Abstract
Nkx2.5, a transcription factor implicated in human congenital heart disease, is required for regulation of second heart field (SHF) progenitors contributing to outflow tract (OFT). Here we define a set of genes (Lrrn1, Elovl2, Safb, Slc39a6, Khdrbs1, Hoxb4, Fez1, Ccdc117, Jarid2, Nrcam, and Enpp3) expressed in SHF containing pharyngeal arch tissue whose regulation is dependent on Nkx2.5. Further investigation implicates Jarid2, which has been implicated in OFT morphogenesis as a direct target of Nkx2.5 regulation. Jarid2 expression was upregulated in SHF mesoderm of Nkx2.5 deficient embryos. Chromatin immunoprecipitation analysis showed Nkx2.5 interaction with consensus binding sites in the Jarid2 promoter in pharyngeal arch cells. Finally, Jarid2 promoter activity and mRNA expression levels were downregulated by Nkx2.5 over-expression. Given the role of Jarid2 as a regulator of early cardiac proliferation, these findings highlight Jarid2 as one of several potential mediators of the critical role played by Nkx2.5 during OFT morphogenesis.
Keywords: congenital heart disease, secondary heart field, chromatin immunoprecipitation, knockout, microarray, double outlet right ventricle
Introduction
Nkx2.5, a vertebrate homologue of the Drosophila tinman gene, is expressed throughout the developing heart during embryogenesis and is a direct regulator of many genes determining the differentiated heart muscle phenotype (Jay and Izumo, 2002). Despite its broad expression, loss of function experiments in mice reveal particularly important roles for Nkx2.5 in SHF regulation and in conduction system development. For example, hearts of Nkx2.5 null embryos display a hypoplastic OFT and a single amorphous chamber of left ventricular character (Lints et al., 1993;Tanaka et al., 1999), indicating an almost complete loss of SHF specification or expansion (Prall et al., 2007). This severe SHF phenotype is partially rescued in mice expressing low levels of Nkx2.5 (approx. 25% of normal): the hearts of these mice exhibit more limited arterial pole defects of OFT rotation, alignment and septation, i.e. double oultlet right ventricle (DORV) and ventricular septal defects (VSD) (Prall et al., 2007). Finally, hearts of mice heterozygous for Nkx2.5 gene deletion, which express 50% of normal Nkx2.5 mRNA levels, develop normally with the exception of a variably penetrant central conduction system hypoplasia and atrial septal defects (ASD) (Biben et al., 2000; Jay et al., 2004; Prall et al., 2007).
Point mutations in the human NKX2-5 gene have been implicated in the occurance of familial, sporadic, syndromic and non-syndromic CHD that includes isolated conotruncal OFT anomalies such as Tetralogy of Fallot, a cardiac alignment defect leading to pulmonary artery stenosis with secondary RV hypertrophy, VSD and overriding aortic valve, as well as other anomalies such as secundum ASD with or without AV delay (Schott et al., 1998; Goldmuntz et al., 2001; McElhinney et al., 2003). Several of the better characterized mutations directly or indirectly affect the ability of NKX2-5 to bind DNA and hence may affect the ability of NKX2-5 to regulate target genes (Kasahara et al., 2000; Kasahara et al., 2001; McElhinney et al., 2003).
Despite the high prevalence of OFT anomalies in CHD and the strong linkage of Nkx2.5 to these defects our knowledge of direct transcriptional targets of Nkx2.5 in the SHF remains scant. Herein we define a set of genes regulated by Nkx2.5 in pharyngeal arch (PA) tissue encompassing the SHF by an analysis that combines new and existing transcriptomic data. Of these Nkx2.5-regulated genes, we establish the transcriptional regulator, Jarid2, as a direct target gene of Nkx2.5 in the SHF.
Results
Identification of Nkx2.5 regulated genes using microarray-based gene expression analysis
To identify Nkx2.5 target genes relevant specifically to the SHF we developed a data analysis schema that would identify genes satisfying three criteria: 1) genes must exhibit differential expression in RNA samples from the cardiothoracic region of E9.5 wild-type mice in comparison to mice heterozygous or homozygous for Nkx2.5-gene deletion; 2) gene transcripts must be elevated in the PA-containing region from wild-type E10.5 mice in comparison to hearts from wild-type E12.5 mice, and 3) the genes must show a response to graded changes in Nkx2.5 expression.
Employing this schema we performed a microarray analysis of PA tissue from wild-type E10.5 mouse embryos versus hearts from E12.5 mouse embryos, identifying 684 genes expressed in the PA but absent in the heart (data not shown). We then examined the comparative expression levels of the resulting 684 genes in the cardiothoracic tissues of E9.5 embryos wild-type, and heterozygous or homozygous for targeted deletion of the Nkx2.5 gene as reported in a publicly available database*. As shown in Fig. 1A, these first two analyses together identified 28 PA expressed genes that displayed a significant increase (7 of 28) or decrease (21 of 28) in expression in Nkx2.5 null versus heterozygote and wild-type embryos.
Figure 1. Identification of Nkx2.5 regulated genes in the SHF.
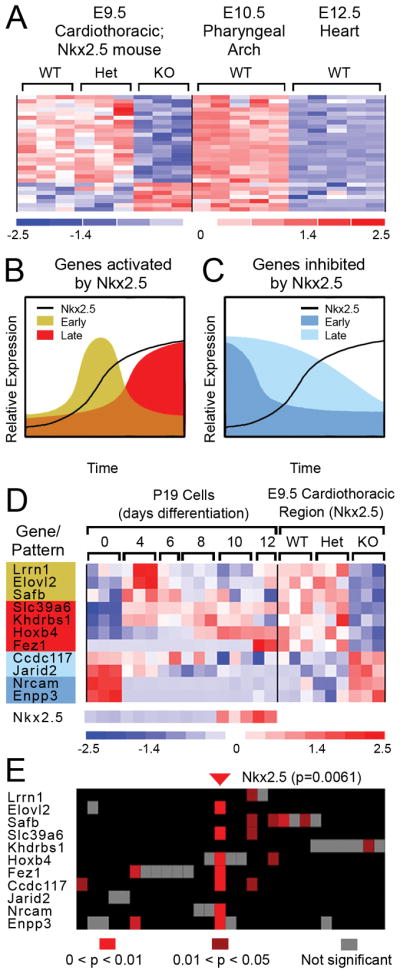
A, Shown is a heatmap of the expression intensities of 28 gene transcripts satisfying two criteria: 1) differential expression in E9.5 cardiothoracic region of wild-type, Nkx2.5-heterozygous and homozygous null mice; and, 2) presence in the PA-containing region from wild-type E10.5 embryos, but absent in hearts from E12.5 embryos. B and C, Schematic depiction of temporal expression profiles of Nkx2.5 targets responding rapidly (“Early”) or slowly (“Late”) to Nkx2.5 induction. D, Eleven of 28 genes shown in A exhibit corroborative expression patterns in differentiating P19CL6 (P19) cells. Genes are color-coded to match the potential patterns shown in B and C. Nkx2.5 expression in differentiating P19 cells is shown in the bottom of the panel; Nkx2.5 data is not available for the 9.5 cardiothoracic regions because of the microarray format used in that study (Affymetrix Mu11k Sub A and Sub B). E, Analysis of promoter sequences (−2000 to +500 relative to transcriptional start) for genes depicted in D detected 27 TF binding sequences (columns correspond to individual motifs). An Nkx2.5 binding element (Nkx2.5_01) was present one or more times in 7 of the 11 sequences analyzed and scored highly significant (p=0.0061).
