Abstract
IMAC in combination with mass spectrometry is a promising approach for global analysis of protein phosphorylation. Nevertheless this approach suffers from two shortcomings: inadequate efficiency of IMAC and poor fragmentation of phosphopeptides in the mass spectrometer. Here we report optimization of the IMAC procedure using 32P-labeled tryptic peptides and development of MS/MS/MS (MS3) for identifying phosphopeptide sequences and phosphorylation sites. The improved IMAC method allowed recovery of phosphorylated tryptic peptides up to ~77% with only minor retention of unphosphorylated peptides. MS3 led to efficient fragmentation of the peptide backbone in phosphopeptides for sequence assignment. Proteomics of mitochondrial phosphoproteins using the resulting IMAC protocol and MS3 revealed 84 phosphorylation sites in 62 proteins, most of which have not been reported before. These results revealed diverse phosphorylation pathways involved in the regulation of mitochondrial functions. Integration of the optimized batchwise IMAC protocol with MS3 offers a relatively simple and more efficient approach for proteomics of protein phosphorylation.
Global analysis of protein phosphorylation will provide insight into mechanisms by which this dynamic post-translational modification modulates diverse cellular processes. Mass spectrometry-based phosphoproteomics is a potentially powerful approach for global profiling and quantification of protein phosphorylation. Such studies usually involve selective isolation of phosphorylated peptides and their subsequent fragmentation in a mass spectrometer to assign the sequence and localize phosphorylation sites. Three strategies have been described for enriching phosphopeptides based on antibodies (1, 2), chemical derivatization (3, 4), or ionic interactions (e.g. IMAC and strong ion exchange chromatography) (5–9). These methods have achieved limited success in enriching phosphopeptides for proteomics studies. Among these approaches, IMAC is the most convenient and holds much potential for the efficient isolation of phosphopeptides (10–13). The method has been used for several global analyses of protein phosphorylation in model organisms and cellular organelles (5–9). Nevertheless further refinement of extant IMAC protocols is required to achieve high reproducibility and efficiency (14).
Poor fragmentation of phosphopeptides in the mass spectrometer represents the second major challenge for phosphoproteomics. The availability of a relatively low energy fragmentation pathway via β-elimination of the phosphate moiety limits fragmentation at peptide bonds that would be informative for identifying the sequence and site(s) of phosphorylation of the peptide. Mass spectrometers under development, such as instruments using electron transfer dissociation (15), might address this problem. However, such mass spectrometers are not currently commercially available. In addition, fragmentation of doubly charged and singly charged peptides in electron transfer dissociation mass spectrometry is compromised. In summary, despite extensive effort in the past several years, efficient proteomics of protein phosphorylation remains a daunting challenge.
Here we report analysis of the phosphoproteome of mitochondria using an improved method that integrates an optimized batchwise IMAC protocol for isolation of phosphopeptides and MS31 for fragmentation of phosphopeptides. The improved IMAC procedure allowed recovery of ~77% of phosphopeptides while retaining few unphosphorylated peptides. MS3 addressed the poor fragmentation of phosphopeptides by generating highly informative fragmentation patterns that allow peptide identification. Analysis of mitochondrial phosphorylation revealed 84 phosphorylation sites from 62 proteins. Most identified phosphorylation sites have not been reported before, providing novel information about mitochondrial regulatory mechanisms. The optimized batchwise IMAC protocol in combination with MS3 offers a relatively simple and more efficient approach for proteomics of protein phosphorylation.
EXPERIMENTAL PROCEDURES
Materials
The reagents used in this work included SelfPack Poros 20 MC beads from Applied Biosystems (Foster City, CA), [γ-32P]ATP from Amersham Biosciences, TFA from Fluka (Buchs, Switzerland), sequencing-grade trypsin from Promega Corp. (Madison, WI), and iodoacetamide and 99.99% acetic acid from Sigma.
Labeling of A431 Whole Cell Lysate Proteins via in Vitro Autophosphorylation
Four 10-cm dishes of A431 cells (80% confluence) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. The cells were washed with cold Dulbecco's PBS twice, and 200 μl of reaction buffer (50 mm HEPES, pH 7.2, 5 mm MgCl2, 100 mm KCl, 1 mm ATP, and protease inhibitor mixture) were added. The cells were harvested by scraping and transferred to 1.5-ml microcentrifuge tubes. The cells were drawn into a sterile 1-ml syringe through a 30-gauge needle and expelled by a steady pressure on the plunger. The process was repeated 12 times. [γ-32P]ATP (0.2 mCi/ml of cell lysate) was added to the lysate and incubated for 1 h at 30 °C.
