Abstract
We have previously established the utility of site-directed spin labeling and electron paramagnetic resonance to determine structural relationships among proteins in intact intermediate filaments. Using this same approach we have introduced spin labels at 21 residues between amino acids 169 and 193 in rod domain 1 of human vimentin. The electron paramagnetic resonance spectra provide direct evidence for the coiled coil nature of the vimentin dimer in this region. This finding is consistent with predictions but has never been demonstrated previously. In a previous study we identified residue 348 in the rod domain 2 as one point of overlap between adjacent dimers in intact filaments. In the present study we defined residue 191 in the rod domain 1 as a second point of overlap and established that the dimers are arranged in an anti-parallel and staggered orientation at this site. Finally, by isolating spin-labeled samples at successive stages during the dialysis that lead to filament assembly in vitro, we have been able to establish a sequence of interactions that occurs during in vitro assembly, starting with the α helix and loose coiled coil dimer formation, then the formation of tetrameric species centered on residue 191, followed by interactions centered on residue 348 suggestive of octamer or higher order multimer formation. A continuation of this strategy revealed that both 191–191 and 348–348 interactions are present in low ionic strength Tris buffers when vimentin is maintained at the “protofilament” stage of assembly.
The intermediate filament (IF)1 protein family consists of greater than 50 members, constituting one of the larger gene families in the human genome (1, 2–5). Although the sizes and primary sequences among IF proteins can vary considerably, they all share a common predicted domain structure consisting of a central rod domain flanked by head and tail domains (see Fig. 1 for a schematic). The central rod domain of IF proteins is ~310 amino acids in length and shows a strong conservation of the predicted secondary structure. The rod domain is divisible into larger “coil” subdomains predicted to be α helical, joined by short “linker” domains whose sequence does not predict α helicity. Further, the coil domains exhibit a heptad repeat pattern characteristic of proteins that assemble into coiled coil dimers. In this heptad repeat, the first and fourth residues (“a” and “d” positions) are largely hydrophobic and form a hydrophobic “stripe” along one side of the helix. This stripe is the apposing interface between two monomers that assemble into a coiled coil dimer (6). Rigorous demonstration that a particular amino acid or region is α helical or coiled coil has been provided by x-ray crystallography of vimentin fragments and by our initial EPR studies (7–10).
FIG. 1. A schematic view of the vimentin molecule showing the domain structure of vimentin.
The central rod domain is predicted to consist of two subdomains, rod 1 and rod 2, that are predicted to be largely α helical. The relative location and amino acid sequence of the region of rod domain 1 studied in this report is shown. The predicted position of each residue in this region in the heptad repeat is indicated by the a–g notation below the sequence.
IF proteins are generally insoluble in physiologic solutions, typically requiring extreme conditions such as 8 m urea or 6 m guanidine to be fully solubilized. Subsequent removal of the urea or guanidine by dialysis allows for the spontaneous reassembly of denatured proteins into native-looking IFs (11–13). Although this feature is beneficial to the study of IF assembly in vitro, it has created great difficulties in efforts to generate IF protein crystals suitable for high-resolution structural studies. Because of the inability to crystallize intact IF proteins some investigators have taken a “divide and conquer” approach by generating crystals of IF protein fragments or of chimeric proteins that include IF fragments. These data have revealed the structure of small regions containing α helical dimers, including the perplexing “stutter” region within rod 2b (7–9). The degree to which the crystal structure of the isolated fragments resembles that of those same regions in intact filaments will need to be confirmed.
The determination of the architecture of IF proteins within intact filaments has also proven difficult to approach experimentally. Our understanding of the architecture of intact filaments and the assembly process required to reach that point has been postulated on the basis of evidence derived from a number of approaches including cross-linking studies, secondary structural predictions, circular dichroism, extrapolation from studies of proteins exhibiting similar predicted secondary structure, sedimentation studies, size exclusion chromatography, and so forth (14–17). These studies collectively have led to a long-standing model of IF assembly that starts with the formation of an in-parallel and in-register coiled coil dimer. Subsequent assembly is predicted to consist of the formation of a tetramer composed of two anti-parallel, staggered dimers.
