Abstract
Site-directed spin labeling and electron paramagnetic resonance were used to probe residues 281–304 of human vimentin, a region that has been predicted to be a non-α-helical linker and the beginning of coiled-coil domain 2B. Though no direct test of linker structure has ever been made, this region has been hypothesized to be flexible with the polypeptide chains looping away from one another. EPR analysis of spin-labeled mutants indicates that (a) several residues reside in close proximity, suggesting that adjacent linker regions in a dimer run in parallel, and that (b) the polypeptide backbone is relatively rigid and inflexible in this region. However, this region does not show the characteristics of a coiled-coil as has been identified elsewhere in the molecule. Within this region, spectra from positions 283 and 291 are unique from all others thus far examined. These positions, predicted to be in a noncoiled-coil structure, display a significantly stronger interaction than the a–d contact positions of coiled-coil regions. Analysis of the early stages of assembly by dialysis from 8 M urea and progressive thermal denaturation shows the close apposition and structural rigidity at residues 283 and 291 occurs very early in assembly and with a relatively sudden onset, well before coiled-coil formation in other parts of the molecule. These features are inconsistent with hypotheses that envision the linkers as flexible regions, or as looping away from one another, and raise the possibility that the linker may be the site at which dimer alignment and/or formation is initiated. Spin labels placed further downstream yield spectra suggesting that the first regular heptad of rod domain 2 begins at position 302. In conjunction with our previous characterization of region 305–336 and the solved structure of rod 2B from 328–405, the full extent of coiled-coil domain in rod 2B is now known, spanning from vimentin positions 302–405.
Intermediate filaments (IFs1) are one of the three major classes of cytoskeletal structures in metazoan cells. Though more than 60 IF genes have been defined in the human genome, most IFs are assembled from just 1 or 2 IF proteins. Which specific IF protein(s) is/are expressed varies by cell type and the state of development/differentiation (1–4).
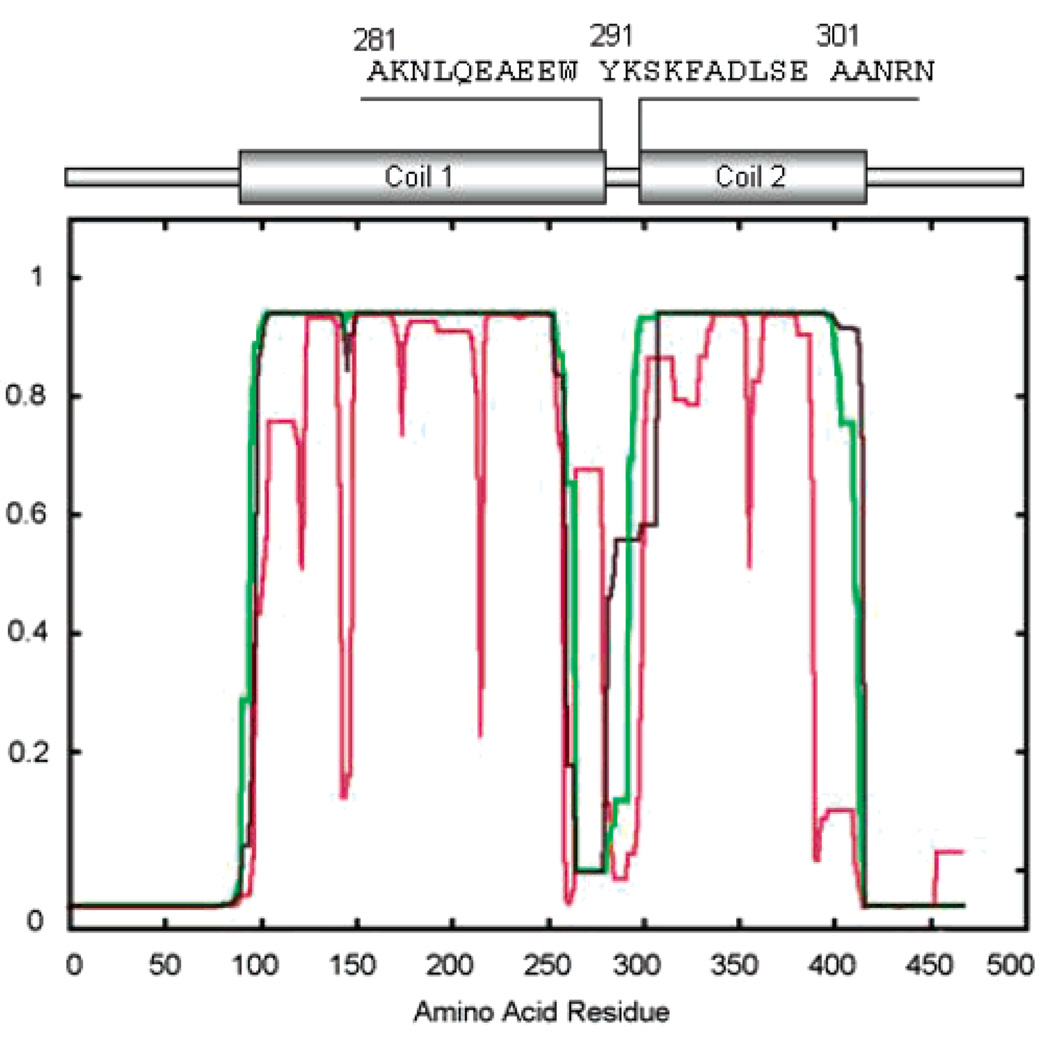
Though there is considerable diversity in the IF family in both size and primary sequence, the vast majority of IF proteins show the conservation of several features: (1) they are localized to and/or can assemble into 8–11 nm IFs, and (2) they exhibit a common predicted domain structure. The predicted domain structure consists of (a) head and tail domains that vary widely in size and primary sequence and show no obvious predicted secondary structure and (b) a central rod domain that is approximately 310 amino acids in length. This rod domain is predicted to consist of extended runs of an α helix containing a heptad repeat pattern, which predicts the formation of coiled-coils (coil domains) separated by short regions called linkers that are not predicted to be α-helical, nor do they show a heptad repeat pattern (1–3, 5–16). However, depending on how parameters are set in programs that make such predictions, the strength of the coiled-coil prediction can vary quite dramatically (Figure 1) (17). Importantly, no direct test of structure has ever been made in these linker regions.