To determine if these 28 genes additionally exhibited dynamic responses to graded changes in Nkx2.5 expression in a heterologous system, we conducted a microarray study of the cardiogenic embryonal carcinoma cell line P19CL6 (Habara-Ohkubo, 1996) during differentiation in vitro. Notably, Nkx2.5 is initially expressed at low levels in proliferating P19CL6 cells but is substantially induced during the late stages of a 12-day differentiation period when beating cardiac myocytes appear (Fig. 1D). Examination of the temporal expression profiles of the 28 genes during differentiation revealed that eleven exhibited patterns consistent with activation/de-repression (seven genes) or downregulation/repression (four genes) in response to increasing Nkx2.5 mRNA expression (Fig. 1B-D). Among the 7 candidates activated/de-repressed in differentiating P19CL6 cells, three exhibited an early, transient activation (i.e., Elovl2, Lrrn1, Safb) while the other four (i.e., Slc39a6, Khdrbs1, HoxB4, Fez1) showed late activation. Of the four genes whose expression decreased concomitant with the induction of Nkx2.5 expression, two showed relatively rapid downregulation (i.e., Nrcam, Enpp3) while the other two showed gradual downregulation (i.e., Ccdc117, Jarid2) during the P19CL6 differentiation period.
Examination of expression profiles for the 11 putative Nkx2.5 target genes in the heterologous system revealed that their responses to changes in Nkx2.5 expression were concordant with the responses observed in cardiothoracic tissue from E9.5 embryos (Fig. 1D). Specifically, all genes activated/de-repressed in vitro in cells expressing high levels of Nkx2.5 were downregulated by loss of Nkx2.5 expression in Nkx2.5 null E9.5 cardiothoracic tissue, while all genes repressed in cells with high Nkx2.5 expression levels were upregulated by loss of Nkx2.5 expression in cardiothoracic tissue of E9.5 Nkx2.5 nulls (Fig. 1D).
Promoter analyses highlight potential direct gene regulation by Nkx2.5
Unbiased in silico promoter analyses using the Transfac Public database of transcription factor (TF) binding sequences detected twenty-seven statistically over-represented TF binding sequences in the 5′ flanking regions of the 11 genes. Of these 27 sites, a binding sequence for Nkx2.5 was found at the highest frequency, constituting 10 of the 27 sites. Furthermore, Nkx2.5 binding sites were the most prevalent in terms of number of promoters containing a given TF site (7 of 11 promoters) and scored at the highest overall significance (p=0.0061) (Fig. 1E). Because the Tranfac Public database uses only two of the several sites reported for Nkx2.5, we manually screened the gene flanking sequences for other known Nkx2.5 binding sequences (Chen and Schwartz, 1995). This analysis revealed additional Nkx2.5 binding sites in the four remaining genes (i.e., Lrrn1, Safb, Khdrbs1 and Jarid2) (see below, and data not shown). Together, these findings support the possibility that these 11 genes represent direct downstream targets of Nkx2.5.
Jarid2 mRNA expression in SHF mesoderm, pharyngeal endoderm, OFT endocardium and cranial mesenchyme is augmented in Nkx2.5 nulls
Of the 11 putative Nkx2.5 targets identified, the nuclear protein Jarid2 has been shown to be critical to OFT morphogenesis (Lee et al., 2000; Takeuchi et al., 2006). We therefore performed a more in-depth evaluation of Jarid2 as a candidate Nkx2.5 target gene. In situ hybridization analysis (ISH) was performed to assess the expression of Jarid2 in cranial and caudal PA regions at a stage when the SHF is actively contributing to morphogenesis of the OFT, and to assess the effect of Nkx2.5 loss of function on Jarid2 expression in this population. ISH of embryos at the E9.5 (19-somite) stage showed modest Jarid2 mRNA expression in pharyngeal endoderm and mesoderm and extending into OFT myocardium and endocardium of both the OFT and right ventricle (Fig. 2A, C). In later stage embryos (29–30 somite/E10.5), Jarid2 ISH signal was more widespread throughout the heart in wild-type mouse embryos, consistent with previously published data regarding Jarid2 expression (Lee et al., 2000) (data not shown). No ISH signal was observed with control sense Jarid2 probes at any stage (data not shown). The PA-specific Jarid2 mRNA expression observed at E9.5 stages is consistent with lacZ expression patterns reported in mice bearing a gene trap insertional mutation of the Jarid2 locus (Takeuchi et al., 1995; Takeuchi et al., 1999). Collectively these data confirm that Jarid2 is expressed in the SHF in regions critical for the morphogenic contribution of OFT progenitors (Waldo et al., 2001; Goddeeris et al., 2007; Park et al., 2008). The apparent overlap of Jarid2 expression with previously published studies of Nkx2.5 expression in this region (Komuro and Izumo, 1993; Lints et al., 1993; Tanaka et al., 1999; Prall et al., 2007) is also consistent with a functional relationship between the two genes in the context of SHF regulation and OFT morphogenesis.