The cross-linking of K5 and K14 keratins in low ionic strength triethanolamine buffers (an in vitro assembly protocol that results in small subunits referred to as “protofilaments”) has produced data supporting the existence of two different tetramer alignments (14, 18). These same data show that both tetramer alignments exist in a hexamer, as well as in the assembled filament. Direct testing of this model, however, has proven difficult to accomplish. Subsequent cross-linking analysis has revealed the importance of residues in rod 1A and linker 2 in stabilizing the A11 alignment (so-called because rod 1 domains of two dimers are apposed in a tetramer) (19). These data also suggest that the A11 alignment is more stable than the A22 alignment (where the rod 2 domains are aligned).
Vimentin exists mainly in the monomeric form in solutions above 6 m urea, assembling into dimers at near 5 m and tetramers at 3 m urea concentrations (17, 20, 21). A cross-linking of the single cysteine in wild type vimentin has been cited as evidence for an A22 tetramer formation in soluble vimentin (17, 21). Cross-linking of proteolytically prepared desmin rods that are filament assembly-incompetent in low ionic strength ethanolamine demonstrates an A11 structure (22). This alignment was also demonstrated in full-length desmin IFs subjected to cross-linking.
We have previously reported the utility of SDSL and EPR in contributing to the understanding of IF architecture, using the human type III IF protein vimentin as a model (10). In this approach a spin label is introduced to specific sites within the protein, and the protein is induced to assemble into intermediate filaments. The resulting filaments are examined by EPR in physiologic solution and in real time. The spectra yields basic information about the secondary structure at the site of the spin label, as well as the proximity of spin labels to one another. In our previous study we placed spin labels at 14 successive residues in vimentin rod domain 2 and generated direct evidence that this region engages in a coiled coil dimer formation. By introducing spin labels at the exterior surface of a dimer we explored the architecture of dimer-dimer interactions and provided evidence that two dimers are oriented in an anti-parallel manner with an overlap at residue 348.
Using the same approach, we have now characterized vimentin structure in rod 1, identified the orientation and point of overlap between dimers in this region, and have deduced a sequence of steps that accompany the in vitro assembly of vimentin filaments. These data describe the structure of the vimentin IF and establish an approach for evaluating how specific mutations impact filament assembly.
MATERIALS AND METHODS
Vimentin characterization, mutation, cloning, expression, purification, and spin labeling were described in detail in a previous report (10). In brief, the spin label is ultimately attached to cysteine residues, so mutations that create cysteines were introduced into the vimentin expression construct (generously provided by Dr. Roy Quinlan, University of Durham, Durham, UK) at specific sites using the Stratagene QuikChange kit. Sequence changes were verified by DNA sequencing. Mutant vimentin was produced by bacterial expression and was purified from inclusion bodies using high/low salt washes and chromatography. Spin labeling was accomplished by incubation of the purified vimentin in 100 µm tris-(2-carboxyethyl)phosphine, hydrochloride (Molecular Probes, Eugene, OR) followed by 500 µm O-87500 (Toronto Research Chemicals, Toronto, Canada). The unincorporated spin label was removed by CM-Sepharose chromatography (10) using an Amersham Biosciences FPLC. The labeled proteins were stored long term at −80 °C.
The filament assembly was performed either as a single step dialysis against filament assembly buffer, or a stepwise process as described by Carter et al. (13). Briefly, labeled proteins were solubilized in 8 m urea, which was then removed by dialysis either in a single step procedure or, where indicated, in a stepwise process through progressively reduced concentrations of urea followed by low ionic strength Tris and then by the addition of NaCl and MgCl2. Filament assembly was verified each time by electron microscopy of negatively stained samples. As reported in Fig. 1 some mutations were not capable of assembling into filaments.
