Abstract
Staphylococcus epidermidis is a highly significant nosocomial pathogen mediating infections primarily associated with indwelling biomaterials (e.g., catheters and prostheses). In contrast to Staphylococcus aureus, virulence properties associated with S. epidermidis are few and biofilm formation is the defining virulence factor associated with disease, as demonstrated by animal models of biomaterial-related infections. However, other virulence factors, such as phenol-soluble modulins and poly-γ-DL-glutamic acid, have been recently recognized that thwart innate immune system mechanisms. Formation of S. epidermidis biofilm is typically considered a four-step process consisting of adherence, accumulation, maturation and dispersal. This article will discuss recent advances in the study of these four steps, including accumulation, which can be either polysaccharide or protein mediated. It is hypothesized that studies focused on understanding the biological function of each step in staphylococcal biofilm formation will yield new treatment modalities to treat these recalcitrant infections.
Keywords: arginine catabolism, biofilm, biofilm maturation, biomaterial-related infections, phenotypic variation, Staphylococcus epidermidis
Staphylococcus epidermidis is a commensal bacterium that colonizes the skin and mucous membranes of mammals and is the most prevalent staphylococcal species found in humans. Epidemiological studies have demonstrated that healthy people carry between 10 and 24 different strains of S. epidermidis at any one time [1]. It has been speculated that one human benefit of S. epidermidis colonization is inhibition of attachment of more virulent bacteria such as Staphylococcus aureus; however, as with the entire human microbiota, we are just beginning to understand these complex interactions [2,3]. Unfortunately, concomitant with advances in medical practice, S. epidermidis has become the most common cause of primary bacteremia and infection of indwelling medical devices, particularly in immunocompromised individuals and neonates. Although sterile site S. epidermidis infections are known to occur (i.e., native valve endocarditis), most infections are associated with a foreign body (i.e., catheter or other biomaterial) [4]. In contrast to S. aureus, which is much more virulent and synthesizes an array of toxins and other virulence factors, the main defined virulence factor associated with S. epidermidis is its ability to form biofilm and colonize biomaterials. Biofilm is defined as a complex interaction of unicellular organisms, typically encased in an extracellular matrix of polysaccharide, protein and nucleic acid. S. epidermidis biofilm is recalcitrant to the deleterious action of antibiotics and impedes the host immune response. Thus, treatment of patients with S. epidermidis biofilm-mediated infections typically involves removal of the offending device and subsequent replacement, causing an increase in morbidity and cost. Fortunately, advances in genetic techniques within the past 10 years have allowed investigators to probe mechanisms of virulence within S. epidermidis, particularly those factors that mediate biofilm formation. This article will focus on those defined factors that allow S. epidermidis to colonize and persist in biomaterial-related infections through the formation of biofilm.
Genome structure & population biology of S. epidermidis
The genomes of two S. epidermidis isolates, ATCC12228 (~2.5 Mb) and RP62A (also known as ATCC35984; ~2.6 Mb), have been fully sequenced [5,6]. Although the core genome is very similar between S. epidermidis and S. aureus, S. epidermidis, as predicted, encodes fewer known and putative virulence factors and pathogenicity islands compared with S. aureus. This paucity of virulence factors is most likely responsible for the lack of invasiveness of S. epidermidis infection. The most notable differences between S. aureus and S. epidermidis include the lack of staphylococcal enterotoxins, leukocidins, α-toxin, protein A and a multitude of adherence factors in the latter. However, S. epidermidis does encode at least one unique virulence factor, a poly-γ-DL-glutamic acid (PGA) capsule that is essential for virulence in Bacillus anthracis [7]. In addition, the genome sequence reflects the ecological niche of S. epidermidis as eight sodium ion/proton exchangers and six osmoprotection transports systems have been identified [5,8]. These systems are predicted to aid in the survival of S. epidermidis on the skin surface as they counteract the osmotic pressure and high salt concentrations. The population structure of S. epidermidis is epidemic in structure and at least nine clonal lineages are disseminated worldwide [9]. One major clonal complex, CC2, represented 74% of isolates worldwide in one study; furthermore, one particular sequence type, ST2, represented 31% of all isolates. Similar results were found in other multilocus sequence typing (MLST) studies [9–12]. However, rapid evolution (and thus, pulsed-field gel electrophoresis [PFGE] patterns) occurs through frequent transfer of mobile genetic elements and recombination; possibly through insertion sequence elements [9]. In fact, it has been widely suggested that S. epidermidis is a reservoir of antibiotic resistance genes and other genomic islands that S. aureus acquires through horizontal transfer [13,14]. Other molecular typing methodologies, including sequence analysis of repeat regions of sdrG/aap genes and multiple-locus variable-number tandem repeat analysis, have been developed which yield similar discriminatory power as MLST or PFGE [15,16]. However, recent molecular epidemiology studies have demonstrated that the combination of PFGE and staphylococcal cassette chromosome mec (SCCmec) typing has the ability to predict clonal complexes as defined by MLST [17].
S. epidermidis biofilm formation
With regard to virulence, S. epidermidis can be viewed as intermediary between apathogenic species, such as Staphylococcus carnosus [18], and the highly virulent S. aureus. Biologically, S. epidermidis is an example of an opportunistic bacterium where it can be considered a symbiont or a pathogen depending on the biological context [2,19]. Several hypotheses have addressed why S. aureus has evolved to be more virulent than S. epidermidis, including the enhanced ability of S. aureus to acquire foreign DNA and enriched immune response in the nares (S. aureus) in contrast to the skin (S. epidermidis) [20]. Currently, however, the most attractive hypothesis suggests that the enhanced virulence in S. aureus is due to the complex transmission pathway when comparing S. aureus and S. epidermidis [20]. In contrast to S. aureus, whose major niche is the nares, S. epidermidis can readily be transferred to the skin of other individuals through common contact or the constant sloughing of skin. Massey et al. proposed a mathematical model that predicts those S. epidermidis isolates that have enhanced virulence will be lost in the population [20]. Therefore, predictably, examination of the proposed virulence factors reveals that S. epidermidis has evolved multiple systems to protect itself against factors of the innate immune system, including antimicrobial peptides and phagocytosis, instead of those factors that assist in mediating invasive infections. Otto has recently published several excellent reviews focusing on the biology of S. epidermidis, phenol-soluble modulins (PSMs), the recently described three-component antimicrobial peptide-sensing system, and other factors that help mediate resistance to the innate immune system [8,21–24]. Therefore, these systems will not be described here. Instead, this article will focus on the literature describing biofilm initiation, accumulation, maturation and dispersal in addition to a brief discussion of PGA. Table 1 lists the identified putative virulence factors of both S. epidermidis RP62A and ATCC12228. Note that ATCC12228 does not encode either icaADBC or bhp (see below), and therefore does not form a detectable biofilm. It should also be noted that only a few of these virulence traits have been rigorously tested in relevant animal models of biomaterial-related infection.