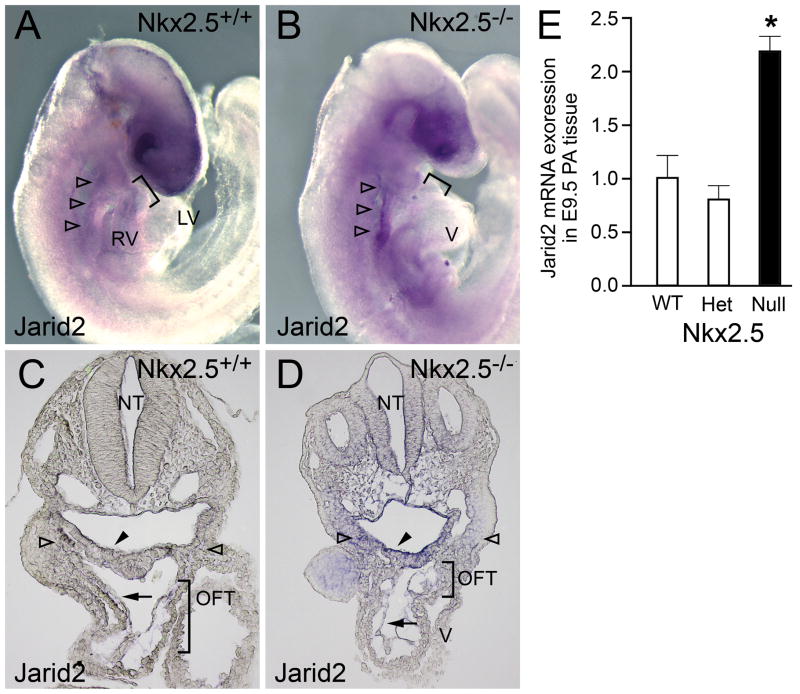
Figure 2. Jarid2 mRNA expression in wild-type and Nkx2.5 null embryos.
Wholemount and sectioned wholemount ISH for Jarid2 mRNA are shown for wild-type Nkx2.5 (A, C) and null Nkx2.5 (B, D) E9.5 embryos. Jarid2 mRNA is detected in SHF-related splanchnic mesoderm (open arrowheads) and pharyngeal endoderm (black arrowheads) as well as in the endocardium (black arrows) and myocardium of the OFT (bracket) in wild-type and Nkx2.5−/− embryos. Staining is more easily detectable in SHF-related splanchnic mesoderm and pharyngeal endoderm as well as OFT endocardium of null embryos. E, results of qRT-PCR for Jarid2 mRNA in combined heart and PA regions of E9.5 embryos wild-type (WT), heterozygous (Het) or mutant null (Null) for Nkx2.5. Results are expressed as fold change relative to WT mRNA levels, and were normalized with respect to qRT-PCR for β-actin mRNA in these samples. Error bars represent standard deviation as calculated according to the ΔΔC(t) method (Allen et al., 2009). *, significant by t-test (P <0.05). Abbreviations: OFT: outflow tract; LV: left ventricle; RV: right ventricle; V: common ventricle; NT: neural tube.
The expression pattern of Jarid2 in Nkx2.5 null embryos at the E9.5 (19-somite) stage was qualitatively similar to the pattern seen in wild-type embryos, occurring in pharyngeal endoderm and mesoderm and OFT myocardium and endocardium (Fig. 2B and D). However, Jarid2 expression was elevated in the null embryos in comparison to the wild-type embryos. The observed expression domain is highly similar, if not identical to the pattern of lacZ expression observed in E9.5 Nkx2.5 homozygous null embryos bearing one Nkx2.5-lacZ knock-in allele (Prall et al., 2007). It was not possible to analyze Jarid2 mRNA expression in Nkx2.5−/ − null embryos at later stages (i.e. after E10.5) due to the lethality resulting from Nkx2.5 deficiency.
Quantitative RT-PCR expression analysis confirmed that Jarid2 expression was increased >2-fold in PA and heart-containing tissue from E9.5 Nkx2.5 nulls as compared to wild-type embryos (Fig. 2E). These quantitative increases in levels of Jarid2 mRNA observed in SHF regions of Nkx2.5 null hearts support a model of repression of Jarid2 by Nkx2.5 in SHF-containing PA regions during OFT morphogenesis.
ChIP analysis shows Nkx2.5 interaction with predicted binding elements in the Jarid2 promoter and in intron 2 in PA cells
We next investigated whether the apparent repression of Jarid2 by Nkx2.5 was through a direct interaction of Nkx2.5 with Jarid2 control regions. A multi-species sequence alignment of the Jarid2 promoter region and nearby genomic flanking regions revealed two conserved Nkx2.5 NK homeodomain binding elements (NKEs) of the consensus TNNAGTG (Chen and Schwartz, 1995) present in the region 5′ to exon 1 (−203) and within the second intron (+1872) (Fig. 3A and Suppl. Fig. 1). We therefore employed chromatin immunoprecipitation (ChIP) analysis to determine whether Nkx2.5 binds to these NKE sites in vivo in cells from SHF. As shown in Fig. 3B, PCR performed on chromatin fragments derived from E9.5 PA tissue and precipitated using Nkx2.5 antibodies generated amplicons containing the Jarid2 −203 NKE as well as the +1872 NKE. As a control, PCR performed with material immunoprecipitated using non-immune rabbit IgG (Control IgG) showed no evidence of these amplicons. Additionally, no amplicons were generated in control reactions performed using primers specific for exon 1 sequences which lack potential Nkx2.5 binding sites using these chromatin samples (data not shown). Reactions performed with E9.5 embryonic heart-derived chromatin fragments precipitated using Nkx2.5 antibodies were also negative, indicating that Nkx2.5 associates with the Jarid2 5′ element (−203) and the intron 2 element (+1872) in PA cells, but not in heart cells at this stage of development. Findings from quantitative PCR analysis using these primers show a 4.5 fold enrichment of the −203 NKE in PA samples, with a more modest (~2-fold) enrichment of the +1872 NKE (Fig. 3C). Again, no apparent enrichment for these elements was detected in chromatin from hearts at this stage. These results suggest that Jarid2 upregulation in the setting of Nkx2.5 deficiency is due to loss of direct repression by Nkx2.5 acting on Jarid2 control regions specifically in SHF cells of the PA.
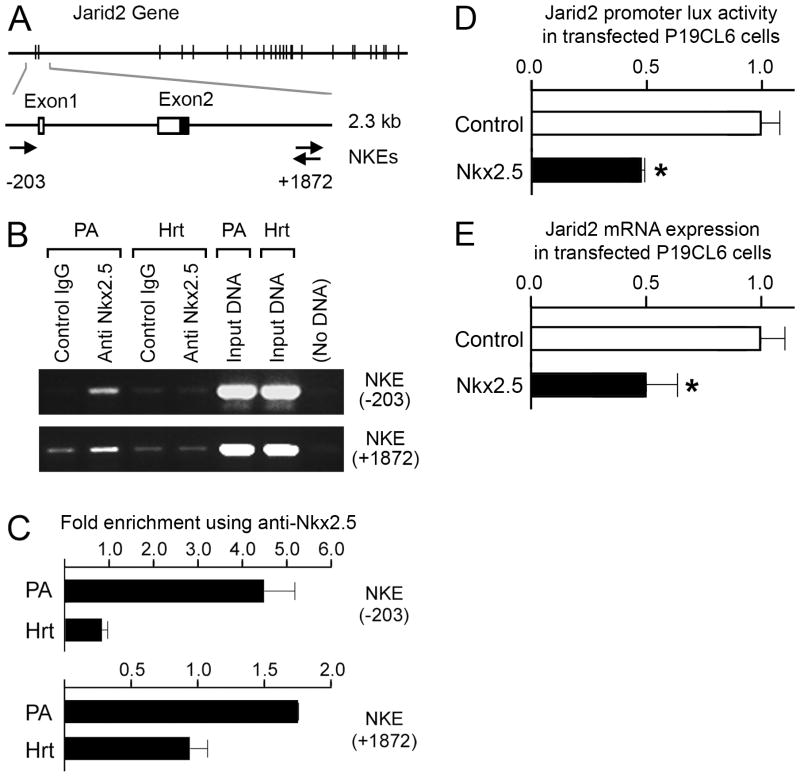
Figure 3. Nkx2.5 binds Jarid2 promoter region in vivo in E9.5 PA cells and represses Jarid2 promoter activity and mRNA expression.