Removal of Free [γ-32P]ATP by Two Cycles of TCA Precipitation
After labeling of the whole cell lysate proteins by autophosphorylation in vitro, remaining free [γ-32P]ATP was removed by two cycles of TCA precipitation as briefly described below. The cytosolic proteins were precipitated by adding volume of 100% TCA followed by a 2-h incubation on ice. After centrifugation at 15,300 × g for 20 min, the supernatant was removed, and the protein pellet was washed twice with cold acetone. The protein pellet was redissolved in 8 m urea, and the TCA precipitation was repeated to remove remaining free [γ-32P]ATP.
Tryptic Digestion of Mouse Liver Mitochondria Preparation
The mitochondrial fraction from mouse liver was prepared and digested as described in the supplemental information.
Methylation
Methylation of carboxylic groups in acidic amino acid residues (Asp and Glu) and at the C termini of peptides was performed as described previously (5). Briefly 100 μg of tryptic digest was dried in a SpeedVac (Thermo Savant, San Jose, CA) for 6 h. Then 50 μl of 2 m methanolic HCl was added to the dried peptides, and the reaction was carried out for 2 h on a shaker. Anhydrous methanol used to prepare methanolic HCl had been distilled in the presence of CaH2. The peptide mixture was thoroughly dried, and the methylation reaction was repeated to ensure complete conversion of peptides to their corresponding methyl esters.
Batchwise IMAC
Poros 20 MC beads were activated according to the manufacturer's instructions (see detailed procedure in the supplemental information). Ten microliters of activated beads were equilibrated with 200 μl of loading solution containing acetonitrile/methanol/water (1:1:1, v/v/v) in 0.1% acetic acid before sample loading. One hundred micrograms of methylated mitochondrial tryptic peptides were dissolved in 20 μl of acetonitrile/methanol/water (1:1:1, v/v/v), and the pH was adjusted to 2.5–3.0 with 2 m NH4HCO3 (pH 8.0). The peptide mixture was centrifuged at 100,000 × g for 20 min to remove insoluble particles, which might have contained peptides. The resulting supernatant was mixed with IMAC beads and incubated at room temperature with shaking for 30 min. The suspension was centrifuged in a microcentrifuge at 13,000 × g for 1 min, and the supernatant was removed. The resulting beads were washed once with 200 μl of washing buffer I (acetonitrile/methanol/water/acetic acid (75:10:14:1, v/v/v/v) containing 100 mm NaCl) and twice with 200 μl of washing buffer II (acetonitrile/water/acetic acid (85:14:1, v/v/v)). Bound phosphopeptides were eluted from the IMAC beads with three 50-μl washes of elution solution (acetonitrile/water/TFA (45:50:5, v/v/v)).
Mass Spectrometry and Protein Sequence Database Searching
Nano-HPLC/LTQ mass spectrometry was carried out as described previously (16) except that a 9-min gradient of 6–90% B buffer (90% acetonitrile, 9.95% water, 0.05% acetic acid) in A buffer (97.95% water, 2% acetonitrile, 0.05% acetic acid) at a flow rate of 0.1 μl/min was used. The LTQ mass spectrometer was operated in a data-dependent mode in which one full MS scan was followed by four pairs of MS2/MS3 scans. Normalized collision energy was set to 22% during MS2 acquisition and 35% during MS3 acquisition. MS3 was automatically triggered when a neutral loss peak of 98, 49, or 32.7 (±2) m/z was detected among the top eight most intense peaks in the MS2 spectrum. The acquired data were used for peptide identification as described in the supplemental information.
RESULTS
The on-line IMAC approach, consisting of a chromatography column harboring consecutively packed IMAC beads and C18 beads, has been used for isolation of phosphopeptides for proteomics analysis. This approach has the advantage of low sample loss but lacks some features of batchwise isolation, such as rapid and efficient sample washing, convenient solvent exchange, and high sample loading capacity. Given that phosphate groups have relatively low binding affinity for IMAC beads, that off rates are potentially high, and that many phosphorylated peptides are present in the proteome at low stoichiometry, we hypothesized that the advantages of batchwise isolation could significantly improve the efficiency of IMAC separations.
Isolation of phosphopeptides from a complex peptide mixture through batchwise IMAC can be divided into four steps: conversion of the carboxylic acid groups of peptides to their corresponding methyl esters (methylation), binding of phosphopeptides to the IMAC beads (loading), removal of unphosphorylated peptides from the beads (washing), and release of phosphopeptides from the beads (elution). Optimization of each step is desired to maximize phosphopeptide recovery while minimizing contamination from unphosphorylated peptides.
