome regulation in mice and humans including the lack of definitive evidence for an Xce interval in humans [20,21].
Our most striking finding is that favourable X-inactivation improves survival. Because we could not measure the X-inactivation index Xi in mice that died prematurely, we have no information on whether Xi related directly to premature death. However, Xi related directly to Xce genotype, which in turn strongly related to premature death, implying that Xi related at least indirectly to premature death.
What is the basis for the relationships between X-inactivation and clinical outcomes in our model? Although Xi trended with Col4a5 mRNA levels, we did not observe correlations between Col4a5 mRNA levels and clinical outcomes. Perhaps this is not surprising. We measured Col4a5 mRNA levels in whole kidneys from animals surviving to 6 months of age, a method that does not account for detailed patterns of cellular mosaicism or spatial distribution of the α5(IV) chain. Baumal et al. described temporal conversion from mosaic to global α5(IV) GBM expression in canine XLAS [22], raising—among other possibilities—those of escape from X-inactivation and/or selection for cells in which the normal allele is active as confounding influences over Col4a5 mRNA levels in females. Moreover, type IV collagen gene expression is subject to complex coordinate regulation in the wake of Alport mutations [23–25]. The availability of XLAS mice will allow future studies that examine Col4a5 mRNA and GBM α5(IV) expression in parallel with dynamics of X-inactivation, starting with younger animals in which X-inactivation effects on these parameters are most directly evident, and correlating to clinical outcomes.
It is possible that X-inactivation effects are also mediated by mechanisms distinct from Col4a5 regulation. Xce is the major regulator of X-inactivation choice [26]; therefore, not just Col4a5 but all X-chromosome alleles on the (B6 × 129) F1 background will be subject to its effects. Xce may also interact with other genetic modifiers including those previously characterized in murine Alport syndrome [25,27]. We cannot exclude the possibility, however unlikely, that differences between Groups 1 and 2 are due to paternal alleles closely linked to Xce but not themselves involved in X-inactivation choice.
In as much as clinical outcomes are related to X-chromosome dosing, our findings suggest that X-inactivation choice can be considered a potential therapeutic target in symptomatic carriers. This group represents a unique population in that a normal copy of the COL4A5 gene is present in every cell although not expressed. The ability to manipulate X-inactivation choice post-natally would provide clinicians with a powerful tool obviating the need for gene delivery. Finally, our findings highlight insights to be gained from clinical studies of female carrier mosaics as these may apply to dosing considerations in gene replacement therapy of genetic diseases arising by loss of function.
Acknowledgments
The authors are grateful to R. Ehlenfeldt for technical assistance and Dr. B. Migeon for comments on a preliminary version of the manuscript. This work was supported by US National Institutes of Health grants DK60695 and DK64273 and with resources and the use of facilities at the Minneapolis VA Medical Center.
Conflict of interest statement. None declared.
References
- 1.Segal Y, Kashtan CE. Genetic abnormalities in glomerular function. In: Alpern RJ, Hebert SC, editors. Seldin and Giebisch's The Kidney: Physiology & Pathophysiology. Boston, MA: Elsevier Inc., Academic Press; 2007. [Google Scholar]
- 2.Perkoff GT. Familial aspects of diffuse renal diseases. Annu Rev Med. 1964;15:115–124. doi: 10.1146/annurev.me.15.020164.000555. [DOI] [PubMed] [Google Scholar]
- 3.Jais JP, Knebelmann B, Giatras I, et al. X-linked Alport syndrome: natural history and genotype-phenotype correlations in girls and women belonging to 195 families: a “European Community Alport Syndrome Concerted Action” study. J Am Soc Nephrol. 2003;14:2603–2610. doi: 10.1097/01.asn.0000090034.71205.74. [DOI] [PubMed] [Google Scholar]
- 4.Gross O, Netzer KO, Lambrecht R, et al. Meta-analysis of genotype-phenotype correlation in X-linked Alport syndrome: impact on clinical counselling. Nephrol Dial Transplant. 2002;17:1218–1227. doi: 10.1093/ndt/17.7.1218. [DOI] [PubMed] [Google Scholar]
- 5.Jais JP, Knebelmann B, Giatras I, et al. X-linked Alport syndrome: natural history in 195 families and genotype–phenotype correlations in males. J Am Soc Nephrol. 2000;11:649–657. doi: 10.1681/ASN.V114649. [DOI] [PubMed] [Google Scholar]