FIGURE 1.
Vimentin Schematic. A traditional schematic of the vimentin molecule is shown, with coil domains flanking the noncoil or linker region. Three variants of Coils version 2.1 (http://www.ch.embnet.org/software/COILS) predictions used to generate such models are shown, revealing the uncertainty of predictions in the putative linker region. Also shown is the amino acid sequence studied in this report and its approximate location in the schematic of vimentin.
No intact IF protein or IF has been crystallized, thus a high-resolution structural map of neither the IF nor the IF protein has been determined. However, a variety of approaches, both theoretical and experimental have contributed to a model of IF structure and assembly. Strelkov and co-workers, for example, have been successful at the crystallization of fragments of the homopolymeric protein vimentin (18, 19). From the solution of these crystals’ structures has come the first detailed pictures of 2 regions of IF coiled-coil structure. The higher order structure of IF proteins within a filament and within assembly intermediates has been approached with cross-linking methodology. From both vimentin and keratin cross-link studies have emerged data that suggest a set of overlapping structures that are found in assembled filaments (20–23).
We have employed site-directed spin labeling (SDSL) and electron paramagnetic resonance (EPR) to characterize the full range of IF structure, yielding data on secondary, tertiary, and quaternary structures, and identifying sites where proteins are in close contact with one another in intact filaments (24–26). Collectively, these data support a model of the IF as beginning with the formation of an in-parallel and in-register dimer, where the central rod domain forms, at least in part, a coiled-coil dimer. Dimers then assemble into antiparallel tetramers with rod domains 1 associated (A11 interaction), followed by a species with rod domains 2 associated in an antiparallel interaction (A22 interaction). Cross-linking and SDSL–EPR have both contributed to establishing relationships between adjacent dimers, providing data on the relative orientation and registry of adjacent dimers.
Most data that have been generated address the structure of the rod domain and its assembly into a coiled-coil dimer, and the relationship between adjacent rod domains in higher order structures. Essentially, nothing is known about the structure of the linker domains, and very little data has been generated that addresses the structure of either head or tail domains.
In this article, we use SDSL-EPR to describe a series of spin-labeled mutants in the linker region of human vimentin (residues 281–304). EPR analysis of these spin-labeled mutants indicates that this region lacks coiled-coil structure, but also shows that several sites in this linker region are positioned extremely close to one another in intact filaments and that the polypeptide backbone in this region is quite rigid. Of considerable interest is evidence that the region from 283–291 assembles into its final structure much earlier than the coiled-coil domains within rod 1B and 2B that we have examined and is highly resistant to thermal denaturation.
MATERIALS AND METHODS
Vimentin characterization, mutation, cloning, expression, purification, and spin labeling were described in detail in previous reports (24–26). In brief, a nitroxide spin label is ultimately attached to a cysteine residue that is targeted to specific sites in vimentin. Cysteine codons were introduced into the vimentin expression construct (generously provided by Dr. Roy Quinlan, University of Durham, Durham U.K.) using the Stratagene QuikChange kit (Stratagene, La Jolla, CA). Sequence changes were verified by DNA sequencing. Mutant vimentin was produced by bacterial expression using pT7 and arabinose inducible BL21(DE3) cells (Invitrogen, Carlsbad, CA). Inclusion bodies were purified from bacterial cell pellets using high/low salt washes and chromatography (27, 28). Spin labeling was accomplished by incubation of the purified vimentin in 100 µM TCEP (Molecular Probes, Eugene, OR) followed by 500 µM O-87500 (Toronto Research Chemicals, Toronto, Canada). The unincorporated spin label was removed by CM sepharose chromatography using an Amersham Pharmacia FPLC (26). Labeled proteins were stored long term at −80 °C.
Filament assembly was performed either as a single step dialysis against the filament assembly buffer or as a stepwise process following the protocol of Carter et al. (29). Briefly, labeled proteins were solubilized in 8 M urea, which was then removed by dialysis, either in a single step procedure or, where indicated, in a stepwise process through progressively reduced concentrations of urea, followed by low ionic strength Tris, followed by the addition of NaCl and MgCl2. Filament assembly was verified by electron microscopy of negatively stained samples.
EPR was conducted on a JEOL X-band spectrometer equipped with a loop gap resonator. Approximately 4 µL of the sample, at a concentration of ~25 µM protein, was placed in a sealed quartz capillary tube. Unless otherwise indicated, spectra were acquired at 20–22 °C with a single 60 s scan over 100 G at a microwave power of 2 mW, and a modulation amplitude optimized to the natural line width of the attached nitroxide. Spectra were normalized to the same number of spins by normalizing each spectrum to the same integrated intensity. To improve the fidelity of the calculation, double integration of each sample was performed following its solubilization in 2% SDS.
RESULTS
Figure 1 shows a conventional schematic of the vimentin molecule identifying coiled-coil domains predicted using Coils version 2.1, according to the method of Lupas et al. (17). When a given region within the central rod domain is not predicted to form coiled-coils, it has been designated a linker. Depending on how parameters in Coils are set, the area designated as the linker can vary considerably in size, location, and strength of coiled-coil prediction. Three variations of Coils predictions are shown in Figure 1. The diagram at the top depicts one model for the location of linker 1–2 (residues 283–290) as well as the amino acid sequence for the region studied in this article (residues 281–304).