Table 1.
Virulence factors of Staphylococcus epidermidis.
| Virulence factor | RP62A† | ATCC12228‡ | Ref. |
|---|---|---|---|
|
Proteases/exoenzymes/extracellular proteins | |||
| Esterase | 1941 | 1929 | [5] |
| 2109 | 2095 | ||
| Serine protease | 2390 (htrA) | 0722 (htrA) | [124] |
| 2401 | 0723 | ||
| Serine V8 protease | 1387 (sspA) | 1543 (sspA) | [125,126] |
| Cysteine protease | 2390 (sspB) | 0184 (sspB) | [125] |
| 2391 (sspC) | 0183 (sspC) | ||
| Lipase | 2336 (lip) | 0245 (lip) | [5,127–129] |
| 0018 (geh) | 2403 (geh) | ||
| 2297 (geh1, gehC) | 0281 (geh1, gehC) | ||
| 2388 (geh2, gehD) | 0185 (geh2, gehD) | ||
| 0309 (lipA) | 0424 (lipA) | ||
| Elastase | 2252 (sepA) | 2219 (sepA) | [130] |
| Thermonuclease | 0891 (nuc) | 1004 (nuc) | [5] |
| Nuclease | 1570 | NP | [5] |
| Zinc metalloprotease | 0829 | 0938 | [5] |
| Clp protease | 0436 (clpP) | 0551 (clpP) | [75] |
| 0564 (clpB) | 0674 (clpB) | ||
| 1238 (clpX) | 1349 (clpX) | ||
| 0165 (clpC) | 0287 (clpC) | ||
| Fatty acid modifying enzyme | Undefined | Undefined | [131] |
| Lantibiotics (epidermin and Pep5) | NP | NP | [132,133] |
|
Toxins/hemolysins | |||
| Phenol-soluble modulins | 0736 (β1) | 0486 (β1) | [92,134,135] |
| 0737 (β1) | 0487 (β1) | ||
| 0738 (β1) | 0489 (β1) | ||
| 0739 (β2) | 0490 (β2) | ||
| 2397 (β1) | 0177 (β) | ||
| 2400 (β1) | 0174 (β1) | ||
| 0083 (α) | 1634 (hld) | ||
| 1489 (hld) | |||
| β-hemolysin | 2544 (hlb) | 0008 (hlb) | [136] |
| Hemolysin III | 1769 | 1760 | [5] |
| Hemolysin | 2258 | 2226 | [5] |
|
Iron acquisition | |||
| Staphyloferrins | 1781 | 1772 | [137] |
| SitA, B, C iron transporter | 0292 (sitA) | 0407 (sitA) | [138] |
| 0291 (sitB) | 0406 (sitB) | ||
| 0290 (sitC) | 0405 (sitC) | ||
|
Surface proteins/adherence/MSCRAMMs | |||
| Staphylococcus epidermidis surface protein A | 1316 (sesA) | 1429 (sesA, fmtB) | [32] |
| S. epidermidis surface protein E | 0719 (sesE) | 0828 (sesE, vsaC) | [32] |
| S. epidermidis surface protein G | 1482 (sesG) | NP | [32] |
| S. epidermidis surface protein H | 1483 (sesH) | 1628 (sesH) | [32] |
| S. epidermidis surface protein I | 1654 (sesI) | NP | [32] |
| S. epidermidis surface protein C | 2264 (sesC) | 2232 (sesC) | [32] |
| Serine-aspartate repeat-containing protein F | 0026 (sdrF)§ | 2395 (sdrF)§ | [45,46] |
| Serine-aspartate repeat-containing protein G | 0207 (sdrG) | 0331 (sdrG)§ | [139] |
| Serine-aspartate repeat-containing protein H | 1487 (sdrH) | 1632 (sdrH) | [140] |
| Autolysin/adhesin | 0100 (aae) | 2319 (aae) | [36] |
| Bifunctional autolysin | 0636 (atlE) | 0750 (atlE) | [35] |
|
Factors that promote intercellular aggregation & biofilm formation | |||
| Polysaccharide intercellular adhesin | 2293 (icaA) | NP | [54] |
| 2294 (icaD) | |||
| 2295 (icaB) | |||
| 2296 (icaC) | |||
| Biofilm-associated protein homolog | 2395 (bhp, sesD) | NP | [33] |
| Accumulation-associated protein | 2398 (aap, sesF) | 0175 (aap) | [28,82,83] |
| Extracellular matrix binding protein | 1011 (ebh) | 1128 (ebhA) | [47] |
| Teichoic acids | 0295 (tagA) | 0410 (tagA) | [141] |
| 0296 (tagH) | 0411 (tagH) | ||
| 0297 (tagG) | 0412 (tagG) | ||
| 0298 (tagB) | 0413 (tagB) | ||
| 0299 (tagX) | 0414 (tagX) | ||
| 0300 (tagD) | 0415 (tagD) | ||
| eDNA | 2117 (cidA) | 2105 (cidA) | [37,38] |
| 0636 (atlE) | 0750 (atlE) | ||
|
Capsule | |||
| Poly-γ-DL-glutamic acid | 2107 (capB) | 2093 (capB) | [7] |
| 2106 (capC) | 2092 (capC) | ||
| 2105 (capA) | 2091 (capA) | ||
| 2103 (capD) | 2089 (capD) | ||
|
Resistance to antimicrobial peptides | |||
| Antimicrobial peptide sensor | 0311 (apsX) | 0426 (aspX) | [24] |
| 0312 (apsR, graR) | 0427 (aspR, graR) | ||
| 0313 (apsS, graS) | 0428 (aspS, graS) | ||
| Multiple peptide resistance factor | 0930 (mprF, fmtC) | 1041 (mprF, fmtC) | [24,142] |
| D-alanylation of teichoic acids | 0518 (dltA) | 0624 (dltA) | [24,143,144] |
| 0519 (dltB) | 0625 (dltB) | ||
| 0520 (dltC) | 0626 (dltC) | ||
| 0521 (dltD) | 0627 (dltD) | ||
| VraFG | 0314 (vraF) | 0429 (vraF) | [24] |
| 0315 (vraG) | 0430 (vraG) | ||
The exact mechanism required to form a functional, mature staphylococcal biofilm is unknown. However, it has been classically viewed as a four-step process: adherence, accumulation, maturation and detachment. A mature S. epidermidis biofilm consists of a variety of adhesive molecules, including polysaccharide intercellular adhesin (PIA), proteinaceous factors (Bhp, Aap and Embp), teichoic acids and extracellular (e) DNA. However, complicating experimental analysis of S. epidermidis biofilm formation is the fact that not all isolates encode factors that are thought to augment biofilm formation. For instance, not all isolates encode icaADBC, the operon responsible for synthesizing PIA. Although a significant amount of S. epidermidis isolates obtained from a defined biomaterial infection encode icaADBC, multiple studies have demonstrated that the majority of commensal S. epidermidis isolates obtained from the skin of healthy individuals do not encode icaADBC [11,25–30]. In fact, a human colonization study demonstrated that an icaADBC mutant outcompeted its isogenic wild-type strain, suggesting synthesis of PIA incurs a fitness cost when colonizing skin [31]. In addition, a study by Bowden et al. reported only 9, 0 and 13% of bacteremia, blood culture contaminant and colonizing skin flora isolates, respectively, encoded bhp, a protein associated with strong biofilm formation in both S. epidermidis and S. aureus [32,33]. Clearly, future studies need to define the function of each factor in the establishment of a mature, functional biofilm.