32P-Labeled phosphopeptides were used to optimize the IMAC protocol. The yield of the IMAC procedure was determined by measuring 32P radioactivity, whereas contamination from unphosphorylated peptides was monitored by HPLC/MS/MS of the eluted fraction and peptide identification via sequence database searching. To make 32P-labeled peptides, proteins from cell lysate are usually in vitro phosphorylated and digested with trypsin. The resulting tryptic peptides are subjected to C18 or C8 chromatography to remove unlabeled 32P. Our control experiments using 32P-labeled ATP revealed that both C18 beads and C8 beads retained more than 1% of the radioactivity that was derived from 32P-labeled ATP presumably from non-proteinaceous molecules (see the supplemental information for details). Given the fact that only a small percentage of 32P in 32P-labeled ATP will be transferred onto proteins during in vitro phosphorylation, it is likely that less than 1% radioactivity of the 32P-labeled ATP would be used for in vitro phosphorylation. Therefore, it was necessary to remove non-proteinaceous 32P-containing molecules by using a method other than C18 or C8 beads before screening IMAC conditions. To restrict the radioactivity present in the sample to polypeptides, proteins phosphorylated in vitro were precipitated twice with TCA and then digested with trypsin. Such precipitation could remove non-proteinaceous small molecules, including those present in 32P-labeled ATP. The tryptic peptides were further subjected to C8 purification to remove small molecules prior to being used for IMAC optimization.
Methylation
Conversion of carboxylic acid groups in peptides to their corresponding methyl esters precludes binding of unphosphorylated peptides to IMAC beads through acidic groups, thereby reducing contamination. The methylation reaction is reversible, and completeness of the reaction is compromised by the presence of water. We used three steps to drive the methylation reaction to completion. First, 100 μg of tryptic digests were dried in a SpeedVac for 6 h to ensure complete removal of water from the sample. Second, the anhydrous methanol used in the methylation reaction was redistilled against CaH2 to remove residual water present in the methanol. Finally the methylation reaction itself was performed twice. After treating tryptic peptides from A431 cells with our methylation procedure, we did not identify unmethylated tryptic peptides by nano-HPLC/MS/MS, indicating a fairly complete methylation reaction (data not shown).
As the IMAC method relies on the interaction between negatively charged groups and the immobilized metal, the chemical modification of extraneous acidic groups serves to enhance the selectivity and sensitivity of the method. Phosphorylation occurs substoichiometrically among its target proteins and is far less prevalent than acidic amino acid side chains. Although Fe3+ shows some inherent selectivity toward the more acidic PO4 groups, competition from abundant carboxylic acid groups would lead to a loss of sensitivity for the method. Our analysis of methylated peptides suggested that the methylation reaction was restricted to carboxyl groups as we did not detect other methylation products. Any losses due to the complete drying of the sample after methylation would be accounted for in the solubility data (Fig. 1A). These data demonstrate that sample losses due to complete drying of the methylated peptides can be minimized by selecting a reconstitution buffer containing organic solvent.
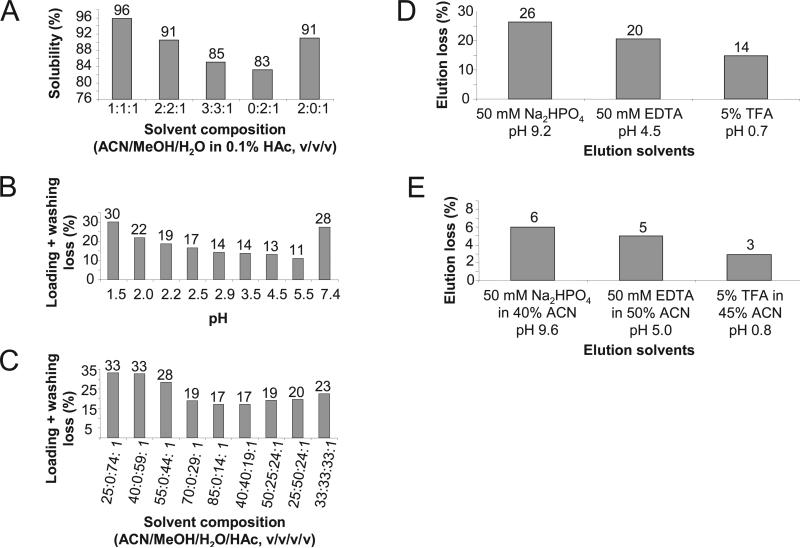
Fig. 1. Optimizing the batchwise IMAC protocol using 32P-labeled peptides.