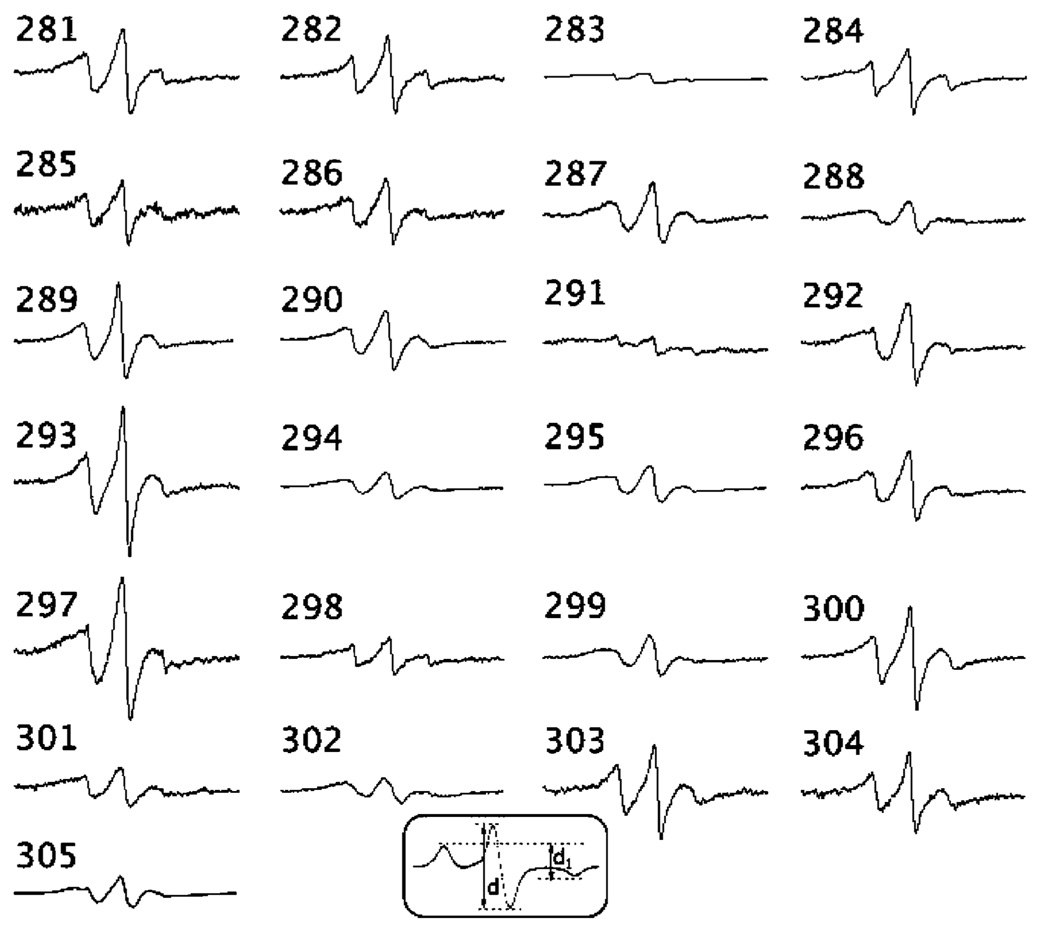
In order to evaluate the residue dynamics within this putative linker region and the proximity between these residues on opposite strands, we made 24 single-cysteine substitutions in the region encompassing positions 281–304. Following assembly of the spin-labeled protein into protofilaments, each mutant was examined by EPR spectroscopy. The EPR spectrum of each spin-labeled mutant is shown in Figure 2. The amplitude of each spectrum is normalized to the same number of spins (see Materials and Methods).
FIGURE 2.
EPR spectra of site-directed spins labels attached to positions 281–305 in human vimentin. Spectra are normalized to the same number of spins by double integration of the SDS-solubilized sample. The initiation of the normal heptad broadening pattern begins with residue 302 (an a position), with 305 being a d position. The spectrum of position 305 was previously published (Hess et al., 2001). Inset: indication of the d1/d line height ratio obtained from frozen spectra.
It has been conventionally thought that the role of linkers is to provide for rod domain flexibility (30). However, our findings reveal a rigid structure throughout the L2 region, suggesting an alternative role for this conserved region of IF structure. This is demonstrated by the fact that none of the sites within this region displays a line shape reflecting a highly flexible backbone. The most distinguishing feature within this set of spectra is the level of spectral broadening. Broadening of the EPR spectrum can arise from an increase in the hyperfine anisotropy contribution, the dipolar coupling between proximal (<2 nm) spin labels, or both. Motions restricted to correlations times approaching 1 µs result in substantial broadening. Although the line shapes of the spectra reflect an immobilized spin-labeled side chain at each position examined, none display the defined splittings at the hyperfine extrema indicative of an absence of motion on the time scale of the experiment (10−10–10−7 s).
Thus, it is evident that dipolar interactions contribute substantially to the broadened spectra within the L2 region. The broadening is especially severe when labels are placed at positions 283, 288, 291, 294, 298, 299, and 302. An evaluation of the broadening as a function of the distance-dependent dipolar interaction was performed by collecting the EPR spectra of frozen samples. In the absence of motion (i.e., frozen in liquid N2), a model-independent semiquantitative analysis of the extent of broadening due to dipolar interaction can be obtained from the d1/d spectral ratio (31, 32) (see inset, Figure 2). Strong dipolar interactions result in line height ratios >0.5 and indicate that the spins are located within 1.5 nm of one another. As shown in Table 1, positions 283 and 291 are distinguished on the basis of large d1/d values (0.63 and 0.71, respectively). The degree of broadening at these sites exceeds that of any other site examined in vimentin thus far by SDSL–EPR (24–26). In these instances, the spin-labeled side chains are very close to their partners on adjacent strands (on the order of 1.0 nm).
Table 1.
Line Height Ratio d1/d for Positions 281–304 in Vimentina
| position | d1/d |
|---|---|
| 281 | 0.43 |
| 282 | 0.38 |
| 283 | 0.63 |
| 284 | 0.44 |
| 285 | 0.43 |
| 286 | 0.40 |
| 287 | 0.45 |
| 288 | 0.46 |
| 289 | 0.39 |
| 290 | 0.43 |
| 291 | 0.71 |
| 292 | 0.38 |
| 293 | 0.41 |
| 294 | 0.48 |
| 295 | 0.44 |
| 296 | 0.43 |
| 297 | 0.43 |
| 298 | 0.51 |
| 299 | 0.50 |
| 300 | 0.43 |
| 301 | 0.48 |
| 302 | 0.59 |
| 304 | 0.39 |
The d1/d ratio (see Figure 2, inset) was measured from spectra collected at −100 °C.