Adherence to biomaterials
The initial step in biofilm formation is the adherence of the bacteria to a foreign body or biomaterial. These initial interactions are nonspecific and hydrophobic in nature [34]. However, specific proteins have been identified that mediate binding to these abiotic surfaces. In S. epidermidis, these include the bifunctional adhesins/autolysins AtlE and Aae [35,36]. Not only do these proteins have specific adherence functions (by binding noncovalently to vitronectin), they may also function to release eDNA, which has recently been demonstrated to be an important adherence/aggregation factor in both S. aureus and S. epidermidis biofilm formation [37,38]. Studies by Mann et al. recently demonstrated that cidA-mediated eDNA release functions in both the initial attachment and accumulation phase of S. aureus [39]. Furthermore, biomaterials are rapidly coated with human serum proteins, including fibronectin, collagen, fibrinogen and vitronectin. Staphylococci have multiple adherence factors, known as ‘microbial surface components recognizing adhesive matrix molecules’ (MSCRAMMs), which are known to bind serum proteins (Table 1) [40]. The most-studied MSCRAMM in S. epidermidis is the fibrinogen-binding protein SdrG. Deletion of sdrG leads to a decrease of adherence to fibrinogen-coated surfaces, and antibodies to SdrG lead to a decrease of S. epidermidis adherence to biomaterials in vivo [41–43]. Recent studies have also demonstrated that SdrG promotes platelet adhesion and aggregation [44]. An additional well-characterized MSCRAMM, SdrF, binds collagen, and anti-SdrF antibodies significantly reduced adherence to ventricular assist devices [45,46]. Embp is a 1.1-MDa protein capable of binding fibronectin [47]. Studies by Christner and colleagues found that a 460-kDa isoform of Embp is capable of binding fibronectin and, in addition, mediating biofilm accumulation in an icaADBC- and aap-negative isolate (see below for discussions of icaADBC and aap) [48]. In addition, Embp-mediated biofilm was recalcitrant to phagocytosis by macrophages, suggesting that, in some isolates, Embp alone is sufficient to form functional, clinically relevant biofilms [48]. Finally, bioinformatic analyses have uncovered multiple putative LPXTG motif-containing cell wall-anchored proteins termed S. epidermidis surface proteins (Table 1) [5,32]. Although the function of these proteins is unclear, it is known that some are S. epidermidis specific and SesH, SesI and SesG may be markers for disease capacity [32,49]. SesC was shown by Shahrooei and colleagues to be highly expressed during biofilm growth [50]. In addition, polyclonal anti-SesC reduced the in vitro biofilm-forming and fibrinogen-binding ability of S. epidermidis RP62A, suggesting SesC may be essential for biofilm formation; Bowden et al. found sesC was present in all isolates tested [32,50].
Accumulation
The synthesis of PIA (termed poly-N-acetyl-glucosamine [PNAG] in S. aureus) is the best-studied mechanism of biofilm accumulation in S. epidermidis [51]. However, clinical studies have demonstrated that clinically relevant S. epidermidis isolates are PIA negative, demonstrating that proteinaceous factors can substitute to function as an accumulative molecule during biofilm formation (see below) [28,52]. The majority of clinical S. epidermidis isolates synthesize PIA [11,25–30], which is a homoglycan composed of β-1,6-linked 2-amino-2-deoxy-D-pyranosyl residues [51,53]. PIA contains negative charges due to partial de-N-acetylation and positive charges due to O-succinylation [53]. PIA is synthesized by the ica operon gene products [54]; the ica operon is composed of four genes: icaA (1238 bp), icaD (305 bp), icaB (869 bp) and icaC (1067 bp). A divergently transcribed repressor, icaR (557 bp), which has homology to the TetR family of transcriptional regulators, is found just upstream of ica [55]. Gerke and colleagues have found that IcaA and IcaD, both membrane proteins, act in concert as a UDP N-acetylglucosaminyltransferase. IcaC, also a membrane protein, is hypothesized to act in translocating polysaccharide synthesized by IcaAD through the cytoplasmic membrane [56,57]. IcaB acts as a deactylase; deacetylation of PIA is required for PIA attachment to the cell surface, as well as biofilm formation, surface colonization, immune system evasion and virulence in a mouse model [57–59]. The importance of PIA in the virulence of S. epidermidis has been demonstrated in two animal models of device-associated infections, a rat catheter model and a mouse foreign-body infection model [60–62]. Secretion of PIA mediates initial adherence to surgical polymers, interbacterial adhesion, and facilitates biofilm formation [51,63]. In addition, PIA mediates biocide resistance and inhibits neutrophil-dependent killing [64,65]. Therefore, clinically, one of the most relevant functions of PIA is as a facilitator of biofilm formation, which increases the persistence of infections and leads to a decreased efficacy of antibiotic-induced bactericidal activity [59,65–67].