EPR was conducted on a JEOL X-band spectrometer equipped with a loop gap resonator (23). Approximately 4 µl of sample, at a concentration of 25–75 µm protein, were placed in a sealed quartz capillary tube. Spectra were acquired at 20–22 °C with a single 60s scan over 100 G at a microwave power of 2 mW, and a modulation amplitude optimized to the natural line width of the attached nitroxide.
RESULTS
Rod 1 Monomer-Monomer Interactions
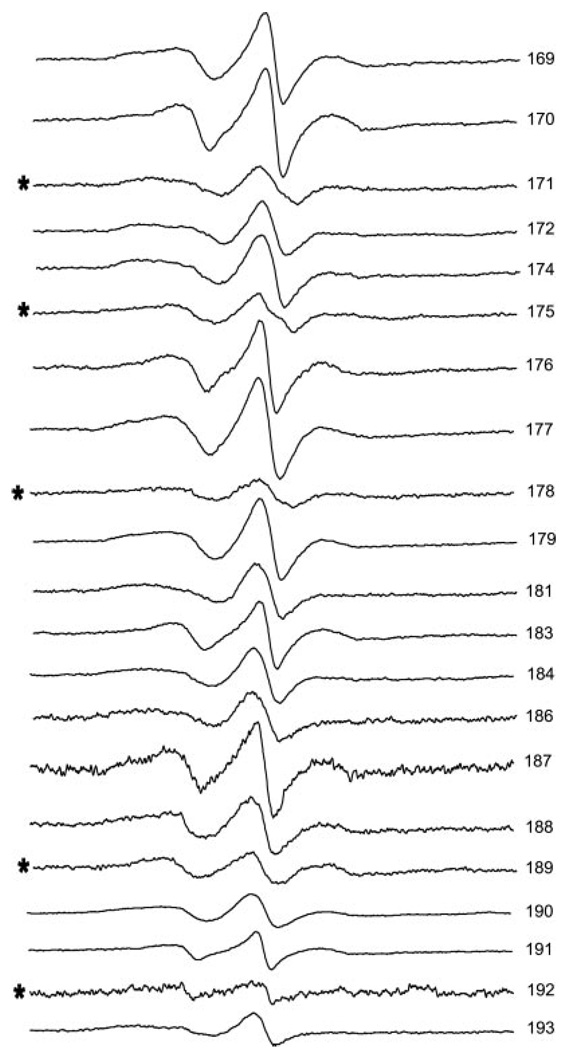
A schematic of the vimentin monomer is presented in Fig. 1. The amino acid sequence at this site is presented. The position of each amino acid in the heptad repeat pattern is indicated by notations of a–g. This heptad repeat is thought to impose a coiled coil assembly of polypeptide chains. Residues within the region of 169–193 that were changed to cysteine, spin labeled, and examined by EPR are presented in Fig. 2. For each position the spectra reflect moderate to strong side chain motion, indicating a region of fixed secondary structure with varied levels of side chain contact.
FIG. 2. Room temperature EPR spectra of 21 different spin-labeled mutants of human vimentin.
The location of the spin label is indicated by the residue number at the left edge of the spectrum. Residues predicted to occupy an “a” or “d” position in the heptad repeat are indicated by *. The spectrum for mutant 192 has been amplified 4×.
The spectra in Fig. 2 are normalized to represent the same number of spins. Here it can be seen that some spectra display severe broadening, indicative of magnetic dipolar interaction between labels situated within 2.0 nm of one another. Modeling of coiled coil dimer interactions predicts that if the dimer is aligned in-parallel and in-register spectra from spin labels in the a and d positions would show evidence of close proximity whereas those in the b, c, e, f, and g positions would show evidence of more distant interactions. As is apparent in Fig. 2, the a/d positions of 171, 175, 178, 189, and 192 (indicated by *) are more broadened than other positions within the range of 169–188. More moderate broadening (discussed below) is seen from positions 190, 191, and 193. Thus the EPR spectra provide direct support for the coiled coil dimer prediction within this region of rod 1B.