Given the fact that several positions display at least moderate dipolar broadening (d1/d > 0.4), this consistent self-interaction of sites along this region suggests a close association between vimentin L2 strands in the assembled filament. This, combined with the low level of backbone flexibility, would suggest a structure of rigid, parallel-running strands.
In prior studies, we have demonstrated a signature patter in EPR spectra that is characteristic of coiled-coils (25, 26). No clear pattern consistent with a–d sites of a heptad repeat are seen in this region until position 302. At this point, the spectra of 302 and more downstream positions are consistent with a heptad repeat pattern in which 302 resembles an a position, and 303 and 304 resemble b and c positions, respectively. As shown previously, position 305 has broadening consistent with a d position. Thus, on the exclusive basis of spectral data, the coiled-coil of rod 2 appears to begin near position 302.
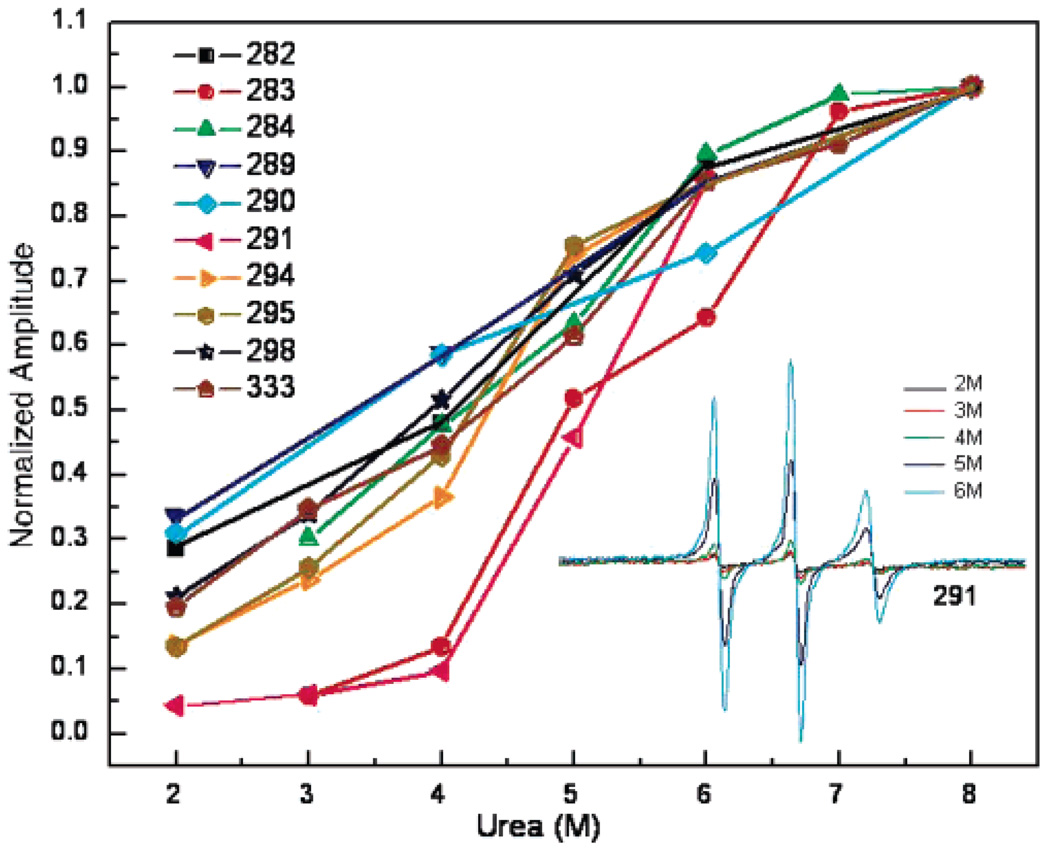
Intermediate filaments provide an advantage in the study of filament assembly because they will form filaments by removal of urea through dialysis. Previously, we have shown that a–d positions such as 333 display a linear increase in structure with urea removal, with dipolar interactions between apposing side chains apparent as the urea concentration approaches 3 M. As the urea level is brought below 3 M, the compaction of the coiled-coil increases, and dipolar broadening continues to increase until the urea level reaches 1 M. We conducted similar studies on the linker region. We compared the spectra from nine different spin-labeled positions in this linker region as a function of urea concentrations ranging from 8 to 2 M. To evaluate the formation of structure at each site, the height of the central (MI = 0) spectral line was plotted as a function of urea (Figure 3). The amplitude of this line can be measured with high sensitivity and, therefore, provides a reliable estimate of spectral broadening. Because spectra are broadened by spin-label immobilization and/or spin–spin interaction, increased structure from both dynamics and assembly contribute to this parameter, although not necessarily in a linear manner. As shown in Figure 3, the removal of urea produces an increase in spectral broadening at residues 282, 284, 289, 290, 294, 295, and 298, similar to the previously examined d position, 333. However, the strongly interacting positions 283 and 291 behave uniquely in this regard, forming tight associations with their partners in 4 M urea. This demonstrates that tight and specific associations are formed within the linker domains early in assembly, preceding even the strong interactions found at a and d positions of the coiled-coil domains.
FIGURE 3.
Sequence of assembly events. Positions 283 and 291 in the vimentin L2 domain display contacts earlier in the assembly from urea. As shown in the inset, dialysis to lower concentrations of urea generates broadened spectra. The broadening reflects restricted side chain and backbone motions that accompany folding and, in the cases of sites close to each other, additional broadening due to magnetic dipolar interactions. The spectral broadening as approximated by the central (mI = 0) EPR line height is plotted as a function of urea concentration.