Transcriptional regulation of the icaADBC operon has been extensively studied and multiple factors function to modulate its expression, including SarA, SarZ, LuxS, ClpP, σB and the tricarboxylic acid (TCA) cycle [68–75]. Both SarA and SarZ are required for icaADBC transcription and subsequent PIA synthesis [68,69,72,73]; loss of SarZ reduces virulence in a mouse foreign-body model. The quorum-sensing system, LuxS, also represses icaADBC transcription and subsequent PIA synthesis; a luxS mutant was more virulent than wild-type in a rat model of biofilm infection [76]. At this point, it is unclear how LuxS and the AI-2 system interacts with the icaADBC operon. The stress sigma factor, σB, also functions in an indirect manner to regulate PIA synthesis [70]. Using transposon mutagenesis, Knobloch and colleagues found that insertion of Tn917 into rsbU repressed icaADBC transcription through an icaR-dependent manner whereby icaR transcription was upregulated in an unknown, indirect manner [70]. Finally, the observation that several environmental stimuli altered PIA synthesis (iron availability, ethanol concentration and anoxic growth conditions) led to the hypothesis that TCA cycle activity and the bacterial metabolic status was linked to PIA synthesis [77]. Subsequent inactivation of aconitase caused derepression of icaADBC transcription and PIA synthesis [71]; however, the metabolic link between the TCA cycle and icaADBC transcription is not known at this time. An additional transcriptional regulator of icaADBC transcription is IcaR, a member of the TetR family of transcriptional regulators [55,69,70,78–81]. icaR is divergently transcribed from icaADBC and negatively regulates icaADBC transcription in early exponential phase [55]. Furthermore, as stated, σB influences icaADBC transcription and PIA biosynthesis by indirectly affecting icaR transcription [70]. X-ray crystallographic analysis of the S. epidermidis IcaR protein and subsequent electrophoretic mobility shift assays demonstrated that two IcaR dimers bind cooperatively to a 28-bp region centered 17 bases upstream of the icaADBC start codon [81]. Consistent with other TetR regulators, the DNA affinity of IcaR was greatly decreased in the presence of streptomycin and gentamicin [81]. Cerca et al. demonstrated that in S. aureus, SarA and σB are required for icaR expression and IcaR does not significantly affect its own transcription [78].
In addition to its significant function in immune evasion and biofilm accumulation, PIA also significantly affects the architecture of a maturing biofilm. As shown in Figure 1, S. epidermidis strains expressing PIA have significant tower formation and 3D structure compared with isogenic strains not expressing PIA. In addition, using flow cell parameters with increased shear stress (i.e., increased fluid flow rate), S. epidermidis isolates that do not synthesize PIA are not able to form biofilms (Figure 1). These data may suggest that S. epidermidis isolates that synthesize PIA are advantageous in infections with a high shear stress (i.e., catheter lumen). Confocal analysis confirms what is observed in the flow cells; towers are much more common and larger than the few towers that are observed in PIA-negative biofilms (Figure 2). Further studies are needed to address what particular factors are responsible for tower formation and subsequent maturation in PIA-positive staphylococcal biofilms.
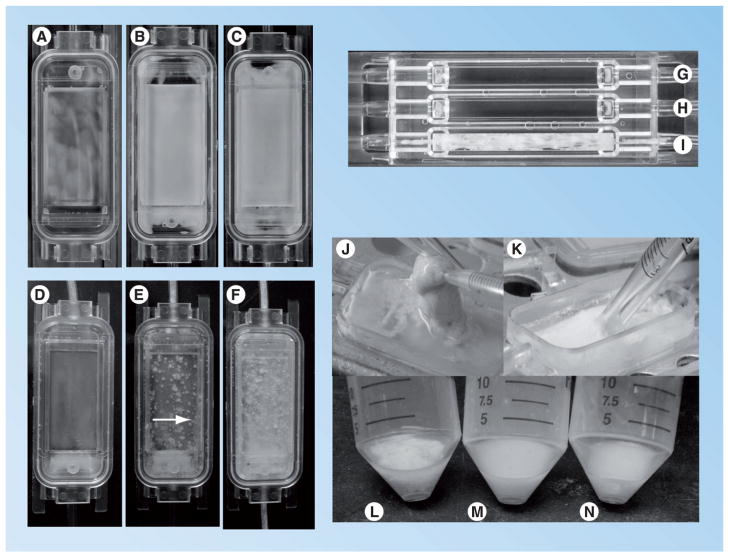
Figure 1. Biofilm formation by Staphylococcus epidermidis 1457 and an isogenic icaADBC mutant as compared in a flow cell.
(A–C) S. epidermidis 1457 icaADBC::dhfr [69] grown in a flow cell for 24 (A), 48 (B) and 72 h (C). (D–F) Staphylococcus epidermidis 1457 [51] grown in a flow cell for 24 (D), 48 (E) and 72 h (F). Note the significant tower formation and 3D structure associated with 1457 as compared with 1457 icaADBC at 48 and 72 h (noted by arrow in (E)). S. epidermidis 1457 sarA::tetM [69] (G), S. epidermidis 1457 icaADBC::dhfr (H) and S. epidermidis 1457 (I) grown in a flow cell with high shear stress. Note the lack of biofilm formation in (G & H) containing S. epidermidis 1457 mutants unable to synthesize polysaccharide intercellular adhesin (polysaccharide intercellular adhesin [PIA]; icaADBC and sarA mutations). Tryptic soy broth at a flow rate of 0.5 ml/min was used in both flow cells shown in (A–F) and (G–I). However, the shear stress was greater in the flow cell shown in panels (G–I) due to the smaller surface area of the material supporting bacterial growth. (J & K) Note the contribution of PIA to biofilm synthesis in 1457 (J) in contrast to 1457 icaADBC::dhfr (K). S. epidermidis 1457 PIA-dependent biofilms can be picked up with a pipette, whereas PIA-independent biofilms in the 1457 background can easily be resuspended with a pipette. (L–N) Note that biofilms from 72 h flow cells (as shown in (C & F)) from 1457 icaADBC::dhfr (M) and 1457 sarA::tetM can easily be resuspended in broth, whereas the biofilm from 1457 (L) is not resuspended upon vortexing.
Figure 2. Confocal microscopy of polysaccharide intercellular adhesin-dependent and -independent biofilms.