A, Mouse Jarid2 has one Nkx2.5 binding element 5′ and proximal to the transcription start site and a second in intron 2. Shown is a schematic diagram of the Jarid2 mouse gene (top of panel) with the region encompassing exons 1 and 2 expanded (2.3 kb). Arrows indicate position and directionality of the Nkx2.5 binding elements (TNNAGTG) (NKE). Numeric positions of the two elements (−203 and +1872) are relative to the transcriptional start site annotated for Jarid2 mRNA (accession no. BC052444.1). B, Data are shown for ChIP assay for binding of Nkx2.5 to Jarid2 promoter proximal and intron 2 regions bearing conserved binding consensus sites for Nkx2.5 in SHF-containing PA regions (PA) and heart (Hrt) of wild-type E9.5 mouse embryos. PCR assay generates amplicons for both 5′ and intron 2 candidate regulatory regions containing NKEs shown in A using antibody against Nkx2.5 (anti-Nkx2.5) versus control total rabbit immunoglobulin (Control IgG) using PA-derived chromatin (PA) but not heart chromatin (Hrt). Controls shown include DNA from non-selected input chromatin (Input, PA and Hrt) and a no-template (No DNA) control for background amplification. C, qPCR quantitation of above ChIP experiments are shown in graphical fashion. Results are expressed as fold enrichment for genomic fragments containing Nkx2.5 binding consensus sites using anti-Nkx2.5 antibody over control IgG. Error bars represent minimum and maximum fold enrichment as calculated by the ΔC(t) method and based upon standard deviation of C(t) values at linear amplification (see Methods). D, Results of promoter-reporter assays comparing activity of control nt −997 to +964 Jarid2 promoter-luciferase (white bar above) to activity of nt −997 to +964 Jarid2 promoter-luciferase in the presence of Nkx2.5 overexpression (black bar below). Results are expressed as normalized fold change relative to WT luciferase levels. E, Results of qRT-PCR for Jarid mRNA in control eGFP-transfected growth-phase P19CL6 cells (white bar above) and eGFP and Nkx2.5 co-transfected growth-phase P19CL6 cells (black bar below) following FACS sorting for eGFP fluorescence. Results are expressed as normalized fold change relative to WT mRNA levels. *, significant by t-test (P <0.05).
Over-expression of Nkx2.5 suppresses Jarid2 promoter activity and represses Jarid2 mRNA expression
To examine the relationship between Nkx2.5 and Jarid2 mRNA expression more directly, we performed transfection assays using an Nkx2.5 over-expression construct and a Jarid2 reporter gene construct. The Jarid2 promoter encompassed nt −997 to +964 relative to the Jarid2 transcriptional initiation site, which notably includes the −203 NKE but not the +1872 NKE. As shown in Fig. 3D, when transfected into P19CL6 cells Nkx2.5 over-expression repressed the Jarid2 reporter activity by 50%. We next examined the consequence of Nkx2.5 over-expression on endogenous Jarid2 expression. Undifferentiated (growth phase) P19CL6 cells normally express high amounts of Jarid2 mRNA. However, over-expression of Nkx2.5 in these cells resulted in approximately a 50% decrease in endogenous Jarid2 mRNA expression (Fig. 3E). These findings in combination with our ChIP data showing direct association of Nkx2.5 to Jarid2 promoter regions further support the hypothesis that Jarid2 is directly and negatively regulated by Nkx2.5.
Discussion
Identification of Nkx2.5 target genes in the SHF
Here we define a set of genes expressed in SHF-containing pharyngeal arch tissue at E10.5 whose regulation is dependent on Nkx2.5 expression. These genes represent candidate mediators of the critical effects that Nkx2.5 has on OFT morphogenesis. The genes identified by our analysis fall into several broad categories reflecting diverse functions in metabolism and ion transport (Slc39a6, Elovl2, Enpp3)(Taylor and Nicholson, 2003; Jakobsson et al., 2006; Rucker et al., 2007), cell structure, growth or adhesion (Lrrn1, Safb, Fez1, Ccdc117, Nrcam) (Grumet et al., 1991; Ishii et al., 1999; Haines et al., 2005; Ivanova et al., 2005), and transcription or RNA metabolism (Khdrbs1, Hoxb4, Jarid2) (Lee et al., 2000; Brend et al., 2003; Richard et al., 2005). Several have been defined as having cardiac-specific function (Enpp3) or expression in early heart or pharyngeal arch precursors (Lrnn1, Safb, Khdrbs1, Hoxb4, Jarid2)(Brend et al., 2003; Haines et al., 2005; Ivanova et al., 2005; Richard et al., 2005). However detailed expression analysis and gain or loss of function studies have not been done for most of these genes, particularly with respect to the SHF.
Nkx2.5 negatively regulates Jarid2 in SHF Mesoderm
One of our identified candidate genes, Jarid2 stood out as having a known function in cardiac development. Jarid2 is a member of the AT-rich interactive domain (ARID)2 family of transcriptional regulators, and is known to be critical to OFT morphogenesis as evidenced by the fact that Jarid2 nulls display DORV and associated VSD. Our findings collectively indicate that Jarid2 is directly and negatively regulated by Nkx2.5: 1) Jarid2 mRNA expression was quantitatively increased in SHF regions of Nkx2.5 null hearts; 2) Nkx2.5 was found to bind NKEs in the Jarid2 promoter in chromatin from E9.5 pharyngeal arch cells; 3) Nkx2.5 over-expression repressed the transcriptional activity of Jarid2 promoter sequences containing an Nkx2.5 consensus binding site; and, 4) Nkx2.5 over-expression repressed endogenous Jarid2 expression in a heterologous cell culture system.