A, effect of solvent on phosphopeptide solubility. B, combined loading and washing loss at different loading pH values. C, combined loading and washing loss at different organic solvent concentrations in washing solutions. D, elution efficiency of phosphopeptides from IMAC beads with buffers containing 50 mm Na2HPO4, 50 mm EDTA, or 5% TFA. E, elution efficiency of phosphopeptides from IMAC beads with buffers containing organic solvent. Experimental procedures for the experiments are described in detail in the supplemental information. HAc, acetic acid; MeOH, methanol.
Loading
After the methylation reaction, the dried peptide mixture was resolubilized for IMAC. High solubility is essential to reduce sample loss. Because the solubility of peptides depends on acidity and organic solvent concentration, a series of acidic solutions with varied organic solvent concentrations was screened. Recovery was determined by soluble 32P radioactivity. Highest solubility (~96%) was achieved in acetonitrile/methanol/water solution (1:1:1, v/v/v) with 0.1% acetic acid (Fig. 1A). This buffer is advantageous for downstream IMAC as the high proportion of organic components (67%) increases ionic interactions between immobilized Fe3+ and phosphate groups.
pH affects the binding affinity between phosphate and Fe3+ by modulating the charge status of the phosphate group. As expected, the proportion of phosphopeptides lost in the IMAC loading and washing steps varied depending on the pH of the buffer (Fig. 1B). The highest recovery was achieved when a relatively high pH (5.5) was used presumably because phosphate carrying a more negative net charge binds more tightly to the IMAC beads. However, such a high pH is likely to invite binding of unmethylated peptides, which might be present in residual amounts, to the IMAC column. As a compromise between recovery and selectivity, a pH of 3.0 was used during enrichment of biological samples.
Based on our data, we would not advise increasing the loading buffer pH as a means of enhancing recovery. The recovery benefit from using a washing buffer pH of 5.5 was modest (11% loss at pH 5.5 versus 14% loss at pH 2.9). On the other hand, carboxylic acids have a pKa of ~5, whereas the phosphate group has a pKa of ~2. Thus it is likely that the benefit of any increased recovery when using a buffer of higher pH would be mitigated by a loss of selectivity.
Washing
The goal of the washing step is to maintain binding of phosphopeptides to the IMAC beads while removing unphosphorylated peptides. High organic solvent composition encourages binding between phosphate groups and Fe3+ while preventing weak hydrophobic interactions between non-phosphopeptides and the IMAC beads. Furthermore salt will disrupt weak, nonspecific interactions between the IMAC beads and polar groups in the amino acids.
The amount of organic solvent, in the form of acetonitrile and methanol in the washing solution, also affected loss of phosphopeptides during washing. Phosphopeptide loss, estimated by monitoring radioactivity in the washing eluate, was reduced to 17% as organic solvent content in the washing solution was increased to 85% (Fig. 1C). To prevent nonspecific binding from ionic or polar interactions, 100 mm NaCl was included in the washing solution, which led to an additional 3% loss of radioactivity (data not shown). A final washing procedure was chosen that provided acceptable phosphopeptide recovery while minimizing contamination from unphosphorylated peptides. The procedure included three washing steps: the first using 75:10:14:1 (v/v/v/v) acetonitrile/methanol/water/acetic acid containing 100 mm NaCl and the second and third using 85:14:1 (v/v/v) acetonitrile/water/acetic acid.
Elution
Fifty millimolar NaH2PO4, 50 mm EDTA, and 5% TFA were tested for elution efficiency with 32P-labeled peptide samples. These reagents can disrupt the interactions between Fe3+ and the phosphate group (NaH2PO4) or between Fe3+ and the iminodiacetic acid group (EDTA and TFA). Between 14 and 26% of phosphopeptides remained bound to the IMAC beads when these elution buffers were used (Fig. 1D). When 40–50% acetonitrile was included in the elution solutions, almost all the radioactivity could be eluted, leaving only 3–6% of the initial radioactivity in the beads (Fig. 1E). Organic solvent prevents phosphopeptides from binding to the IMAC beads through hydrophobic interactions. Based on these results, a solvent containing 5% TFA, 45% acetonitrile, and 50% water was selected as the elution buffer.
Protocols for on-line IMAC-HPLC do not include organic solvents in the elution buffer because a high organic content would prevent some phosphopeptides from binding to the downstream C18 beads. To demonstrate this issue, 32P-labeled phosphopeptides were allowed to bind to C18 beads pre-equilibrated with buffers containing varying concentrations of acetonitrile (see the supplemental information for the experimental procedure). We observed an inverse correlation between the binding capacity of the C18 beads and the concentration of acetonitrile in the buffer (Supplemental Fig. S2). Therefore, performing IMAC and HPLC consecutively in a single column necessitates loss of a significant portion of phosphopeptides due to the inherent inability to include organic solvent in the elution solution.