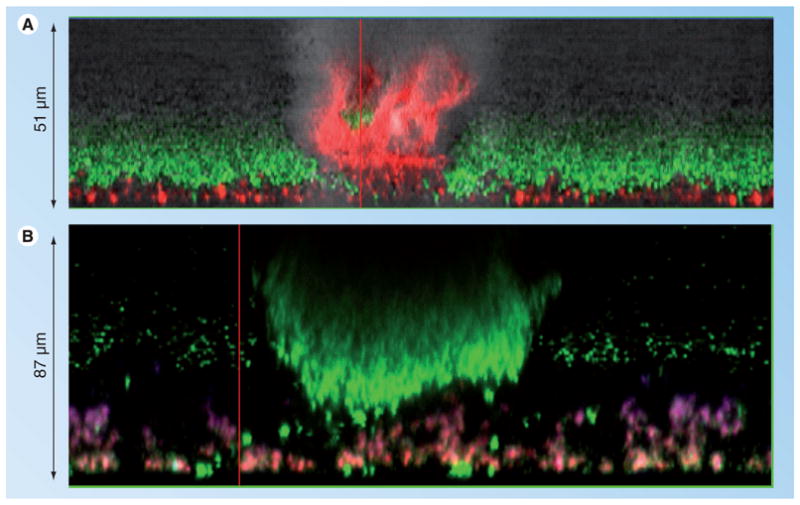
Staphylococcus epidermidis 1457 icaADBC::dhfr (A) and 1457 (B) were grown in Lab-Tek™ borosilicate coverglass systems for 24 h in tryptic soy broth and stained with wheat germ agglutinin (WGA), Syto9 and Toto-3. WGA (purple stain) was used to identify polysaccharide intercellular adhesin, Syto-9 (green stain) was applied to identify viable cells in the biofilm, and Toto-3 (red stain) was used to stain both dead cells and eDNA. Note that, in contrast to towers found in 1457, the towers in 1457 icaADBC::dhfr are comprised of dead cells and/or eDNA. 1457 towers are much more common, larger and contain live cells. In addition, note the WGA staining of polysaccharide intercellular adhesin in 1457 (B).
Although PIA is a highly significant factor in S. epidermidis biofilm formation and maturation, S. epidermidis strains have been isolated from clinically relevant infections that do not encode icaADBC and thus do not synthesize PIA [28]. Biofilm accumulation in these isolates is protein mediated as they are protease sensitive but resistant to polysaccharide-degrading enzymes [28]. Biofilm accumulation proteins in S. epidermidis include Embp (discussed previously) and two other specific proteins, Aap and Bhp (Bap in S. aureus). Aap is a 220-kDa LPXTG protein containing an N-terminal A domain and a B domain of variable number 128-bp repeat [82,83]. Aap is processed by both bacterial and host proteases into its active form and is a fibrillar protein that is extruded from the cell in localized tufts [82,83]. Aap accumulation is mediated by Zn2+-dependent dimerization of B domains on neighboring cells, whereas the A domain mediates adherence to corneocytes, implicating a further role in adherence to skin [84,85]. The function of Aap in a PIA-dependent biofilm is unknown; however, polyclonal antibodies against Aap in RP62A (a strain that synthesizes PIA) inhibited biofilm formation up to 87%, suggesting a role of Aap in initial adherence or early biofilm maturation [86].
In contrast to Aap, which is found in approximately 90% of S. epidermidis isolates, Bhp is encoded in approximately 15–45% of isolates, depending on the study [28,32]. In S. aureus, bap is even less frequently encountered; a recent study by Vautor et al. found that bap was not encoded in 262 S. aureus isolates obtained from various animal and human sources [87]. It is possible that Bap is closely linked to genetic backgrounds that are coadapted with bovine niches [33]. The S. epidermidis bhp gene is 8226 bp in length, encoding a protein with a predicted molecular mass of 284.4 kDa [33]. Tormo and colleagues demonstrated that bhp induces biofilm formation and accumulation in the absence of PIA [33]. Interestingly, although bap is encoded within a pathogenicity island in S. aureus (SaPIbov2), bhp in S. epidermidis is not associated with a mobile element [33,88]. Studies in S. aureus have demonstrated that bap transcription is SarA and σB dependent, and formation of a Bap-dependent biofilm is sensitive to the staphylococcal proteases Aur and SspA [89,90], suggesting a complex interaction between the metabolic state of the bacterium and protein-dependent biofilm formation.
Maturation
It is well established that bacteria growing within a biofilm are unique from those growing exponentially in the planktonic phase. Microarray studies have demonstrated that both S. epidermidis and S. aureus growing in a biofilm state have unique transcriptional responses compared with cells growing exponentially [91–93]. For example, these experiments demonstrate that staphylococci growing in a biofilm shift their physiology towards anaerobic or microaerobic metabolism and downregulate protein, cell wall and DNA synthesis. Although these experiments have been extremely helpful in defining the ‘average’ transcriptional response of biofilm growth (as all cells growing in a biofilm were examined), it is also well established that cells growing within a biofilm have spatial and temporal responses to their immediate environment (e.g., nutrient and oxygen availability and interactions with metabolic waste) (Figure 3) [94]. For example, Rani and colleagues have recently demonstrated that S. epidermidis growing within a biofilm consists of at least four metabolic states: aerobic growth, anaerobic growth, dormant cells and dead cells [95]. It is hypothesized that these defined physiological states found within a biofilm allow for tolerance to antibiotics; therefore, it follows that disruption of the ability of a particular biofilm region (e.g., anaerobic state) to develop may enhance the ability of antibiotics to clear biofilm-mediated infections. One specific operon that is consistently upregulated within biofilm populations of both S. epidermidis and S. aureus, in contrast to cells growing in a planktonic form, is the arginine deiminase operon (ADI) [91,92,96]. Many eubacteria utilize the ADI pathway to catabolize arginine under microaerobic or anaerobic conditions to generate ammonia and ATP [97]. When growing under anoxic conditions, arginine can serve as a sole carbon source [98]. There are several examples in the literature suggesting that arginine metabolism is important during the metabolic shift to anaerobic growth [91,92,99–101]. The ADI pathway is typically comprised of four genes found in an operon structure [98,102–107]. The first gene in the pathway is arcD, an arginine/ornithine antiporter, which facilitates the entry of arginine into the cell. When the ADI pathway is active, ornithine accumulates in the culture medium. Next, arcA (arginine deiminase) deiminates arginine, generating citrulline and ammonia. Citrulline is phosphorolyzed by ornithine transcarbamylase (arcB), resulting in carbamoylphosphate and ornithine. Carbamate kinase (arcC) finally transfers the phosphate from carbamoylphosphate to ADP, yielding 1 mol of ATP per mol of arginine. The resulting carbamate is chemically broken down to CO2 and ammonia. Analysis of the two publicly available S. epidermidis genomes (ATCC12228 [6] and RP62A [5]) demonstrates that at least one complete copy of the ADI gene cluster is found in these genomes. Surprisingly, the ATCC12228 genome contains two complete ADI gene clusters. One gene cluster, which contains argR1 through arcR1, and a second gene cluster containing arcC1 and arcB3, are identical to the gene clusters found in the RP62A genome. However, ATCC12228 contains a unique gene cluster, argR2 through arcR2, which is not found in the RP62A genome. This unique argR2–arcR2 gene cluster is contained on a 34-kb genomic island termed arginine catabolic mobile element (ACME) II in ATCC12228 [108]. This island is similar to ACME I, a genomic island found in S. aureus USA300, that also contains the ADI gene cluster [108]. This arc gene cluster within ACME I, which has been used as a genetic marker for identification of the USA300 background in molecular epidemiological studies [109], has been postulated to function as a virulence factor in S. aureus. First, Streptococcus pyogenes’ arginine deiminase functions to inhibit human peripheral blood mononuclear cell proliferation and may help the organism survive low pH (due to production of ammonia) and control pH homeostasis [110,111]. Second, depletion of L-arginine by arginine deiminase would decrease the production of nitric oxide (which is synthesized through L-arginine), a molecule used in the adaptive and innate immune responses against microbial infections [108]. Third, the catabolism of arginine may function to increase the pH of the extracellular milieu on the skin, allowing S. aureus to better colonize skin surfaces and, thus, have a greater ability to cause skin and soft tissue infections [101]. Finally, Diep and colleagues have recently shown that deletion of the entire ACME I element decreased virulence and fitness in a rabbit bacteremia model [112]. Overall, these data suggest that the generation of ATP through arginine catabolism is an important aspect of biofilm maturation. In addition, through generation of ammonia, induction of arcABDCR may be important for pH homeostasis within certain biofilm niches. Zhu and colleagues have addressed the function of arginine utilization in S. aureus UAMS-1 (USA200 background) by construction of an arcD1 mutant [101]. It was determined that the loss of arginine metabolism had no phenotypic effect on biofilm formation (although PIA production was reduced) or virulence in a mouse foreign-body infection model. However, it is important to note that UAMS-1 contains only the native copy of arcABDCR and does not contain the ACME island. In addition, it was also noted that the UAMS-1 arcD mutant accumulated significantly less ammonia in the culture medium under both flow cell and planktonic cultures. These data suggest that arginine metabolism has a significant function in overall amino acid metabolism and, possibly, pH homeostasis within a biofilm. The function of both ADI operons in S. epidermidis is unknown to date.