The potential relevance of Nkx2.5 regulation of Jarid2 to OFT development is underscored by their coincident expression in the SHF region at E9.5 within the pharyngeal mesoderm and pharyngeal endoderm proximal to the aortic sac. Intercommunication between these tissues and adjacent pharyngeal ectoderm is critical for the SHF and its morphogenic contribution of OFT progenitors (Waldo et al., 2001; Goddeeris et al., 2007; Park et al., 2008). Published Nkx2.5 in situ hybridization studies (Komuro and Izumo, 1993; Lints et al., 1993) confirm that Nkx2.5 is also expressed in pharyngeal mesoderm and endoderm at this stage. Furthermore, the expression pattern of lacZ in embryos (E9.5) homozygous for a Nkx2.5-lacZ knock-in (Prall et al., 2007) is highly similar if not identical to the Jarid2 expression pattern we observed in the Nkx2.5 nulls (E9.5). Thus, the overlap of Jarid2 mRNA expression with Nkx2.5 expression domains in wild-type and Nkx2.5 null embryos, respectively, are consistent with a close functional relationship between Nkx2.5 and Jarid2. These observations, in addition to our data demonstrating both the physical association of Nkx2.5 with Jarid2 promoter regions in SHF-containing PA cells and the ability of over-expressed Nkx2.5 to repress Jarid2 promoter activity and mRNA expression in cell assays reinforce our conclusion that Nkx2.5 is a direct negative regulator of Jarid2 in the SHF.
Jarid2 expression in the SHF: implications for OFT morphogenesis
The augmentation of Jarid2 expression observed in Nkx2.5 nulls highlights an apparent need to normally suppress Jarid2 in the cardiogenic tissues (i.e., SHF). Jarid2 nulls display DORV and associated VSD; however, the significance of Jarid2 over-expression in the SHF is not yet known. Jarid2, like its other jmjC domain-containing family members, exhibits histone modifying capabilities, and suppresses cardiomyocyte proliferation via its ability to repress cyclinD via a histone methyltransferase activity (Shirato et al., 2009). Secondary to this repression, Jarid2 nulls display increased cardiac myocyte proliferation in some genetic backgrounds (Takeuchi et al., 1999; Toyoda et al., 2003; Ohno et al., 2004). Given that reduced proliferation in the SHF is observed in both Nkx2.5 null mutants and Nkx2.5 hypomorphs (Prall et al., 2007), it is plausible that over-expression of Jarid2 in the SHF contributes to the decreased SHF precursor proliferation observed Nkx2.5 nulls. Previous studies of Jarid2 nulls did not assess proliferation rates of OFT cardiomyocytes or SHF progenitors at stages during OFT morphogenesis (i.e., E8.5–10.5), so it remains to be established whether or not Jarid2 over-expression leads to reduced proliferation in the SHF and the subsequent and RV/OFT hypoplasia and DORV.
As well as potentially regulating proliferation of SHF progenitor cells, modulation of Jarid2 expression levels may play a yet broader role in the SHF. Several recent studies have highlighted a major role for Jarid2 in mediating the recruitment of histone methylase-containing PRC2 repressor complexes to multiple loci in embryonic stem (ES) cells. Undifferentiated ES cells, like the P19CL6 EC cells used in our study, maintain high levels of Jarid2 expression that decrease upon the initiation of in vitro differentiation. In the absence of Jarid2, this systematic gene regulation is disrupted with delayed and defective development of the resultant embryoid bodies (Peng et al., 2009; Shen et al., 2009; Li et al., 2010; Pasini et al., 2010). It is likely that Jarid2-mediated function in SHF cells will involve a distinct set of target genes and activator/repressor complexes as compared to ES cells. Nevertheless, these studies raise the interesting possibility that Jarid2 expression systematically regulates SHF progenitor specification or their rate of differentiation, potentially through recruitment to target loci via cardiac specifying TFs like Nkx2.5.
Experimental Procedures
Embryo and cell harvest, RNA extraction and probe preparation
SHF and hearts were collected by dissection from wild-type mouse embryos at E10.5 and E12.5. P19CL6 cells were harvested after 4, 6, 8, 10 or 12 days in differentiation medium (Habara-Ohkubo, 1996; Monzen et al., 1999; Monzen et al., 2008)RNA was prepared from individual tissue isolates and from P19CL6 cells by Trizol extraction and purification with the RNeasy Mini kit (Qiagen, Valencia, CA). Following RNA sample assessment, for mouse tissue samples, total RNA (10 ng) of each sample (n=5) was converted into biotin-labeled cDNA suitable for hybridization to Affymetrix GeneChips (Affymetrix, Santa Clara, CA) using the WT-Ovation™ Pico amplification kit and the FL-Ovation™ cDNA Biotin V2 kit (NuGen, San Carlos, CA). For P19CL6 cells, 2 μg of each sample (n=3 for each time point) were converted into biotin-labeled cRNA using the SuperScript II cDNA synthesis kit (Invitrogen, Carlsbad, CA) and the Enzo HighYield in vitro transcription amplification kit (Enzo Life Sciences, Farmingdale, NY). Tissue samples were hybridized to Affymetrix Mouse Genome 430A GeneChips; P19CL6 samples were hybridized to Affymetrix Mouse 430 2.0 GeneChips. Hybridization, post hybridization washing, staining and fluorescence scanning were performed using Affymetrix instrumentation in accordance with manufacturer recommendations.
Microarray data processing
DNA microarray hybridization data (processed by MAS4 algorithm) for Nkx2.5 wild-type, heterozygous and homozygous cardiothoracic regions from E9.5 embryos was obtained directly from the CardioGenomics website*. DNA microarray hybridization data for P19CL6 cells was normalized using the ArrayQuest webtool (Argraves et al., 2005) to implement the Bioconductor (Gentleman et al., 2004) build of GCRMA(Wu et al., 2004). Affymetrix detection (presence and absence) scores and normalized hybridization values for E10.5 SHF and E12.5 heart samples were obtained using the Bioconductor MAS5 algorithm. Raw and processed expression data were deposited at NCBI Gene Expression Omnibus (accession GSE17936). Normalized hybridization data and detection call data were analyzed with dChip software (Li and Wong, 2001). Criteria for potential Nkx2.5 targets were: 1) differential expression between wild-type and Nkx2.5 null E9.5 SHF, defined by fold change >2, p<0.05 (Student’s t-test, unpaired); 2) majority ‘present’ detection scores for E10.5 SHF and majority ‘absent’ detection scores for wild-type E12.5 heart.