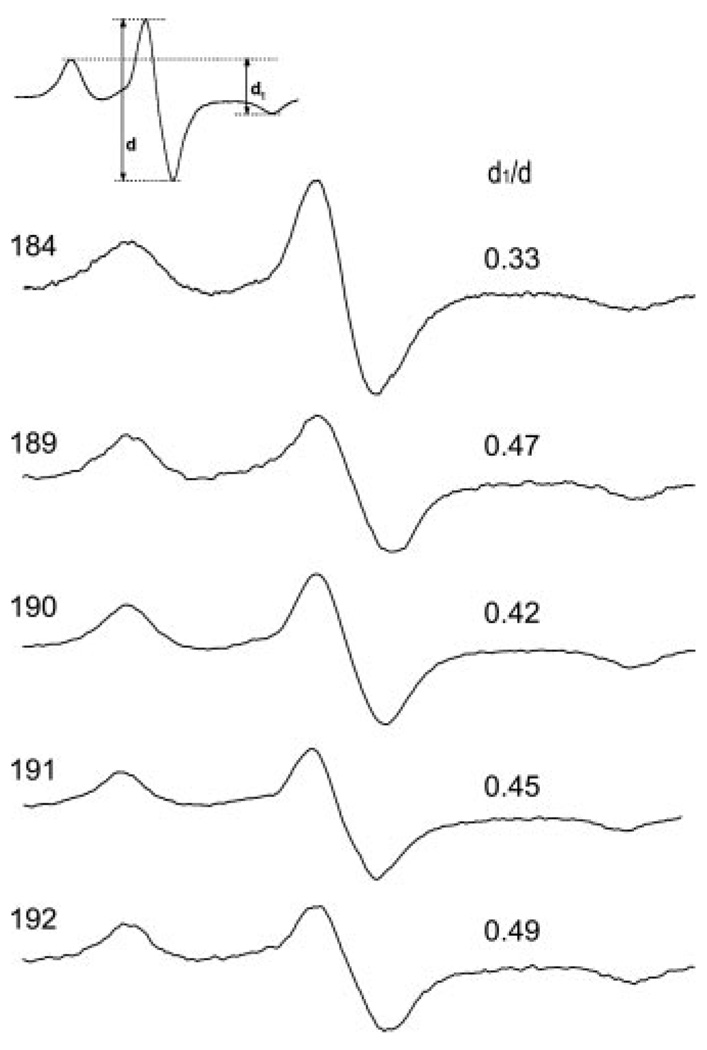
Because the spectral shapes shown in Fig. 2 depend on both the side chain mobility and the proximity of nitroxides on neighboring subunits, the analysis for the strength of the dipolar interaction can be simplified by freezing the sample to arrest all motions influencing the nitroxide line shape. The −100 °C spectra for vimentin samples harboring spin labels at positions 184, 189, 190, 191, and 192 are given in Fig. 3. A model-independent evaluation of the magnetic interaction strength can be obtained from the broadening parameter d1/d (24) whose value for spins >2.0 nm apart is ~0.3 but increases as labels are brought into closer proximity. As suggested by the room temperature spectra in Fig. 2, labels located at position 184 do not show appreciable interaction (d1/d = 0.33), whereas labels located at position 192 show the strongest interaction (d1/d = 0.50). Position 192 is a predicted “d” position and as such, would be expected to have a d1/d ratio approaching 0.5 (10). The ratio of 0.5 is undoubtedly because of the intradimer proximity (d-d interaction in a coiled coil) plus the proximity of another spin-labeled dimer. Also consistent with the room temperature data, the non-a/d positions of 190 and 191 show dipolar interaction indicating that these positions approach one another in intact filaments.
FIG. 3. EPR spectra of frozen vimentin mutants spin-labeled at the indicated position.
Samples containing more proximal labels display greater broadening. The extent of broadening is revealed by the qualitative parameter d1/d, which is larger for labels experiencing greater dipolar interaction.