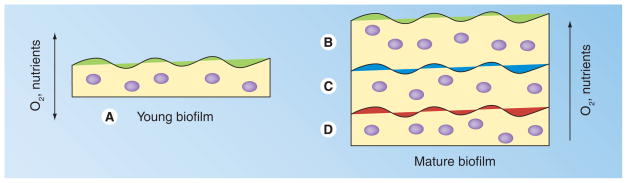
Figure 3. Temporal and spatial heterogeneity in a staphylococcal biofilm.
(A) Young biofilm replete with oxygen and nutrient substrate. By contrast, mature biofilm has cells that have access to both oxygen and substrate (B), substrate but no oxygen (C) and no oxygen or substrate (D), generating metabolic heterogeneity. In media containing a readily catabolizeable substrate such as glucose and in addition a separate carbon source such as amino acids or peptides, the upper regions of the biofilm (B) would have access to the glucose, whereas more microaerobic regions (C) would have access to a secondary carbon source such as amino acids/peptides. Adapted from [94].
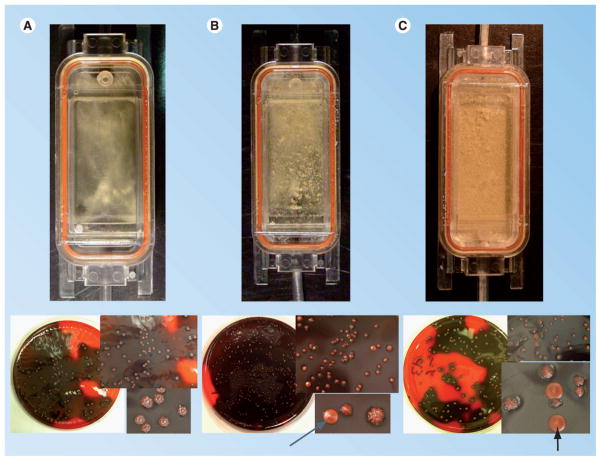
Finally, multiple studies have demonstrated that S. epidermidis undergoes a phenomenon whereby a certain proportion of the population does not produce PIA/biofilm. This observation, termed phenotypic variation (or phase variation), can be detected on media called Congo red agar [113]. Colonies that produce PIA grow as crusty, irregular colonies, whereas PIA-negative colonies are smooth and creamy (Figure 4). It should be noted that several different types of phenotypic variants are found on Congo red agar, which are termed ‘intermediate’. These colonies form an intermediate level of biofilm in comparison to crusty and smooth colonies. Handke et al. demonstrated that phenotypic variation occurs at a fairly high frequency (10−3–10−4) in almost all strains of S. epidermidis when grown in media for an extended period of time (5–7 days) [114]. As shown in Figure 4, phenotypic variation is a consequence of biofilm maturation and is observed at a high frequency after tower formation is observed. Ziebuhr and colleagues demonstrated that in some cases (30%), phenotypic variation is mediated by the insertion of IS256 into the ica operon [115]. However, the mechanisms of phenotypic variation in the remaining 70% of isolates are unknown. Handke et al. subcategorized smooth phenotypic variants (non-IS256 variants) into three classes. Class I phenotypic variants are those in which the transcription of icaADBC is downregulated and thus little PIA is synthesized. Class II phenotypic variants are those that produce the same amount of icaADBC transcript as wild-type, but do not produce any PIA/biofilm. Class III phenotypic variants are those variants in which large regions of the chromosome are deleted, including icaADBC. Some, but not all, class I and class II phenotypic variants are able to revert back to wild-type biofilm-forming capability after extended incubation in tryptic soy broth. In addition, DNA sequencing of three phenotypic variants of S. epidermidis SE5 (SE5 PV2, SE5 PV3 and SE5 PV10) suggested that SE5 PV2 (class I mutant) was an rsbU mutant while SE5 PV3 and SE5 PV10 (both class II mutants) were icaA and icaD mutants, respectively [114]. In a similar manner, Boles et al. isolated multiple colony phenotypes from a Pseudomonas aeruginosa biofilm population [116]. Furthermore, although a dinB (DNA PolIV) knockout did not have an effect on variant generation within a biofilm, a recA mutant grown in a biofilm did not generate ‘mini’ or ‘wrinkly’ variants. These investigators linked this phenomenon to a well-known hypothesis in ecological disciplines, the insurance hypothesis, stating that the presence of diverse subpopulations increases the range of conditions in which the community as a whole can thrive. This hypothesis predicts that RecA-dependent mutagenesis is utilized within a biofilm structure to ensure the diversity of the population is sufficient to survive and proliferate under adverse conditions.
Figure 4. Link between biofilm maturation and phenotypic variation.