In summary, to boost IMAC efficiency we systematically optimized conditions at each step of the procedure: methylation, loading, washing, and elution. The optimized procedure gave a phosphopeptide recovery of ~77%, which takes into account 20% combined loss at the loading and washing steps and 3% loss during the elution step.
MS3 for Identifying Phosphopeptides and Mapping Phosphorylation Sites
Protein identification and mapping modification sites within tryptic peptides are typically performed using MS/MS spectra to search protein sequence databases. Quality of the MS/MS spectra plays a key role in such analysis. An ideal MS/MS spectrum results from fragmentation of multiple peptide bonds, leading to masses of diverse daughter ions that are specified by the peptide sequence and identity of post-translational modifications. Efforts to identify phosphopeptides are hindered by the ready loss of H3PO4 via β-elimination, a relatively low energy fragmentation pathway that occurs upon CID. Thus, MS/MS spectra of phosphopeptides usually contain very limited mass information that is indicative of the peptide sequence (Fig. 2).
Fig. 2. MS3 for peptide identification and mapping phosphorylation sites.
A, MS2 fragmentation (parent ion m/z, 656.56) of phosphopeptide IQELKG-pSQER (where pS is phosphoserine) (carboxylesterase 5, gi|18381028) shows a dominant neutral loss peak at m/z 606.93 and few fragment masses providing sequence information. B, MS3 spectrum after fragmentation of the neutral loss peak shows a rich fragmentation pattern for peptide identification and phosphorylation site determination. Asterisks indicate water loss peaks.
To address this problem, we developed three stages of mass spectrometry for phosphopeptides. In this strategy, the first stage (MS1) determines the mass of the peptide, and the second (MS2) removes the phosphate group using low collision energy (22% collision energy level) to form dehydroala-nine (from phosphoserine) or dehydroamino-2-butyric acid (from phosphothreonine). The third stage (MS3) is fragmentation by CID of the resulting dephosphorylated peptide ions, which behave similarly to unphosphorylated peptides in the mass spectrometer. The information obtained from MS3 can be used to identify peptides and localize phosphorylation sites (Fig. 2). The LTQ mass spectrometer, which has a high ion-trapping capacity, makes such a strategy practical.
It is worth mentioning that multiply phosphorylated peptides identified through database searching using MS3 spectra may be false positives. For example, the presence of two dehydroalanines does not necessarily signify two phosphoserine sites in the original peptide as one of the dehydroalanines may be produced from a serine water loss event during the MS1 stage (in-source fragmentation). This phenomenon is described further in the supplemental information.
Application of Batchwise IMAC Method and MS3 to Mitochondrial Phosphoproteome Analysis
The mitochondrion is a subcellular organelle present in all eukaryotic cells. Best known for its role in oxidative phosphorylation and metabolism, the organelle regulates multiple cellular processes including programmed cell death, oxidation of carbohydrates and fatty acids, redox homeostasis, and other catabolic and anabolic pathways. Mitochondrial dysfunction has been implicated in various diseases such as diabetes, Alzheimer disease, Parkinson disease, and cancer. Emerging evidence, especially from the increasing number of kinases, phosphatases, and phosphoproteins identified in mitochondria, suggests that reversible phosphorylation plays an important role in mitochondrial function (17). Nevertheless the prevalence of protein phosphorylation in mitochondria and the mechanism through which protein phosphorylation regulates diverse mitochondrial functions remain largely unknown.
We applied the optimized IMAC protocol and MS3 to study the phosphoproteome of mitochondria from mouse liver. Mitochondrial proteins (100 μg) were digested with trypsin and enriched for phosphopeptides using IMAC. The resulting peptide mixture was analyzed by nano-HPLC/LTQ mass spec-trometry using a data-dependent MS3 mode. The MS3 spectra were used to search the National Center for Biotechnology Information non-redundant (NCBI nr) mouse database using the Mascot searching algorithm. Manual validation of all hits resulted in identification of 84 phosphorylation sites from 62 phosphoproteins (see Supplemental Table 1 for the peptide list and Supplemental Fig. S1 for the MS3 spectra with assigned daughter ions). The raw MS/MS data for each peptide is available upon request. Only 10 unphosphorylated peptides were identified using the combined MS2 and MS3 data, suggesting little contamination from non-phosphopeptides.