Dimer-Dimer Overlap in Rod Domain 1
In a previous report we introduced spin labels along the exterior surface of a dimer, and established in intact filaments that two dimers overlap at residue 348 in rod domain 2 (see Fig. 4 for orientation). To determine whether these dimers were parallel or anti-parallel we mixed samples containing spin labels located upstream and downstream of position 348 to distinguish a parallel from anti-parallel associations of the dimers. We reasoned that spin labels placed downstream from residue 348, at residue 334, would align opposite 334 if the dimers were aligned in-parallel but would oppose a residue located an equivalent distance upstream if the dimers were in an anti-parallel arrangement. These experiments established that residues 334 and 359 were directly apposed providing evidence for an anti-parallel and staggered overlap for two adjacent dimers at this site. However, a cross-section of an intermediate filament is postulated to contain ~16 dimers, thus a given dimer has the potential to interact with several other dimers.
FIG. 4. Schematic view of three vimentin dimers.
SDSL EPR establishes that a given vimentin dimer interacts with rod domain 2 of one dimer (position 348) and rod domain 1 of another dimer (position 191). Both interactions are anti-parallel.
The overlap of two dimers at residue 348 in rod 2 presages an interaction between the free rod 1 domains. We sought evidence for such an overlap by introducing spin labels along the exterior surface of the dimer in the rod 1 region (positions 169–193). This analysis revealed that residues 187, 188, 190, 191, and 193 (none of which are in a or d positions of the heptad) show significant broadening because of magnetic interaction, with 191 yielding the strongest interaction. In addition, position 192 shows the strongest self-interaction for any residue yet examined in vimentin. Thus the broadening centered near position 191 indicates an interaction between dimers or higher level oligomers, consistent with the A11 alignment shown in Fig. 4. We concluded that the severe broadening seen at position 192 arises from a combination of the d-d interaction from two parallel coiled coil monomers, as well as interaction of the labels located on separate dimers overlapping near position 191 (discussed below).
To establish the orientation of dimers that overlap at residue 191 we used the same approach described previously for the overlap at 348 (10). In the present study, equal molar amounts of two separately labeled mutants were combined and examined by EPR spectroscopy. These mixtures contained one protein with a spin label attached upstream of position 191 and a second mutant protein with the label attached an equivalent distance downstream of residue 191. The observed interaction in frozen spectra from mixtures of single-labeled 184 or 188 with an equal amount of vimentin labeled at 197 or 193, respectively, resulted in a higher d1/d value than any of the single mutants alone (Table I). This thus confirms that rod 1 of one dimer overlaps near residue 191 with another dimer and that these two regions are arranged in an anti-parallel orientation. The precise points of overlap between one vimentin dimer and two of its neighbors and the orientation of those neighbors have now been established in intact filaments in physiologic solution.
TABLE I.
Observed interaction in frozen spectra
| Spin-labeled residue | d1/d |
|---|---|
| 184 | 0.33 |
| 188 | 0.38 |
| 193 | 0.38 |
| 197 | 0.40 |
| 184 + 197 | 0.48 |
| 188 + 193 | 0.50 |
Sequencing of in Vitro Assembly Steps
The data reported thus far have been performed on the end stage of filament assembly procedures, i.e. on intact filaments. We decided to explore whether the various events of assembly that have been documented could be placed in sequence by gathering spectra from intermediates of assembly that emerged during dialysis to progressively lower urea concentrations. We selected different mutants, because each could serve as an “indicator” for specific phases of the assembly process. First, we examined residue 333, located in a d position in rod domain 2, to observe both the assumption of α helicity as well as the formation of the coiled coil dimer. We also chose two non-a/d positions in the central rod domain to reveal the stage where overlap between dimers occurs. Residues 191 and 348 were selected to report the formation of the dimer-dimer interactions centered in rod 1 and rod 2, respectively.