Flow cell biofilms of Staphylococcus epidermidis 1457 were grown in tryptic soy broth for 24 (A), 48 (B) and 72 h (C), and plated onto Congo red agar. Phenotypic variants (as noted by arrows) are readily observed coincident with tower formation and biofilm maturation. Note no phenotypic variants or towers were observed after 24 h of growth (authors’ observation and [114]).
Dispersal
As a staphylococcal biofilm matures, individual cells or intact sections of biofilm can ‘slough off’ and metastasize to other organ systems. Little is known regarding dispersal and detachment mechanisms; however, in both S. epidermidis and S. aureus, this process is agr dependent [117–119]. S. epidermidis agr mutants showed increased bio-film development and colonization in a rabbit model [119]. It is hypothesized that the increased biofilm thickness in agr mutants is due to the loss of δ-toxin and other phenol-soluble modulins [8]. These molecules act as surfactants that inhibit noncovalent interactions of bacteria at the surface of the biofilm. Complementing these observations, agr is most transcriptionally activated at the fluid/biofilm interface [119]. In addition, agr also regulates biofilm formation in S. aureus; addition of AIP-mediated dramatic detachment of S. aureus biofilms [117]. Detachment was related to increased protease activity in biofilm effluent and was related to increased expression of Aur metalloprotease and the SplABCDEF serine proteases [117]. It is unclear what function these proteases may have in the detachment of a PIA- or Aap/Bhp-dependent biofilm in S. epidermidis.
Poly-γ-DL-glutamic acid
Poly-γ-DL-glutamic acid is an extracellular anionic polymer produced by B. anthracis and a few other human pathogens, including Leptospira interrogans and Fusobacterium nucleatum [120–122]. PGA, which is encoded by plasmid pXO2 in B. anthracis isolates, is a significant virulence factor in B. anthracis as it inhibits phagocytosis [121]. Surprisingly, genome sequencing studies have revealed that S. epidermidis encodes in the core genome a highly conserved capBCAD operon [5]. The capBCAD operon is conserved in most staphylococcal species closely related to S. epidermidis, including Staphylococcus capitis, Staphylococcus caprae, Staphylococcus haemolyticus, Staphylococcus warnerii, Staphylococcus saccharolyticus and Staphylococcus hominis [7]. The operon is also found in the human pathogen S. lugdunensis, but is not found in S. aureus [7]. PGA appears to protect S. epidermidis against high salt concentrations in addition to mediating resistance to antimicrobial peptides and phagocytosis, both components of the innate immune system [7]. Therefore, the expression of PGA may be advantageous for those staphylococci that reside in high salt environments such as skin. Surprisingly, even though capBCAD isogenic mutants were significantly less virulent than wild-type in a mouse foreign-body model, the capBCAD mutant had no apparent phenotypic defect regarding biofilm formation in a PIA-producing S. epidermidis isolate (strain 1457) [7]. The interaction (if any) between PIA and PGA, both antiphagocytic, is unknown. However, these data suggest that PIA-dependent biofilms require PGA to become highly recalcitrant to the action of the innate immune system.
Future perspective
We are only beginning to understand the biology of commensal bacteria such as S. epidermidis and their function in the maintenance of human health. Current microbiome studies will allow us to first understand and identify the ‘players’ that utilize the human skin as their ecological niche. It is unclear whether each species has a particular role in maintenance of and integrity of the skin structure or, possibly, immune development [2]. Second, the utilization of newer technologies will enable investigators to further probe each stage of biofilm formation to identify new strategies to inhibit biofilm formation on biomaterials. Many strategies, some very successful, have addressed inhibiting the initial adherence step of biofilm formation. Although vaccines against staphylococcal targets have proven to be problematic, multivalent vaccines against S. epidermidis should certainly target factors that are important for adherence to biomaterials. A virtually untapped area of staphylococcal biofilm research has been the identification of factors that are responsible for biofilm maturation. These maturation processes may include tower formation or the shift from a primarily aerobic metabolism to a microaerobic/anaerobic condition. It is hypothesized that inhibition of maturation may facilitate increased phagocytosis by the innate immune system or susceptibility to antibiotics. Newer technologies, such as laser capture microdissection microscopy, are needed to address the heterotypic nature of biofilms and study the single cell/regional response(s) to the nutrient and oxygen gradients that are generated by biofilms [123]. It is hypothesized that phenotypic variation is a byproduct of maturation and metabolic status of the biofilm. Importantly, future studies need to address the function/role of PIA-, Aap-, Bhp- or Embp-dependent S. epidermidis biofilms. Are all of these biofilms clinically relevant and recalcitrant to the innate immune system and the bactericidal action of antibiotics? Finally, further studies are required to address the interaction of PGA and other biofilm accumulation factors (PIA, Aap, Embp and Bhp) as the loss of PGA appears to dampen the anti-innate immune system properties of a PIA-dependent biofilm [7].
Executive summary.
Staphylococcus epidermidis
Staphylococcus epidermidis is a commensal bacterium living on the skin of humans. It is a significant cause of biomaterial-related infections.
Biofilm synthesis is a primary virulence factor. Staphylococcal biofilm is recalcitrant to host innate immune response and antibiotic treatment; therefore, treatment of S. epidermidis infections frequently requires removal of offending device.
Genome structure of S. epidermidis
The genomes of two S. epidermidis isolates have been sequenced, ATCC12228 and RP62A.
The genome sequence reflects the ecological niche (skin) of S. epidermidis by encoding genes related to osmoprotection.
In contrast to Staphylococcus aureus, S. epidermidis produces few virulence factors. Most are related to biofilm synthesis or resistance to host innate immune system.
Most clinical S. epidermidis isolates are part of a large clonal complex defined as CC2.
S. epidermidisbiofilm formation
Lack of virulence of S. epidermidis in contrast to S. aureus may be related to ease of transmission from host to host.
Many virulence factors produced by S. epidermidis, including phenol-soluble modulins and the three-component antimicrobial peptide-sensing system, help to mediate resistance to the innate immune system.
Biofilm formation in staphylococci is typically viewed as a four-step mechanism: adherence, accumulation, maturation and detachment.
A factor complicating experimental analysis of S. epidermidis biofilm formation is that not all isolates encode factors demonstrated to augment biofilm formation, including icaADBC, aap, embp and bhp.
Adherence to biomaterials is mediated by both nonspecific and specific interactions. Specific adhesins include the bifunctional adhesins/autolysins AtlE/Aae and the MSCRAMM proteins SdrG, SdrF and Embp.
The best studied accumulation factor is polysaccharide intercellular adhesin (PIA) synthesized by gene products of the icaADBC operon. icaADBC is transcriptionally regulated by multiple factors, demonstrating that PIA synthesis is tied to metabolic status of the bacterium.