Classification of Identified Phosphoproteins and Phosphorylation Sites
The 62 phosphoproteins identified were categorized into nine functional groups: metabolism, apoptosis, cell cycle, cell maintenance, transcription/translation, structural, chaperones, stress response, and unknown (Supplemental Fig. S3). Of the 84 identified phosphorylation sites, 78 (93%) were at serine residues, and six (7%) were at threonine (Supplemental Fig. S3).
To seek a phosphorylation motif within these phosphopeptides, relative frequencies of occurrence of amino acids at positions flanking the phosphorylation sites were calculated and plotted on a site density map (Supplemental Fig. S3). Arginine was favored at the –3 position (basophilic motif), and glutamic acid was favored at the +3 position (acidophilic).
The unique phosphorylation sites and proteins identified in this preliminary analysis of the mitochondrial phosphoproteome offer further evidence of the important role of phosphorylation events for regulating mitochondrial proteins. Of the 62 proteins identified in this study, only four were previously reported as mitochondrial phosphoproteins. The remaining 58 represent novel identifications and suggest that phosphorylation has a greater impact on mitochondrial function than is currently appreciated.
Although the direct regulatory effects of few mitochondrial phosphorylation events have been well characterized, the consequence of phosphorylation has been elucidated for two of the proteins identified in this analysis: pyruvate dehydrogenase E1 and the β subunit of the F1-ATP synthase. Phosphorylation of three serine residues within pyruvate dehydrogenase has been described; all three were identified in this analysis. Phosphorylation at any of the three sites is sufficient to inhibit the enzymatic activity of the pyruvate dehydrogenase complex (18). Phosphorylation of the β subunit of the F1-ATP synthase leads to recruitment of mitochondrial 14-3-3 proteins, which bind to the phosphorylated region and inhibit ATP synthesis (19). Despite the inhibitory nature of these phosphorylation events, phosphorylation may also stimulate the function of mitochondrial proteins. For example, phosphorylation of an 18-kDa subunit of oxidative phosphorylation complex 1 has been shown to increase respiration rate (20). Thus, the specific role of phosphorylation in the regulation of most of the proteins identified in this study awaits further characterization.
Comparison of the newly discovered phosphorylation sites with consensus sequences reported for various protein kinases suggests that the greatest proportion of mitochondrial proteins identified in this study are potential targets of the calcium/calmodulin-dependent kinase family (20 of 62) (21). This prediction is reasonable given the central role of calcium in regulating signaling cascades and enzymatic activities both in mitochondria and throughout the cell. Potential PKA substrates were also well represented among the identified phosphopeptides (Table I). PKA is found in mitochondria and plays a role in mediating signaling cascades in response to elevated cAMP levels. Furthermore experimental evidence suggests a role for mitochondrially localized PKA activity in enhancing the cellular respiration rate and inhibiting apoptosis (20). Translocation of activated PKC isoforms to mitochondria has been found to be proapoptotic and inhibitory toward mitochondrial function (20). A number of potential PKC substrates were identified in our study, including proteins involved in apoptosis and oxidative stress pathways and proteins overex-pressed in response to chemotherapy. Of these potential substrates, VDAC is a known target of PKC, its phosphorylation having cardioprotective effects following ischemia in perfused rat hearts (22). Akt is a protein kinase implicated as a mediator of extracellular and intracellular signaling events. Its effects range from inhibiting apoptosis to transducing insulin signaling cascades and regulating the cell cycle. Activation of Akt can lead to its translocation to mitochondria, and mitochondrial proteins have been demonstrated to be Akt substrates (20, 23). Our screen identified four peptides that conform to the Akt consensus motif. Although the identity of the kinase responsible for a given modification requires experimental confirmation, these observations based on consensus sequences further suggest a regulatory influence of phosphorylation events in mitochondria.
Table I.
Scansite (25) screening of kinase phosphorylation and binding motifs at high stringency (0.2%) and medium stringency (1.0%) within identified phosphorylation sites
| Type | Hits |
||
|---|---|---|---|
| 0.2% | 1.0% | ||
| Casein kinase 2 | Acidophilic | 10 | 16 |
| Akt kinase | Basophilic | 4 | 4 |
| Clk2 kinase | Basophilic | 1 | 2 |
| Protein kinase A | Basophilic | 6 | 14 |
| PKC ε | Basophilic | 1 | 3 |
| CaMKa II | Basophilic | 2 | 2 |
| DNA PKb | DNA damage kinase group | 1 | 1 |
| ATMc kinase | DNA damage kinase group | 1 | 2 |
| Cdk5 kinase | Proline-dependent | 3 | 4 |
| 14-3-3 mode 1 | 14-3-3 mode 1 | 1 | 7 |
CaMK, calcium/calmodulin-dependent kinase.