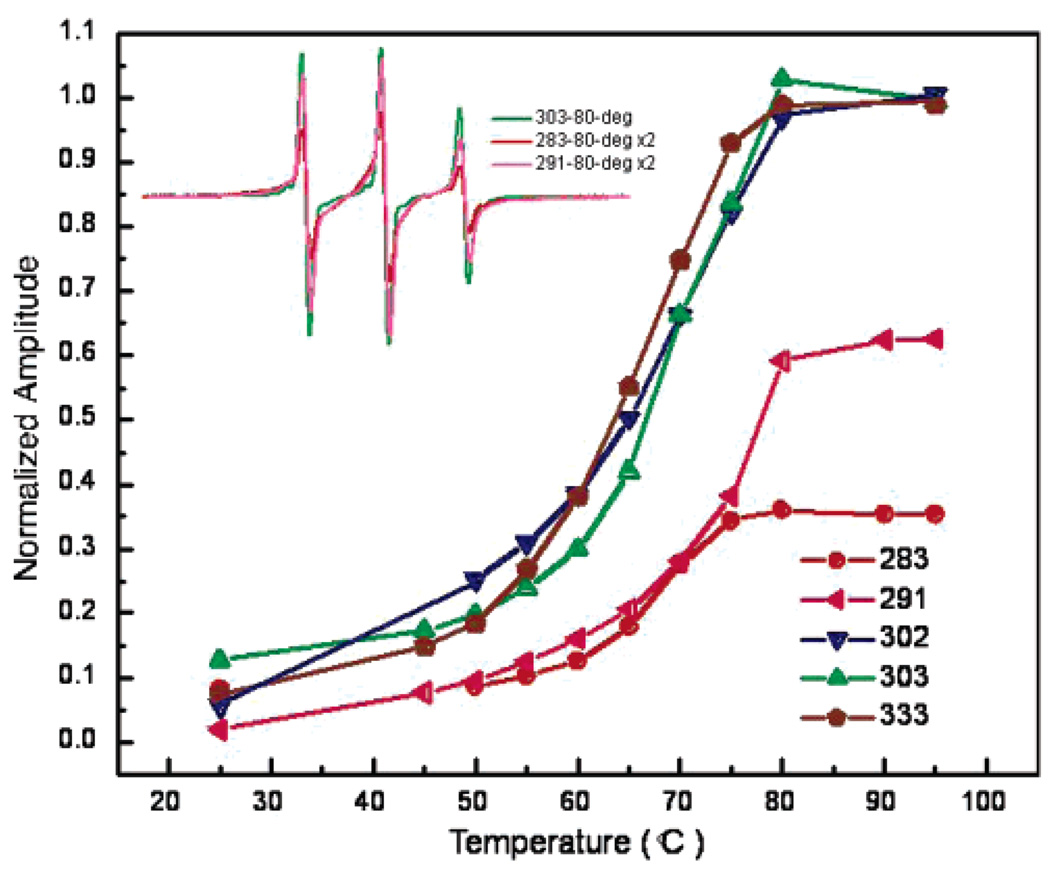
In addition to probing the onset of structure at positions upon the removal of urea, heat denaturation of vimentin was carried out to determine the thermostability of sites within the L2 region. Thus, samples of vimentin filaments containing spin labels within the L2 region (and position 333 for comparison) were scanned over successively higher temperatures in the range 25–95 °C. Plotted in Figure 4 are the central line height amplitudes of spin-labeled vimentin as a function of temperature. While the spectra from each site become similarly more narrow with increasing temperature, reflecting the increased dynamics of the spin-labeled side chain, positions 283 and 291 retain significant broadening through 95 °C. Because by 80 °C the spectra of vimentin spin labeled at 283 and 291 lack features reflecting 2° or 3° structure, the retained spectral broadening at these temperatures is attributed to a spin-coupling relaxation (33). The association of side chains located at residues 283 and 291 at high temperatures provides further evidence that this region serves as a core structural element that may serve to tether vimentin chains and establish proper alignment within the protein dimer, a fundamental step in filament assembly.
FIGURE 4.
Spectral broadening at positions 283 and 291 is maintained at high temperature. Shown is the temperature dependence of the normalized spectral amplitude for spin-labeled side chains within the vimentin L2 domain. The melt curves were obtained by scanning vimentin containing a spin label at the indicated position at successively higher temperatures, starting from 25 °C. For each position, amplitudes were normalized to the spectral amplitude of the same amount of sample in 8 M urea.
DISCUSSION
In our earlier applications of SDSL–EPR to the study of the structure and assembly of the intermediate filament protein vimentin, we focused on regions of the molecule that were predicted to adopt an α-helical, coiled-coil structure. Our EPR data revealed coiled-coil structure within rod 2B in intact filaments (26) and were consistent with data derived from crystallized dimeric fragments from rod domain 2B (19). Thus, EPR and crystallography data verified the long standing hypothesis of coiled-coil structure within rod domain 2B. Using an identical approach, we have used EPR spectroscopy to show that amino acids 179–193 adopt a coiled-coil structure as well, consistent with the prediction of this region in rod1B as coiled-coil (25). In addition, we have also demonstrated the ability of SDSL–EPR to identify interactions between coiled-coil dimers and with this approach have identified regions within rod 1B and rod 2B that are closely apposed in the A11 and A22 tetramers, respectively (25, 26).
In this article, we focused on a region predicted to contain at least two different structures, the non-α-helical linker 2 and the beginning of α-helical rod domain 2B (34–37). The existence of each region and the precise beginning of rod 2B has long been hypothesized, and models have been made utilizing structures predicted by computer (38). However, no direct evidence exists to identify either the beginning of rod domain 2B or the structure of the linker region. EPR spectra suggest that the nonhelical linker domain occupies a region much larger than that predicted and that rod domain 2B does not appear to adopt a regular coiled-coil pattern until approximately position 302, more than one heptad later (downstream) from the predicted start of rod 2B at position 291. The data presented here show the noncoiled-coil region to be a rigid domain, with two positions (283 and 291) demonstrating extreme proximity. These positions also demonstrate a very early adoption of structure and resistance to denaturation, suggesting that the nonhelical linker domain may be a region of importance for early assembly events. This may suggest that the linker region serves as a trigger site, which initiates and/or aligns the earliest stages of IF assembly.