- 6.Migeon BR. Non-random X chromosome inactivation in mammalian cells. Cytogenet Cell Genet. 1998;80:142–148. doi: 10.1159/000014971. [DOI] [PubMed] [Google Scholar]
- 7.Guo C, Van Damme B, Vanrenterghem Y, et al. Severe Alport phenotype in a woman with two missense mutations in the same COL4A5 gene and preponderant inactivation of the X chromosome carrying the normal allele. J Clin Invest. 1995;95:1832–1837. doi: 10.1172/JCI117862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vetrie D, Flinter F, Bobrow M, et al. X inactivation patterns in females with Alport's syndrome: a means of selecting against a deleterious gene? J Med Genet. 1992;29:663–666. doi: 10.1136/jmg.29.9.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakanishi K, Iijima K, Kuroda N, et al. Comparison of α5(IV) collagen chain expression in skin with disease severity in women with X-linked Alport syndrome. J Am Soc Nephrol. 1998;9:1433–1440. doi: 10.1681/ASN.V981433. [DOI] [PubMed] [Google Scholar]
- 10.Massella L, Onetti Muda A, Faraggiana T, et al. Epidermal basement membrane α5(IV) expression in females with Alport syndrome and severity of renal disease. Kidney Int. 2003;64:1787–1791. doi: 10.1046/j.1523-1755.2003.00251.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Zhang H, Ding J, et al. Correlation between mRNA expression level of the mutant COL4A5 gene and phenotypes of XLAS females. Exp Biol Med (Maywood) 2007;232:638–642. [PubMed] [Google Scholar]
- 12.Rheault MN, Kren SM, Thielen BK, et al. Mouse model of X-linked Alport syndrome. J Am Soc Nephrol. 2004;15:1466–1474. doi: 10.1097/01.asn.0000130562.90255.8f. [DOI] [PubMed] [Google Scholar]
- 13.Cattanach BM, Williams CE. Evidence of non-random X chromosome activity in the mouse. Genet Res. 1972;19:229–240. doi: 10.1017/s001667230001449x. [DOI] [PubMed] [Google Scholar]
- 14.Johnston PG, Cattanach BM. Controlling elements in the mouse. IV. Evidence of non-random X-inactivation. Genet Res. 1981;37:151–160. doi: 10.1017/s0016672300020127. [DOI] [PubMed] [Google Scholar]
- 15.Courtier B, Heard E, Avner P. Xce haplotypes show modified methylation in a region of the active X chromosome lying 3' to Xist. Proc Natl Acad Sci U S A. 1995;92:3531–3535. doi: 10.1073/pnas.92.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Diggle P. Analysis of Longitudinal Data. New York, NY: Oxford University Press; 2002. [Google Scholar]
- 18.Team, RDC . R: A Language and Environment for Statistical Computing. Vienna, Austria; 2008. [Google Scholar]
- 19.Grunfeld JP, Noel LH, Hafez S, et al. Renal prognosis in women with hereditary nephritis. Clin Nephrol. 1985;23:267–271. [PubMed] [Google Scholar]
- 20.Disteche CM, Filippova GN, Tsuchiya KD. Escape from X inactivation. Cytogenet Genome Res. 2002;99:36–43. doi: 10.1159/000071572. [DOI] [PubMed] [Google Scholar]
- 21.Bolduc V, Chagnon P, Provost S, et al. No evidence that skewing of X chromosome inactivation patterns is transmitted to offspring in humans. J Clin Invest. 2008;118:333–341. doi: 10.1172/JCI33166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumal R, Thorner P, Valli VE, et al. Renal disease in carrier female dogs with X-linked hereditary nephritis. Implications for female patients with this disease. Am J Pathol. 1991;139:751–764. [PMC free article] [PubMed] [Google Scholar]
- 23.Thorner PS, Zheng K, Kalluri R, et al. Coordinate gene expression of the α3, α4 and α5 chains of collagen type IV. Evidence from a canine model of X-linked nephritis with a COL4A5 gene mutation. J Biol Chem. 1996;271:13821–13828. doi: 10.1074/jbc.271.23.13821. [DOI] [PubMed] [Google Scholar]
- 24.Miner JH, Sanes JR. Molecular and functional defects in kidneys of mice lacking collagen α3(IV): implications for Alport syndrome. J Cell Biol. 1996;135:1403–1413. doi: 10.1083/jcb.135.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang JS, Wang XP, Miner JH, et al. Loss of α3/α4(IV) collagen from the glomerular basement membrane induces a strain-dependent isoform switch to α5/α6(IV) collagen associated with longer renal survival in Col4a3−/− Alport mice. J Am Soc Nephrol. 2006;17:1962–1969. doi: 10.1681/ASN.2006020165. [DOI] [PubMed] [Google Scholar]
- 26.Chadwick LH, Pertz LM, Broman KW, et al. Genetic control of X chromosome inactivation in mice: definition of the Xce candidate interval. Genetics. 2006;173:2103–2110. doi: 10.1534/genetics.105.054882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews KL, Mudd JL, Li C, et al. Quantitative trait loci influence renal disease progression in a mouse model of Alport syndrome. Am J Pathol. 2002;160:721–730. doi: 10.1016/S0002-9440(10)64892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]