In silico promoter analysis
Candidate Nkx2.5 targets identified by microarray analysis were analyzed to find those having significant Nkx2.5 binding site in their promoters. Promoter sequences were obtained by automated retrieval from the Ensembl database using the Toucan web tool (Aerts et al., 2003; Aerts et al., 2005) configured to find genomic sequence encompassing the transcription start site (−2000 to +500). These sequences were reviewed manually to ensure that accurate segments of the gene were obtained. Canonical TF binding sites within the promoter sequences were identified with the PAINT v3.5 webtool (Vadigepalli et al., 2003) using the TransFac Public database of TF binding sites and the following settings for variable parameters: 1) minimize false positives; 2) core similarity threshold = 1.0; and 3) find binding sites on complementary strands. Over-representation of TF binding sites was evaluated using PAINT, which calculates a significance score (p-value) by hypergeometric distribution that reflects the likelihood that a TF site occurs by random chance, using as reference a list of TF binding site occurrences (approximately 30,000 total) found in the extant mouse promoter database based on comparable promoter length and identical settings for variable parameters; significance was assessed at p<0.05.
Quantitative RT-PCR
cDNA was prepared from total RNA from pools of 4–6 hearts and PA regions from E9.5 embryo wild-type, heterozygous and null for Nkx2.5 using the iScript cDNA synthesis kit and quantitative RT-PCR (qRT-PCR) for Jarid2 performed using the iQ SYBR green/iCycler amplification system (Bio-Rad, Hercules, CA) and Jarid2 primers: 5′-AGG AGA CTG GAA GAG GCA CA-3′ (nucleotide positions 1497–1516) and 5′-GCT TGT TTG CCC AGC ATA TT-3′ (nucleotide positions 1701–1720)(based on accession no. NM 021878). Control reactions were performed using β-actin primers 5′-CGG GAC CTG ACA GAC TAC CTC -3′ (nucleotide positions 2126–2152) and 5′-AAC CGC TCG TTG CCA ATA -3′ (nucleotide positions 2352–2343) (based on accession no. NC000071.5)(Integrated DNA Technologies, Coralville, IA). Relative quantification and standard deviations were calculated using the ΔΔC(t) method as previously described (Allen et al., 2009). qPCR assays were performed in triplicate and results presented are representative of three independent assays.
Candidate gene validation by in situ hybridization
In situ hybridization (ISH) of wild-type and Nkx2.5 null E9.5 mouse embryos was performed using antisense and sense (control) digoxigenin-labeled riboprobes according to established protocols (Hogan, 1994). Probes were reverse transcribed from a linearized form of a full-length Jarid2 cDNA clone in pCMV-SPORT6 (Thermo Scientific/Open Biosystems, Huntsville, AL) using the DIG RNA Labeling Kit Sp6/T7 (Roche, Indianapolis, IN). Hybridization was performed at 63°C. For section analysis, whole-mount stained embryos were Paraplast embedded, sectioned and mounted in Permount (Sigma, St. Louis MO) prior to digital photography.
Tissue harvest and chromatin immunoprecipitation(ChIP)
PA-containing regions and hearts were dissected from 4 wild-type E9.5 embryos (FVB strain). ChIP was performed using reagents from the Upstate (Millipore) ChIP Assay Kit (Millipore, Billerica MA) according to manufacturer’s protocols. Samples were sheared with a Misonix 3000 sonicator using 10 x 10 sec pulses on power setting 10 with 10 second cooling intervals. Chromatin shearing was assessed by agarose gel electrophoresis of de-crosslinked DNA input samples and chromatin yield was quantified by protein assay determination using the BCA protein assay kit (Pierce/Thermo Scientific, Rockford, IL). ChIP was performed using the H-114 anti-Nkx2.5 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or control normal rabbit IgG (Dako, Denmark) in ChIP dilution buffer at a protein concentration of 1μg/μl and an antibody concentration of 4μg/ml overnight at 4oC. Purified genomic fragments were subject to 37–39 cycles of PCR amplification with Taq polymerase (Qiagen) using the following primers and annealing temperatures: Jarid2 promoter: 5′-AAA AGG GAG TTG AGT GAC AGG A-3′ and 5′-CCC TTG ATC TTC TGG AAG TTG T-3′ amplifying nt (−)212–(−)124 relative to transcription start site (based on accession no. BC052444); 59oC; Jarid2 intron 2: 5′-TGG TTT CTA GTT TGA GGG GAA A-3′and 5′-ACC TAA CCA TCA CAA CCC AAT C-3′ amplifying nt (+)1602–(+)1729 relative to the transcription start site; 59oC; Jarid2 exon 2: 5′-CCC GTG GTC AGA AGA GAG AG-3′ and 5′-GGC ACA GAA AGA CTC CAT CC-3′ amplifying nt (+)92,495–(+)92,652 relative to the transcription start site; 61oC. qPCR of ChIP-recovered amplicons was accomplished using the above primers and the iQ SYBR green/iCycler amplification system. Fold enrichment was calculated by a ΔC(t) method by comparison of cycle number of linear amplification for anti-Nkx2.5 immunoprecipitated genomic DNA vs. genomic DNA immunoprecipitated by control Ig. Triplicate assays were performed and results expressed as mean fold difference calculated as 2−ΔC(t) and maximum and minimum fold differences calculated as 2− (ΔC(t)+SD) and 2− (ΔC(t)-SD) respectively; SD=(S12 + S22)1/2 where S1 is the standard deviation of the C(t) values obtained for amplification of genomic fragments from ChIP material recovered with anti-Nkx2.5 antibody and S2 is the standard deviation of the C(t) values obtained for amplification of genomic fragments from ChIP material recovered with control IgG. Results shown are representative of two independent ChIP experiments.
Jarid2 promoter assay
A 1955bp 5′ flanking region of the Jarid2 locus representing nt −997 to +964 relative to the transcriptional start site (based on accession no. BC052444) was cloned by PCR from BAC clone RP24–125d22 (CHORI, Oakland, CA) using LongAmp Taq polymerase (NEB, Beverly, MA) and oligonucleotides 5′-CAT CTC GAG TCT CGG TCG CGG ACA -3′ (forward) and 5′-ATG CCA TGG TGA GAT CCA AAT GCT GAT TG -3′ (reverse). The resulting promoter fragment was digested with XhoI and NcoI and ligated into the pGL3-Control luciferase vector (Promega, Madison, WI) digested with XhoI and NcoI, therefore replacing the SV40 promoter region with the Jarid2 promoter fragment. Promoter assays were performed in P19CL6 cells as described (Lee et al., 2004) using 25 ng of Jarid2 promoter reporter, 5ng of control TK-Renilla luciferase reporter (Promega) and 25 ng of either control pCS2 expression vector or pCS2-Nkx2.5 (a kind gift from S. Izumo) using 1μl FuGene6 (Roche) per 24 well plate well. Following 24 hours of incubation, samples were lysed and assayed for normalized luciferase activity using a dual luciferase assay system (Promega). Results show mean and standard deviation of duplicate well assays normalized to Renilla luciferase activity and are representative of 3 independent assays. P-values were calculated using Student’s unpaired t-test.