Protein kinase.
Ataxia telangiectasia mutated.
A protein can be modified by other post-translational modifications in addition to phosphorylation; these modifications may work in concert to modulate protein function. We identified proteins in this analysis that were also identified in a separate proteomics analysis of lysine-acetylated proteins (24). Proteins identified in both studies include VDAC1 and sterol carrier protein 2. One might expect transport proteins like these to be regulated by multiple signaling cascades, allowing fine tuning of mitochondrial function to cellular requirements. Such multiple layers of modifications are reminiscent of the histone code where ubiquitination, phosphorylation, methylation, and acetylation of histone tails all contribute to the degree of exposure of DNA and transcription of genes. Given the anabolic and catabolic functions of the mitochondrion, its critical role in maintaining cellular viability, and the convergence of many distinct signaling cascades on the organelle, regulation of its constituent proteins through multiple layers of post-translation modifications seems likely.
To assess the potential conservation of the phosphorylation events identified in this study, protein sequence homology searching was conducted using the NCBI PSI-BLAST tool (www.ncbi.nlm.nih.gov/BLAST/). The protein sequences were submitted for homology searching, and conservation of the identified phosphorylation site among homologous human proteins was determined. We found the modified residue to be conserved among 76% of the identified sites. This result suggests that phosphorylation of mitochondrial proteins may represent a regulatory pathway conserved between mouse and human.
DISCUSSION
In this work, we addressed two major challenges of proteomics of protein phosphorylation: efficient isolation of phosphopeptides and obtaining informative fragmentation. Our optimized batchwise IMAC protocol allowed a phosphopeptide recovery of ~77%. The high number of singly phosphorylated peptides and low number of unphosphorylated peptides identified when mitochondrial proteins were analyzed demonstrate the value of our protocol.
Batchwise IMAC offers four main advantages over the on-line IMAC-HPLC method. First, the IMAC beads can be washed quickly, reducing loss of phosphopeptides from the beads. Second, exchanging buffer solutions is very easy in the batchwise method. Third, organic solvent (up to 50% acetonitrile) can be included in the elution buffer to prevent hydrophobic interactions between phosphopeptides and the IMAC beads, increasing recovery of the phosphopeptides. Such high organic solvent concentrations cannot be used in the on-line method because they would prevent a significant portion of phosphopeptides from binding to the downstream C18 beads. Finally the batchwise IMAC protocol allows high sample loading, holding the potential to identify phosphopeptides present in low abundance.
Our optimized procedure is not compatible with the on-line experiment as our IMAC elution buffer contains a high concentration of ACN that weakens the binding of phosphopeptides to C18 beads. Specifically in evaluating the samples loss of 32P-containing tryptic peptides onto a C18 column, we screened buffers consisting of 25% acetonitrile, 1% acetic acid, 100 mm NaCl (IMAC loading/washing buffer from Ref. 5); 50 mm Na2HPO4 (IMAC elution buffer from Ref. 5); or 45% acetonitrile, 5% TFA (optimized elution buffer from our procedure). We found an inverse correlation between organic solvent content and phosphopeptide binding. The 45% acetonitrile solution retained only 20% of the phosphopeptides (Supplemental Fig. S2). In conclusion, by including organic solvent (ACN) in our IMAC elution buffer, we boost the elution efficiency of phosphopeptides from IMAC beads.
It should be noted that 100-μg samples were used for optimization of the IMAC procedure. We also successfully applied the procedure in small scale to isolate phosphorylated peptides from tryptic digest (data not shown). In this case, only 0.5 μl of IMAC beads was used (see protocol in the supplemental information). The purpose of this study was to develop a method capable of extending the limit of detection for phosphopeptide analysis. This was accomplished through increasing the capacity of the isolation procedure via an optimized batchwise enrichment.
Because of the liability of the phosphate group, MS2 spectra of phosphopeptides usually contain little sequence information, making it difficult to identify the peptides and to localize phosphorylation sites. This problem is avoided in MS3 in which the MS2 fragment showing a loss of H3PO4 is used as the parent ion for further fragmentation. The fragments that have lost H3PO4 contain dehydroalanine (if the phosphate was originally on serine) or dehydroamino-2-butyric acid (if the phosphate was originally on threonine) and fragment similarly to unphosphorylated peptides. The resulting fragmentation patterns are rich in structural information for peptide identification and localization of the previously phosphorylated sites.