Spectra from mutant 333, a d position in rod 2, showed that the first indications of backbone stabilization occur at ~6 m urea and continue to undergo progressive constraint through 1 m urea (Fig. 5). This demonstrates that the initiation of the α helix formation occurs at ~6 m urea with a progressive stabilization of the backbone of the helix as dialysis progresses. The first significant line shape change occurs at the high field (m I = +1) line at 5 m urea, suggesting the presence of some loosely associated dimer at this urea level. This agrees with centrifugation and cross-linking measurements, which suggest that a dimer of vimentin is formed at 5 m urea (17, 20) Interestingly, although the dipolar broadening is not substantially increased as the urea concentration is lowered from 2 m, the tight packing of the position 333 side chain side is not accomplished until the urea is lowered to 1 m. The 1 m spectrum of position 333 features the broad splittings indicative of a strongly immobilized side chain and is identical to the spectrum of the sample in the absence of urea (10)
FIG. 5. Influence of urea on the EPR Spectrum of vimentin spin labeled at residue 333 of rod domain 2.
This residue is predicted to occupy a “d” position in the heptad and thus to serve as an indicator of both α helix formation and coiled coil formation. Spectra of assembly intermediates were taken from samples harvested during stepwise dialysis, and are normalized for the protein concentration in each sample. The concentration of urea at which the spectrum was taken is indicated in the figure.
Fig. 6 shows the results of experiments to probe the association of dimers from non-a/d positions in rods 1 and 2. Significant evidence for dimer-dimer interaction at residue 191 of rod domain 1 emerges first in the transition to 4 m urea, whereas evidence of dipolar broadening at position 348 is not clear until 2 m urea. Thus these results show that with urea-solubilized vimentin, dimer formation occurs earlier at the A11 alignment (as reported by the label in rod 1) than at the A22 alignment. A dimer-dimer interaction of residue 348 emerges in the transition to 2 m urea.
FIG. 6. EPR spectra of vimentin spin-labeled at residue 191 (upper) and residue 348 (lower) at a range of urea concentrations.
Samples removed from a stepwise dialysis were scanned by EPR, and the spectra were plotted to represent a normalized concentration of vimentin. The molar urea concentration against which each sample was equilibrated is indicated. Residue 348 of rod domain 2 is predicted to occupy an “e” position of the heptad and has been shown previously to be close to the point of overlap formed by adjacent vimentin dimers. The dipolar interaction at residue 191 of rod domain 1 (predicted to occupy a “c” position in the heptad) indicates that this residue is near a second point of overlap between adjacent vimentin dimers. Dipolar broadening at 191 occurs at higher urea concentrations, suggesting that the A11 association occurs more readily than the A22 association.
DISCUSSION
In a previous report we provided direct evidence of an α helical coiled coil formation in the rod domain 2 of human vimentin and that these regions were aligned in-parallel and in exact register (10). Identification of the pattern of spectra recorded as spin labels are moved to successive positions in the heptad allowed us to conclude that additional amino acids were in an α helical coiled coil conformation. Last, we demonstrated that spin labels on the exterior of the helix could be used to identify dimer-dimer interactions. We now provide similar data relevant to the structure of monomer-monomer interaction in rod 1B domain. Specifically we provided evidence that residues 169 to 193 of rod domain 1B are arranged as a coiled coil dimer and that the dimers are aligned in-parallel and in-register.
SDSL EPR is particularly powerful in the study of IFs because of the constraints on structure imposed by an elongated and rigid central rod domain. These constraints facilitate modeling and the prediction of sites of overlap between molecules that can be directly tested by SDSL EPR. This is especially relevant to IF proteins, because the number of possible options for interaction is restricted by the rigid structure of the rod domain.
By introducing spin labels along the external surface of the dimer in rod 1 we have identified a point of overlap between the rod 1 of one dimer and that of an adjacent dimer as occurring near residues 191-2. Using mixtures of different mutants we established that this staggered alignment is also anti-parallel. This arrangement is similar to the overlap we detected between adjacent rod 2 domains centered near residue 348 (10). Thus the point of overlap between one vimentin dimer and two of its neighbors has now been defined. These results are consistent with some of the proposed structures defined by cross-linking studies. Specifically, the data presented here are consistent with the alignments identified in desmin by Geisler et al. (22), as proposed by Refs. 16 and 20, and referred to as A11 (interactions between rod 1 domains) and A22 (interactions between rod 2 domains).