Of particular interest in this linker region are five charged residues, three of which are very highly (but not absolutely) conserved across all Type I–III IF proteins (Figure 1, residues 286, 288, 294). EPR data suggests that these charge residues are likely to act to stabilize interchain interactions rather than intrachain interactions because spin labels placed at residue 283 of the linker region continue to show spin–spin interaction at temperatures and urea concentrations that should cause chain separation in the absence of interchain charge stabilization.
In addition to the usual requirements for coiled coil formation (heptad repeat, non-polar a,d positions), an additional region called the trigger sequence has been found to be critical for assembly (39–41). Characterization of small peptides and larger recombinant regions demonstrates that trigger sequences possess the ability to form α-helices, and these helices, once formed, are hypothesized to lead to the zipping up of the entire coiled-coil (41). Although the name implies that these regions trigger coiled-coil formation, the order of assembly within IFs has not been demonstrated (39, 41). An examination of IF sequences for trigger sequences has resulted in the identification of two regions with sequence similarity and, more importantly, functional requirements for coiled-coil formation. As characterized by Wu et al. (40), using a k5/14 assembly pair, coiled-coil triggers are located within rod 1B and rod 2B. These locations flank the early assembling region we have identified within linker 2. Assuming a conservation of structural elements between type I and II keratins and type III vimentin, the action of linker 2 as an early assembly facilitator could easily be incorporated into the model by Kammerer as an interaction that brings two polypeptide chains together and, thus, facilitates the transition from two autonomous helical folding units to a parallel in-register alignment of two monomers (step i to ii of the model of Kammerer et al. (41).
The first solved X-ray structure of vimentin fragments revealed the coiled-coil nature of the carboxy terminus of rod 2B, establishing α-helical coiled-coil structure from positions 328–405 (18, 19). Our initial EPR experiments showed that a region of rod domain 2B, positions 305–336, was found in a coiled-coil structure, and the spectra identified a highly ordered (rigid) conformation throughout the entire region (26). These data were sufficient to identify coiled-coil structure from position 305 through 323, but did not specifically identify the start of rod domain 2B. In this study, we have used SDSL–EPR to identify the start of rod domain 2B as position 302 of vimentin (an a position of the heptad), a position that is in phase with our previous data and supports the existence of a continuous coiled-coil from position 302 through 337. The EPR spectra reveal a region with considerable rigidity due to extensive protein packing. Together with the crystal structure of domain 2B, positions 328–411, the entire rod domain 2B can be identified as a coiled-coil structure from 302–405.
The earliest descriptions of keratin amino acid sequence identified the region later termed linker 2 (37) as a break in the predicted α-helical structure (34, 36). Initial predictions favored a beta turn for the structure of the region linking rod 2A with 2B. However, as more sequences were determined, an analysis of amino acid sequences across several types of IFs resulted in the identification of an eight amino acid sequence as linker 2 and the prediction that linker 2 was α-helical but not coiled-coil (7, 42). Subsequent computer modeling suggested a structure for linker 2 that resulted in the crossing of peptide chains within a dimer such that rod domain 2A of one chain was axially in line with the rod domain 2B of the other polypeptide chain in the dimer (38).
Both EPR and X-ray crystallography have been able to demonstrate specific structure for specific regions of the vimentin molecule. However, the structure of nonhelical linkers has not been described, only modeled (38). Thus, our characterization of positions 281–302 of vimentin represents the first data to examine previous predictions with regard to helical versus nonhelical structure and rigid versus flexible structure. We chose position 281 as the starting point for our characterization because the region preceding it has an amino acid pattern consistent with the possibility of adopting a coiled-coil structure and has been predicted to form a coiled-coil rod 2A structure that we are currently exploring. Spectra from positions 281–301 do not reveal evidence of a coiled-coil structure such as we have documented for domains 1B and 2B (25, 26). Instead of a pattern of a and d positions producing spectra that indicate rigid structure and spin–spin interaction, we see spectra indicating close interactions at numerous positions.
Spectra from residues 283 and 291 are unique in that they represent positions at which the spin labels are closer than any other position thus examined. They are also notable because they have been identified on the basis of computer predictions as the beginning of linker 2 (position 283) and rod 2B (position 291) (43). Across the different families of IF proteins, sequence conservation within linker 2 has been interpreted as evidence for conservation of structure, most likely an α-helical structure (42). Position 291, tyrosine, has been designated as the beginning of rod 2B, occupying a d position and has been further described as being the site of a conserved aromatic amino acid, Y or F, in all IF chain types. The identification of 291 as a d position is consistent with the heptad repeat pattern we have identified beginning with 302 as an a position. However, the EPR spectra we present here do not agree with these predictions because the intervening positions do not display the typical coiled-coil pattern such as that we have identified with rod 1B and rod 2B. Such a signature pattern for the coiled-coil is, however, seen beginning at position 302.
Positions 283 and 291 occupy the flanking positions of the conserved Linker 2 motif (N-R-X-D/E-A-E–E-W-Y). On the basis of cross-linking experiments in keratin filaments that suggest a compact arrangement of these residues, North et al. constructed a model of the L2 core as overlapping loops of the two strands (36). Our results confirm the rigid nature of this region as proposed in the North model; however, the multiple sites displaying dipolar interaction suggest a simpler parallel association of two loops in the L2 core. The fact that the conserved linker motif is distinguished by charged and aromatic residues provides clues for probing the basis of this region’s extreme stability. First, the π–π stacking of aromatic residues (Trp290, Tyr291, and Phe295) may play an important role in the early association of vimentin. Such interactions have been implicated in the assembly of amyloid fibrils (42). Second, salt bridges represent a general feature of stabilization in proteins of thermophilic organisms (44, 45). Further studies will exploit the stability markers of spin-labeled 283 and 291 to determine the consequence of side chain substitution within the L2 region on vimentin assembly and stability.