In vivo Nkx2.5 over-expression assays
P19CL6 cells were cultured in non-differentiating conditions as previously described (Habara-Ohkubo, 1996) in αMEM with high glutamine (GibcoBRL-Invitrogen) with 10% ES cell-grade FBS (GibcoBRL-Invitrogen) and 1x Pen-strep. Following seeding overnight in 60mm dishes at a density of approx. 1 x 105 cells/ml, cells were transfected with 1ug pCS2 Nkx2.5 and 1ug pGreenLantern eGFP expression plasmid (Gibco-BRL) using FuGene (Roche) according to manufacturer’s protocol. Cells were harvested by trypsinization following 24 hrs incubation and subject to FACS sorting on a BD FACS-Aria2 flow cytometry cell sorter (BD Biosciences, San Jose, CA) optimized for eGFP fluorescence wavelengths. RNA was collected from control and Nkx2.5 co-expressing eGFP (+) cell samples using Trizol extraction (Invitrogen), followed by isopropanol precipitation, resuspension, then DNAse digestion and re-purification using RNeasy mini columns (Qiagen). cDNA was synthesized from purified RNA samples and subject to qPCR for Jarid2 and control β-actin as described above. Results shown are representative of three independent experiments; standard deviation and significance score calculation are as above; P-values were calculated using Student’s unpaired t-test.
Supplementary Material
Sequence alignments are shown for Nkx2.5 binding consensus sites conserved in mouse, rat, human, orangutan, dog and horse Jarid2 genes at −203 and +1872 relative to the mouse transcriptional start site. Sequences are shown below a schematic rendering of a 2.3 kb region of the Jarid2 locus spanning the promoter proximal 5′ flanking region and exons 1 and 2. Black portion of exon2 indicates translated region. Arrows over sequence blocks highlight position and direction of the conserved consensus sites (TNNAGTG). Conserved sequences at +1872 contain two overlapping consensus Nkx2.5 binding sites in opposing orientations. Dashed lines in orangutan and dog sequences at +1872 indicate gaps in sequence alignment of their cognate Jarid2 sequences at this position.
Acknowledgments
This work was supported by NIH NCRR grant P20 RR016434 (KLH, JLB and WSA) and by HL095067 (WSA). We thank Dr. Marion A. Cooley for advice with in situ hybridization methodology, Dr. Donald R. Menick for advice and reagents related to anti-Nkx2.5 ChiP methodology, Saurin Jani for bioinformatics assistance, Dr. Richard Peppler for FACS sorting assistance and Marie M. Lockhart for technical assistance.
Footnotes
http://www.cardiogenomics.org; Genomics of Cardiovascular Development, Adaptation, and Remodeling. NHLBI Program for Genomic Applications, Harvard Medical School.
References
- Aerts S, Thijs G, Coessens B, Staes M, Moreau Y, De Moor B. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts S, Van Loo P, Thijs G, Mayer H, de Martin R, Moreau Y, De Moor B. TOUCAN 2: the all-inclusive open source workbench for regulatory sequence analysis. Nucleic Acids Res. 2005;33:W393–396. doi: 10.1093/nar/gki354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen DL, Bandstra ER, Harrison BC, Thorng S, Stodieck LS, Kostenuik PJ, Morony S, Lacey DL, Hammond TG, Leinwand LL, Argraves WS, Bateman TA, Barth JL. Effects of spaceflight on murine skeletal muscle gene expression. J Appl Phys. 2009;106:582–595. doi: 10.1152/japplphysiol.90780.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argraves GL, Jani S, Barth JL, Argraves WS. ArrayQuest: a web resource for the analysis of DNA microarray data. BMC Bioinformatics. 2005;6:287. doi: 10.1186/1471-2105-6-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, Barnett L, Koentgen F, Robb L, Feneley M, Harvey RP. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2–5. Circ Res. 2000;87:888–895. doi: 10.1161/01.res.87.10.888. [DOI] [PubMed] [Google Scholar]
- Brend T, Gilthorpe J, Summerbell D, Rigby PW. Multiple levels of transcriptional and post-transcriptional regulation are required to define the domain of Hoxb4 expression. Development. 2003;130:2717–2728. doi: 10.1242/dev.00471. [DOI] [PubMed] [Google Scholar]
- Chen CY, Schwartz RJ. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddeeris MM, Schwartz R, Klingensmith J, Meyers EN. Independent requirements for Hedgehog signaling by both the anterior heart field and neural crest cells for outflow tract development. Development. 2007;134:1593–1604. doi: 10.1242/dev.02824. [DOI] [PubMed] [Google Scholar]
- Goldmuntz E, Geiger E, Benson DW. NKX2.5 mutations in patients with tetralogy of fallot. Circulation. 2001;104:2565–2568. doi: 10.1161/hc4601.098427. [DOI] [PubMed] [Google Scholar]
- Grumet M, Mauro V, Burgoon MP, Edelman GM, Cunningham BA. Structure of a new nervous system glycoprotein, Nr-CAM, and its relationship to subgroups of neural cell adhesion molecules. J Cell Biol. 1991;113:1399–1412. doi: 10.1083/jcb.113.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habara-Ohkubo A. Differentiation of beating cardiac muscle cells from a derivative of P19 embryonal carcinoma cells. Cell Struct Funct. 1996;21:101–110. doi: 10.1247/csf.21.101. [DOI] [PubMed] [Google Scholar]
- Haines BP, Gupta R, Jones CM, Summerbell D, Rigby PW. The NLRR gene family and mouse development: Modified differential display PCR identifies NLRR-1 as a gene expressed in early somitic myoblasts. Dev Biol. 2005;281:145–159. doi: 10.1016/j.ydbio.2005.01.030. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Press; New York: 1994. [Google Scholar]