In this study, we have extended our previous uses of EPR to include a study of the order of assembly of vimentin molecules during filament assembly. Using samples dialyzed in different urea concentrations, we obtained spectra that reflect several stages in the assembly process: 1) monomer assumption of α helicity, 2) monomer-monomer assembly into a coiled coil dimer, and 3) two different dimer-dimer interactions (residues 191 and 348). The capacity to separate out these different interactions demonstrates the advantage of methods suitable for observing the dynamic rearrangement of proteins in solution.
How closely in vitro assembly from urea-containing buffers resembles IF assembly in vivo remains to be established. Other investigators have examined the structure of vimentin and keratins in denaturing (with urea or guanidine) and non-denaturing (low ionic strength, non-physiologic) media and have drawn parallels between in vitro assembly intermediates and soluble subunits isolated from cells (20, 21, 25). Implicit in all in vitro assembly studies is the assumption that the creation of native-looking filaments implies concordance between the in vitro assembly pathway and the in vivo pathway. This postulates that the assembly from urea is simply a mechanism for slowing the in vivo assembly process.
If the assembly process described in this report faithfully mimics the assembly of filaments in vivo, then it is reasonable to postulate that the assembly of filaments proceeds from a dimer, to a tetramer centered on the 191 residue, then to an octamer or higher order oligomer of two or more tetramers centered on the 348 residue. Left to be determined is the three-dimensional structure of the tetramers and octamers/multimers we have identified. The interactions revealed by EPR can be represented in planar form but in truth may be incorrectly represented in a two-dimensional figure.
SDSL EPR has been able to demonstrate the existence of A11 and A22 interactions between vimentin molecules in solution in both urea-containing buffers and in low ionic strength Tris buffers where filament assembly is incomplete. We have also collected spectra from assembled filaments, and verified that the A11 and A22 interactions are present in assembled filaments and exhibit nearly the same spectra, although evidence for a more rigid structure and or more protein-protein packing/interactions can be deduced from very slight EPR line shape changes between Tris samples and filament samples.
Although a complete description of IF assembly remains an open question, SDSL EPR brings a new method to the investigation of IF structure and assembly. Following the validation of these methods with the homopolymeric IF protein vimentin, it is possible to extend this approach to the study of other classes of IF proteins, such as the cytokeratins, to determine the degree to which the assembly process defined for vimentin is conserved among all IF gene family members. Our data also open a new avenue of exploration, the characterization of the impact of individual mutations on the process of IF assembly. It is now possible to study, for example, the epidermolysis bullosa simplex-type mutations that eliminate IF assembly in epidermal cytokeratins. Such mutations could be characterized for interference with the assembly of monomers into dimers, or dimers into tetramers and so on, providing more information about the mechanisms behind the failure to form filaments.
Footnotes
The abbreviations used are: IF, intermediate filament; EPR, electron paramagnetic resonance; SDSL, site-directed spin labeling.