By analyzing the mitochondrial phosphoproteome using our optimized batchwise IMAC conditions in combination with the MS3 strategy, we discovered numerous novel phosphorylation sites. The data presented demonstrate that our approach is much more efficient than previously reported methods for proteomics of protein phosphorylation.
Acknowledgment
We thank Steve McKnight for helpful suggestions.
Footnotes
This work was supported by Robert A. Welch Foundation Grant I-1550 and National Institutes of Health Grant CA107943. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The abbreviations used are: MS3, MS/MS/MS; MS2, MS/MS; MS1, MS; PKA, cAMP-dependent protein kinase; PKC, protein kinase C; VDAC, voltage-dependent anion channel.
REFERENCES
- 1.Hinsby AM, Olsen JV, Mann M. Tyrosine phosphoproteomics of fibroblast growth factor signaling: a role for insulin receptor substrate-4. J. Biol. Chem. 2004;279:46438–46447. doi: 10.1074/jbc.M404537200. [DOI] [PubMed] [Google Scholar]
- 2.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 3.McLachlin DT, Chait BT. Improved β-elimination-based affinity purification strategy for enrichment of phosphopeptides. Anal. Chem. 2003;75:6826–6836. doi: 10.1021/ac034989u. [DOI] [PubMed] [Google Scholar]
- 4.Tao WA, Wollscheid B, O'Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat. Methods. 2005;2:591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 5.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 6.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Kokubu M, Ishihama Y, Sato T, Nagasu T, Oda Y. Specificity of immobilized metal affinity-based IMAC/C18 tip enrichment of phosphopeptides for protein phosphorylation analysis. Anal. Chem. 2005;77:5144–5154. doi: 10.1021/ac050404f. [DOI] [PubMed] [Google Scholar]
- 8.Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol. Cell. Proteomics. 2004;3:1093–1101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Trinidad JC, Specht CG, Thalhammer A, Schoepfer R, Burlingame AL. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics. 2006;5:914–922. doi: 10.1074/mcp.T500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Cao P, Stults JT. Rapid Commun. Mass Spectrom. 2000;14:1600–1606. doi: 10.1002/1097-0231(20000915)14:17<1600::AID-RCM68>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 11.Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal. Chem. 1999;71:2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 12.Haydon CE, Eyers PA, Aveline-Wolf LD, Resing KA, Maller JL, Ahn NG. Identification of novel phosphorylation sites on Xenopus laevis Aurora A and analysis of phosphopeptide enrichment by immobilized metal-affinity chromatography. Mol. Cell. Proteomics. 2003;2:1055–1067. doi: 10.1074/mcp.M300054-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Stupak J, Zheng J, Keller BO, Brix BJ, Fliegel L, Li L. Open tubular immobilized metal ion affinity chromatography combined with MALDI MS and MS/MS for identification of protein phosphorylation sites. Anal. Chem. 2004;76:4223–4232. doi: 10.1021/ac035231d. [DOI] [PubMed] [Google Scholar]
- 14.Nuehse TS, Stensballe A, Jensen ON, Peck SC. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Kwon SW, Kim SC, Zhao Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 2005;4:998–1005. doi: 10.1021/pr049754t. [DOI] [PubMed] [Google Scholar]
- 17.Pagliarini DJ, Dixon JE. Mitochondrial modulation: reversible phosphorylation takes center stage? Trends Biochem. Sci. 2006;31:26–34. doi: 10.1016/j.tibs.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Holness MJ, Sugden MC. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem. Soc. Trans. 2003;31:1143–1151. doi: 10.1042/bst0311143. [DOI] [PubMed] [Google Scholar]
- 19.Bunney TD, van Walraven HS, de Boer AH. 14-3-3 protein is a regulator of the mitochondrial and chloroplast ATP synthase. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4249–4254. doi: 10.1073/pnas.061437498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horbinski C, Chu CT. Kinase signaling cascades in the mitochondrion: a matter of life or death. Free Radic. Biol. Med. 2005;38:2–11. doi: 10.1016/j.freeradbiomed.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 21.Kennelly PJ, Krebs EG. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 22.Uecker M, Da Silva R, Grampp T, Pasch T, Schaub MC, Zaugg M. Translocation of protein kinase C isoforms to subcellular targets in ischemic and anesthetic preconditioning. Anesthesiology. 2003;99:138–147. doi: 10.1097/00000542-200307000-00023. [DOI] [PubMed] [Google Scholar]
- 23.Bijur GN, Jope RS. Rapid accumulation of Akt in mitochondria following phosphatidylinositol 3-kinase activation. J. Neurochem. 2003;87:1427–1435. doi: 10.1046/j.1471-4159.2003.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 25.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]