With the delineation of rod 2B, and the identification of a rigid structure within the linker 2 domain, EPR continues to reveal details about the structure and assembly of intermediate filaments. Some features have been predicted for many years but only experimentally tested now, whereas other details such as the starting point of rod 2B are slightly different from long-standing predictions. Although EPR cannot produce as detailed a structure as X-ray crystallography can, the careful application of spin labels has again been demonstrated as providing useful information. Continued examination of the vimentin molecule should reveal the full extent of coiled-coil domains and interactions between domains in proximity to each other.
Footnotes
This work was supported by grant DAMD D17-02-1-0664 to J.H., grants NIH RO1 NEI EY08747 and NIH EY015560 to P.G.F., and March of Dimes Grant 5-FY02-202 to J.C.V. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR-12088-01 from the National Center for Research Resources, National Institutes of Health.
Abbreviations: SDSL, site directed spin labeling; EPR, electron paramagnetic resonance; IF, intermediate filament.
REFERENCES
- 1.Parry DA, Steinert PM. Intermediate filaments: molecular architecture, assembly, dynamics and polymorphism. Q. Rev. Biophys. 1999;32:99–187. doi: 10.1017/s0033583500003516. [DOI] [PubMed] [Google Scholar]
- 2.Parry DA, Steinert PM. Intermediate filament structure. Curr. Opin. Cell Biol. 1992;4:94–98. doi: 10.1016/0955-0674(92)90064-j. [DOI] [PubMed] [Google Scholar]
- 3.Herrmann H, Harris JR. Intermediate Filaments. New York: Plenum; 1998. [Google Scholar]
- 4.Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu. Rev. Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 5.Albers K, Fuchs E. The molecular biology of intermediate filament proteins. Int. Rev. Cytol. 1992;134:243–279. doi: 10.1016/s0074-7696(08)62030-6. [DOI] [PubMed] [Google Scholar]
- 6.Conway JF, Parry DA. Structural features in the heptad substructure and longer range repeats of two-stranded alpha-fibrous proteins. Int. J. Biol. Macromol. 1990;12:328–334. doi: 10.1016/0141-8130(90)90023-4. [DOI] [PubMed] [Google Scholar]
- 7.Conway JF, Parry DAD. Intermediate filament structure: 3. Analysis of sequence homologies. Int. J. Biol. Macromol. 1988;10:79–98. [Google Scholar]
- 8.Coulombe PA, Bousquet O, Ma L, Yamada S, Wirtz D. The ‘ins’ and ‘outs’ of intermediate filament organization. Trends Cell Biol. 2000;10:420–428. doi: 10.1016/s0962-8924(00)01828-6. [DOI] [PubMed] [Google Scholar]
- 9.Coulombe PA, Maq L, Yamada S, Wawersik M. Intermediate filaments at a glance. J. Cell Sci. 2001;114:4345–4347. doi: 10.1242/jcs.114.24.4345. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs E, Hanukoglu I. Unraveling the structure of the intermediate filaments. Cell. 1983;34:332–334. doi: 10.1016/0092-8674(83)90367-7. [DOI] [PubMed] [Google Scholar]
- 11.Goldman JE, Steinert PM. Cellular Molecular Biology of Intermediate Filaments. New York: Plenum Press; 1990. [Google Scholar]
- 12.Herrmann H. Structure, Assembly, and Dynamics of Intermediate Filaments. In: Herrmann H, Harris JR, editors. Intermediate Filaments. Vol. 31. New York: Plenum Press; 1998. pp. 319–355. [PubMed] [Google Scholar]
- 13.Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu. Rev. Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 14.Parry DA, Steven AC, Steinert PM. The coiled-coil molecules of intermediate filaments consist of two parallel chains in exact axial register. Biochem. Biophys. Res. Commun. 1985;127:1012–1018. doi: 10.1016/s0006-291x(85)80045-0. [DOI] [PubMed] [Google Scholar]
- 15.Parry DAD, Steinert PM. Intermediate Filament Structure. Austin, TX: R. G. Landes Company; 1995. [Google Scholar]
- 16.Strelkov S, Herrmann H, Aebi U. Molecular architecture of intermediate filaments. BioEssays. 2003;25:243–251. doi: 10.1002/bies.10246. [DOI] [PubMed] [Google Scholar]
- 17.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 18.Strelkov SV, Herrmann H, Geisler N, Wedig T, Zimbelmann R, Aebi U, Burkhard P. Conserved segments 1A and 2B of the intermediate filament dimer: their atomic structures and role in filament assembly. EMBO J. 2002;21:1255–1266. doi: 10.1093/emboj/21.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strelkov SV, Herrmann H, Parry DD, Aebi U. Atomic structure of the consensus motif of the 2B rod domain of the intermediate filament chain vimentin. J. Invest. Dermatol. 2000;114 [Google Scholar]
- 20.Quinlan RA, Franke WW. Molecular interactions in intermediate-sized filaments revealed by chemical cross-linking. Heteropolymers of vimentin and glial filament protein in cultured human glioma cells. Eur. J. Biochem. 1983;132:477–484. doi: 10.1111/j.1432-1033.1983.tb07386.x. [DOI] [PubMed] [Google Scholar]
- 21.Quinlan RA, Hatzfeld M, Franke WW, Lustig A, Schulthess T, Engel J. Characterization of dimer subunits of intermediate filament proteins. J. Mol. Biol. 1986;192:337–349. doi: 10.1016/0022-2836(86)90369-4. [DOI] [PubMed] [Google Scholar]
- 22.Steinert PM, Marekov LN, Fraser RD, Parry DA. Keratin intermediate filament structure. Crosslinking studies yield quantitative information on molecular dimensions and mechanism of assembly. J. Mol. Biol. 1993;230:436–452. doi: 10.1006/jmbi.1993.1161. [DOI] [PubMed] [Google Scholar]
- 23.Steinert PM, Marekov LN, Parry DA. Diversity of intermediate filament structure. Evidence that the alignment of coiled-coil molecules in vimentin is different from that in keratin intermediate filaments. J Biol. Chem. 1993;268:24916–24925. [PubMed] [Google Scholar]