REFERENCES
- 1.Hanukoglu I, Fuchs E. Cell. 1983;33:915–924. doi: 10.1016/0092-8674(83)90034-x. [DOI] [PubMed] [Google Scholar]
- 2.Steinert PM, Parry DA. Annu. Rev. Cell Biol. 1985;1:41–65. doi: 10.1146/annurev.cb.01.110185.000353. [DOI] [PubMed] [Google Scholar]
- 3.Albers K, Fuchs E. Int. Rev. Cytol. 1992;134:243–279. doi: 10.1016/s0074-7696(08)62030-6. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs E, Hanukoglu I. Cell. 1983;34:332–334. doi: 10.1016/0092-8674(83)90367-7. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs E, Weber K. Annu. Rev. Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 6.Herrmann H. In: Intermediate Filaments. Herrmann H, Robin J, editors. Vol. 31. New York: Plenum Press; 1998. pp. 319–355. [Google Scholar]
- 7.Strelkov SV, Herrmann H, Geisler N, Wedig T, Zimbelmann R, Aebi U, Burkhard P. EMBO J. 2002;21:1255–1266. doi: 10.1093/emboj/21.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrmann H, Strelkov SV, Feja B, Rogers KR, Brettel M, Lustig A, Haner M, Parry DA, Steinert PM, Burkhard P, Aebi U. J. Mol Biol. 2000;298:817–832. doi: 10.1006/jmbi.2000.3719. [DOI] [PubMed] [Google Scholar]
- 9.Strelkov SV, Herrmann H, Geisler N, Lustig A, Ivaninskii S, Zimbelmann R, Burkhard P, Aebi U. J. Mol. Biol. 2001;306:773–781. doi: 10.1006/jmbi.2001.4442. [DOI] [PubMed] [Google Scholar]
- 10.Hess JF, Voss JC, FitzGerald PG. J. Biol. Chem. 2002;277:35516–35522. doi: 10.1074/jbc.M206500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinert PM, Idler WW, Zimmerman SB. J. Mol. Biol. 1976;108:547–567. doi: 10.1016/s0022-2836(76)80136-2. [DOI] [PubMed] [Google Scholar]
- 12.Renner W, Franke WW, Schmid E, Geisler N, Weber K, Mandelkow E. J. Mol. Biol. 1981;149:285–306. doi: 10.1016/0022-2836(81)90303-x. [DOI] [PubMed] [Google Scholar]
- 13.Carter JM, Hutcheson AM, Quinlan RA. Exp. Eye Res. 1995;60:181–192. doi: 10.1016/s0014-4835(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 14.Steinert PM, Marekov LN, Fraser RD, Parry DA. J. Mol. Biol. 1993;230:436–452. doi: 10.1006/jmbi.1993.1161. [DOI] [PubMed] [Google Scholar]
- 15.Steinert PM, Marekov LN, Parry DA. J. Biol. Chem. 1993;268:24916–24925. [PubMed] [Google Scholar]
- 16.Quinlan RA, Hatzfeld M, Franke WW, Lustig A, Schulthess T, Engel J. J. Mol. Biol. 1986;192:337–349. doi: 10.1016/0022-2836(86)90369-4. [DOI] [PubMed] [Google Scholar]
- 17.Rogers KR, Herrmann H, Franke WW. J. Struct. Biol. 1996;117:55–69. doi: 10.1006/jsbi.1996.0069. [DOI] [PubMed] [Google Scholar]
- 18.Steinert PM, Marekov LN, Parry DA. Biochemistry. 1993;32:10046–10056. doi: 10.1021/bi00089a021. [DOI] [PubMed] [Google Scholar]
- 19.Mehrani T, Wu KC, Morasso MI, Bryan JT, Marekov LN, Parry DA, Steinert PM. J. Biol. Chem. 2001;276:2088–2097. doi: 10.1074/jbc.M007260200. [DOI] [PubMed] [Google Scholar]
- 20.Herrmann H, Haner M, Brettel M, Muller SA, Goldie KN, Fedtke B, Lustig A, Franke WW, Aebi U. J. Mol. Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann H, Aebi U. Curr. Opin. Struct. Biol. 1998;8:177–185. doi: 10.1016/s0959-440x(98)80035-3. [DOI] [PubMed] [Google Scholar]
- 22.Geisler N, Schunemann J, Weber K. Eur. J. Biochem. 1992;206:841–852. doi: 10.1111/j.1432-1033.1992.tb16992.x. [DOI] [PubMed] [Google Scholar]
- 23.Hubbell WL, Froncisz W, Hyde JS. Rev. Sci. Instrum. 1987;58:1879–1886. [Google Scholar]
- 24.Kokorin AI, Zamaraev KI, Grigorian GL, Ivanov VP, Rozantsev EG. Biofizika. 1972;17:34–41. [PubMed] [Google Scholar]
- 25.Soellner P, Quinlan RA, Franke WW. Proc. Natl. Acad. Sci. U. S. A. 1985;82:7929–7933. doi: 10.1073/pnas.82.23.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]