- Ishii H, Baffa R, Numata SI, Murakumo Y, Rattan S, Inoue H, Mori M, Fidanza V, Alder H, Croce CM. The FEZ1 gene at chromosome 8p22 encodes a leucine-zipper protein, and its expression is altered in multiple human tumors. Proc Natl Acad Sci USA. 1999;96:3928–3933. doi: 10.1073/pnas.96.7.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova M, Dobrzycka KM, Jiang S, Michaelis K, Meyer R, Kang K, Adkins B, Barski OA, Zubairy S, Divisova J, Lee AV, Oesterreich S. Scaffold attachment factor B1 functions in development, growth, and reproduction. Mol Cell Biol. 2005;25:2995–3006. doi: 10.1128/MCB.25.8.2995-3006.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, Kupershmidt S, Roden DM, Schultheiss TM, O’Brien TX, Gourdie RG, Berul CI, Izumo S. Nkx2–5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113:1130–1137. doi: 10.1172/JCI19846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay PY, Izumo S. Elucidating the molecular and genetic interactions responsible for congenital heart disease. Pediatr Res. 2002;51:127. doi: 10.1203/00006450-200202000-00001. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Lee B, Schott JJ, Benson DW, Seidman JG, Seidman CE, Izumo S. Loss of function and inhibitory effects of human CSX/NKX2.5 homeoprotein mutations associated with congenital heart disease. J Clin Invest. 2000;106:299–308. doi: 10.1172/JCI9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H, Usheva A, Ueyama T, Aoki H, Horikoshi N, Izumo S. Characterization of homo- and heterodimerization of cardiac Csx/Nkx2.5 homeoprotein. J Biol Chem. 2001;276:4570–4580. doi: 10.1074/jbc.M004995200. [DOI] [PubMed] [Google Scholar]
- Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Evans S, Ruan TY, Lassar AB. SMAD-mediated modulation of YY1 activity regulates the BMP response and cardiac-specific expression of a GATA4/5/6-dependent chick Nkx2.5 enhancer. Development. 2004;131:4709–4723. doi: 10.1242/dev.01344. [DOI] [PubMed] [Google Scholar]
- Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE. Jumonji, a nuclear protein that is necessary for normal heart development. Circ Res. 2000a;86:932–938. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993b;119:969. doi: 10.1242/dev.119.3.969. [DOI] [PubMed] [Google Scholar]
- McElhinney DB, Geiger E, Blinder J, Benson DW, Goldmuntz E. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol. 2003;42:1650–1655. doi: 10.1016/j.jacc.2003.05.004. [DOI] [PubMed] [Google Scholar]
- Monzen K, Ito Y, Naito AT, Kasai H, Hiroi Y, Hayashi D, Shiojima I, Yamazaki T, Miyazono K, Asashima M, Nagai R, Komuro I. A crucial role of a high mobility group protein HMGA2 in cardiogenesis. Nature Cell Biol. 2008;10:567–574. doi: 10.1038/ncb1719. [DOI] [PubMed] [Google Scholar]
- Monzen K, Shiojima I, Hiroi Y, Kudoh S, Oka T, Takimoto E, Hayashi D, Hosoda T, Habara-Ohkubo A, Nakaoka T, Fujita T, Yazaki Y, Komuro I. Bone morphogenetic proteins induce cardiomyocyte differentiation through the mitogen-activated protein kinase kinase kinase TAK1 and cardiac transcription factors Csx/Nkx-2.5 and GATA-4. Mol Cell Biol. 1999;19:7096–7105. doi: 10.1128/mcb.19.10.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno T, Nakajima K, Kojima M, Toyoda M, Takeuchi T. Modifiers of the jumonji mutation downregulate cyclin D1 expression and cardiac cell proliferation. Biochem Biophys Res Comm. 2004;317:925–929. doi: 10.1016/j.bbrc.2004.03.131. [DOI] [PubMed] [Google Scholar]
- Park EJ, Watanabe Y, Smyth G, Miyagawa-Tomita S, Meyers E, Klingensmith J, Camenisch T, Buckingham M, Moon AM. An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development (Cambridge, England) 2008;135:3599–3610. doi: 10.1242/dev.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, Stennard FA, Wise N, Schaft D, Wolstein O, Furtado MB, Shiratori H, Chien KR, Hamada H, Black BL, Saga Y, Robertson EJ, Buckingham ME, Harvey RP. An Nkx2–5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128:947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S, Torabi N, Franco GV, Tremblay GA, Chen T, Vogel G, Morel M, Cleroux P, Forget-Richard A, Komarova S, Tremblay ML, Li W, Li A, Gao YJ, Henderson JE. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet. 2005;1:e74. doi: 10.1371/journal.pgen.0010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucker B, Almeida ME, Libermann TA, Zerbini LF, Wink MR, Sarkis JJ. Biochemical characterization of ecto-nucleotide pyrophosphatase/phosphodiesterase (E-NPP, E.C. 3.1.4.1) from rat heart left ventricle. Mol Cell Biochem. 2007;306:247–254. doi: 10.1007/s11010-007-9576-5. [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. Congenital heart disease caused by mutations in the transcription factor NKX2–5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, Shinkai Y, Takeuchi T. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3–K9. J Biol Chem. 2009;284:733–739. doi: 10.1074/jbc.M804994200. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Kojima M, Nakajima K, Kondo S. jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mechanisms Of Development. 1999;86:29–38. doi: 10.1016/s0925-4773(99)00100-8. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn. 2006;235:2449–2459. doi: 10.1002/dvdy.20851. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Taylor KM, Nicholson RI. The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim Biophys Acta. 2003;1611:16–30. doi: 10.1016/s0005-2736(03)00048-8. [DOI] [PubMed] [Google Scholar]
- Toyoda M, Shirato H, Nakajima K, Kojima M, Takahashi M, Kubota M, Suzuki-Migishima R, Motegi Y, Yokoyama M, Takeuchi T. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev Cell. 2003;5:85–97. doi: 10.1016/s1534-5807(03)00189-8. [DOI] [PubMed] [Google Scholar]
- Vadigepalli R, Chakravarthula P, Zak DE, Schwaber JS, Gonye GE. PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. Omics. 2003;7:235–252. doi: 10.1089/153623103322452378. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry R, Gentleman R, Murillo F, Spencer F. A Model Based Background Adjustment for Oligonucleotide Expression Arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignments are shown for Nkx2.5 binding consensus sites conserved in mouse, rat, human, orangutan, dog and horse Jarid2 genes at −203 and +1872 relative to the mouse transcriptional start site. Sequences are shown below a schematic rendering of a 2.3 kb region of the Jarid2 locus spanning the promoter proximal 5′ flanking region and exons 1 and 2. Black portion of exon2 indicates translated region. Arrows over sequence blocks highlight position and direction of the conserved consensus sites (TNNAGTG). Conserved sequences at +1872 contain two overlapping consensus Nkx2.5 binding sites in opposing orientations. Dashed lines in orangutan and dog sequences at +1872 indicate gaps in sequence alignment of their cognate Jarid2 sequences at this position.