- 24.Hess J, Budamagunta M, FitzGerald P, Voss J. Characterization of structural changes in vimentin bearing an EBS-like mutation using site directed spin labeling and electron paramagnetic resonance. J. Biol. Chem. 2005;280(3):2141–2146. doi: 10.1074/jbc.M412254200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hess JF, Budamagunta MS, Voss JC, FitzGerald PG. Structural characterization of human vimentin rod 1 and the sequencing of assembly steps in intermediate filament formation in vitro using SDSL and EPR. J. Biol. Chem. 2004;279(43):44841. doi: 10.1074/jbc.M406257200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hess JF, Voss JC, FitzGerald PG. Real-time observation of coiled-coil domains and subunit assembly in intermediate filaments. J. Biol. Chem. 2002;277:35516–35522. doi: 10.1074/jbc.M206500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagai K, Thøgersen HC, Luisi BF. Refolding and crystallographic studies of eukaryotic proteins produced in Escherichia coli. Biochem. Soc. Trans. 1988;16:108–110. doi: 10.1042/bst0160108. [DOI] [PubMed] [Google Scholar]
- 28.Nagai K, Thøgersen HC. Synthesis and sequencespecific proteolysis of hybrid proteins produced in Escherichia coli. Methods Enzymol. 1987;153:461–481. doi: 10.1016/0076-6879(87)53072-5. [DOI] [PubMed] [Google Scholar]
- 29.Carter JM, Hutcheson AM, Quinlan RA. In vitro studies on the assembly properties of the lens proteins CP49, CP115: coassembly with alpha-crystallin but not with vimentin. Exp. Eye Res. 1995;60:181–192. doi: 10.1016/s0014-4835(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 30.Mucke N, Wedig T, Burer A, Marekov LN, Steinert PM, Langowski J, Aebi U, Herrmann H. Molecular and biophysical characterization of assembly-starter units of human vimentin. J. Mol. Biol. 2004;340:97–114. doi: 10.1016/j.jmb.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 31.Kokorin AI, Zamaraev KI, Grigorian GL, Ivanov VP, Rozantsev EG. Measurement of the distance between paramagnetic centers in solid solutions of nitrosyl radicals, biradicals and spin-labelled proteins. Biofizika. 1972;17:34–41. [PubMed] [Google Scholar]
- 32.Likhtenshtein GI. Biophysical Labeling Methods in Molecular Biology. New York: Cambridge University Press; 1993. [Google Scholar]
- 33.McHaourab HS, Oh KJ, Fang CJ, Hubbell WL. Conformation of T4 lysozyme in solution. Hinge-bending motion and the substrate-induced conformational transition studied by site-directed spin labeling. Biochemistry. 1997;36:307–316. doi: 10.1021/bi962114m. [DOI] [PubMed] [Google Scholar]
- 34.Hanukoglu I, Fuchs E. The cDNA sequence of a Type II cytoskeletal keratin reveals constant and variable structural domains among keratins. Cell. 1983;33:915–924. doi: 10.1016/0092-8674(83)90034-x. [DOI] [PubMed] [Google Scholar]
- 35.Hanukoglu I, Tanese N, Fuchs E. Complementary DNA sequence of a human cytoplasmic actin. Interspecies divergence of 3′ non-coding regions. J. Mol. Biol. 1983;163:673–678. doi: 10.1016/0022-2836(83)90117-1. [DOI] [PubMed] [Google Scholar]
- 36.Hanukoglu I, Fuchs E. The cDNA sequence of a human epidermal keratin: divergence of sequence but conservation of structure among intermediate filament proteins. Cell. 1982;31:243–252. doi: 10.1016/0092-8674(82)90424-x. [DOI] [PubMed] [Google Scholar]
- 37.Steinert PM, Parry DA, Racoosin EL, Idler WW, Steven AC, Trus BL, Roop DR. The complete cDNA and deduced amino acid sequence of a type II mouse epidermal keratin of 60,000 Da: analysis of sequence differences between type I and type II keratins. Proc. Natl. Acad. Sci. U.S.A. 1984;81:5709–5713. doi: 10.1073/pnas.81.18.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.North AC, Steinert PM, Parry DA. Coiled-coil stutter and link segments in keratin and other intermediate filament molecules: a computer modeling study. Proteins. 1994;20:174–184. doi: 10.1002/prot.340200207. [DOI] [PubMed] [Google Scholar]
- 39.Steinmetz MO, Stock A, Schulthess T, Landwehr R, Lustig A, Faix J, Gerisch G, Aebi U, Kammerer RA. A distinct 14 residue site triggers coiled-coil formation in cortexillin I. EMBO J. 1998;17:1883–1891. doi: 10.1093/emboj/17.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu KC, Bryan JT, Morasso MI, Jang SI, Lee JH, Yang JM, Marekov LN, Parry DA, Steinert PM. Coiled-coil trigger motifs in the 1B and 2B rod domain segments are required for the stability of keratin intermediate filaments. Mol. Biol. Cell. 2000;11:3539–3558. doi: 10.1091/mbc.11.10.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kammerer RA, Schulthess T, Landwehr R, Lustig A, Engel J, Aebi U, Steinmetz MO. An autonomous folding unit mediates the assembly of two-stranded coiled coils. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13419–13424. doi: 10.1073/pnas.95.23.13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parry D, Fraser RD. Primary and Secondary Sequence Structure of IF Protein Chains and Modes of Molecular Aggregation. In: Goldman RD, Steinert PM, editors. Cellular and Molecular Biology of Intermediate Filaments. New York: Plenum Press; 1985. pp. 175–204. [Google Scholar]
- 43.Conway J, Parry DD. Intermediate filament structure 3. Int. J. Biol. Macromol. 1988;10 [Google Scholar]
- 44.Sanchez-Ruiz JM, Makhatadze GI. To charge or not to charge? Trends Biotechnol. 2001;19:132–135. doi: 10.1016/s0167-7799(00)01548-1. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Nussinov R. Close-range electrostatic interactions in proteins. ChemBioChem. 2002;3:604–617. doi: 10.1002/1439-7633(20020703)3:7<604::AID-CBIC604>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]