S. epidermidis strains have also been isolated from clinically relevant infections that do not synthesize PIA, suggesting that other factors can replace PIA in the biofilm accumulation phase. Aap and Bhp are two identified proteins that can function in this role.
Little is known regarding the maturation phase of staphylococcal biofilm synthesis, but some data suggest that arginine catabolism is important. Phenotypic variation may be a by-product of biofilm maturation and tower formation.
Dispersal of both S. aureus and S. epidermidis biofilms is agr dependent. However, in S. epidermidis, dispersal is related to synthesis of phenol-soluble modulins and δ-toxin, whereas S. aureus dispersal is related to protease production.
Poly-γ-DL-glutamic acid
Poly-γ-DL-glutamic acid is a virulence factor not found in S. aureus that protects S. epidermidis against high salt concentrations in addition to mediating resistance to antimicrobial peptides and phagocytosis.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work was supported by a grant from the National Institutes of Health, National Institute of Allergy and Infectious Disease P01 AI083211. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
- 1.Kloos WE, Musselwhite MS. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol. 1975;30(3):381–385. doi: 10.1128/am.30.3.381-395.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7(12):887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl Environ Microbiol. 2003;69(1):18–23. doi: 10.1128/AEM.69.1.18-23.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4▪.Rogers KL, Fey PD, Rupp ME. Coagulase-negative staphylococcal infections. Infect Dis Clin North Am. 2009;23(1):73–98. doi: 10.1016/j.idc.2008.10.001. Review discussing clinical aspects of Staphylococcus epidermidis. [DOI] [PubMed] [Google Scholar]
- 5▪.Gill SR, Fouts DE, Archer GL, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187(7):2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. First report of the genome sequence of a biofilm-forming S. epidermidis isolate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6▪.Zhang YQ, Ren SX, Li HL, et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol Microbiol. 2003;49(6):1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. First report of the genome sequence of S. epidermidis. [DOI] [PubMed] [Google Scholar]
- 7▪.Kocianova S, Vuong C, Yao Y, et al. Key role of poly-γ-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest. 2005;115(3):688–694. doi: 10.1172/JCI23523. Description of poly-γ-DL-glutamic acid and its function in S. epidermidis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8▪.Otto M. Staphylococcus epidermidis – the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7(8):555–567. doi: 10.1038/nrmicro2182. Comprehensive review of S. epidermidis biology and pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9▪.Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol. 2007;189(6):2540–2552. doi: 10.1128/JB.01484-06. Manuscript demonstrating that S. epidermidis has a clonal population structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wisplinghoff H, Rosato AE, Enright MC, Noto M, Craig W, Archer GL. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob Agents Chemother. 2003;47(11):3574–3579. doi: 10.1128/AAC.47.11.3574-3579.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J Clin Microbiol. 2005;43(9):4751–4757. doi: 10.1128/JCM.43.9.4751-4757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang XM, Noble L, Kreiswirth BN, et al. Evaluation of a multilocus sequence typing system for Staphylococcus epidermidis. J Med Microbiol. 2003;52(Pt 11):989–998. doi: 10.1099/jmm.0.05360-0. [DOI] [PubMed] [Google Scholar]
- 13.Miragaia M, de Lencastre H, Perdreau-Remington F, et al. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS ONE. 2009;4(11):e7722. doi: 10.1371/journal.pone.0007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mongkolrattanothai K, Boyle S, Murphy TV, Daum RS. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob Agents Chemother. 2004;48(5):1823–1836. doi: 10.1128/AAC.48.5.1823-1836.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson A, Koskiniemi S, Gottfridsson P, Wistrom J, Monsen T. Multiple-locus variable-number tandem repeat analysis for typing of Staphylococcus epidermidis. J Clin Microbiol. 2006;44(1):260–265. doi: 10.1128/JCM.44.1.260-265.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francois P, Hochmann A, Huyghe A, et al. Rapid and high-throughput genotyping of Staphylococcus epidermidis isolates by automated multilocus variable-number of tandem repeats: a tool for real-time epidemiology. J Microbiol Methods. 2008;72(3):296–305. doi: 10.1016/j.mimet.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Miragaia M, Carrico JA, Thomas JC, Couto I, Enright MC, de Lencastre H. Comparison of molecular typing methods for characterization of Staphylococcus epidermidis: proposal for clone definition. J Clin Microbiol. 2008;46(1):118–129. doi: 10.1128/JCM.01685-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenstein R, Nerz C, Biswas L, et al. Genome analysis of the meat starter culture Bacterium Staphylococcus carnosus TM300. Appl Environ Microbiol. 2009;75(3):811–822. doi: 10.1128/AEM.01982-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosebury T. Microorganisms Indigenous to Man. McGraw Hill; New York, NY, USA: 1962. [Google Scholar]
- 20▪.Massey RC, Horsburgh MJ, Lina G, Hook M, Recker M. The evolution and maintenance of virulence in Staphylococcus aureus: a role for host-to-host transmission? Nat Rev Microbiol. 2006;4(12):953–958. doi: 10.1038/nrmicro1551. Interesting discussion of the unique niches occupied by S. epidermidis and Staphylococcus aureus and their relationship to transmission and virulence. [DOI] [PubMed] [Google Scholar]
- 21.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otto M. Bacterial sensing of antimicrobial peptides. Contrib Microbiol. 2009;16:136–149. doi: 10.1159/000219377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Queck SY, Otto M. Staphylococcus epidermidis and other coagulase-negative staphylococci. In: Lindsay JA, editor. Staphylococcus Molecular Genetics. Caister Academic Press; Norfolk, UK: 2008. [Google Scholar]
- 24.Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci USA. 2007;104(22):9469–9474. doi: 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho SH, Naber K, Hacker J, Ziebuhr W. Detection of the icaADBC gene cluster and biofilm formation in Staphylococcus epidermidis isolates from catheter-related urinary tract infections. Int J Antimicrob Agents. 2002;19(6):570–575. doi: 10.1016/s0924-8579(02)00101-2. [DOI] [PubMed] [Google Scholar]
- 26.Frebourg NB, Lefebvre S, Baert S, Lemeland JF. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J Clin Microbiol. 2000;38(2):877–880. doi: 10.1128/jcm.38.2.877-880.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galdbart JO, Allignet J, Tung HS, Ryden C, El Solh N. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J Infect Dis. 2000;182(1):351–355. doi: 10.1086/315660. [DOI] [PubMed] [Google Scholar]
- 28.Rohde H, Burandt EC, Siemssen N, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28(9):1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 29.Ziebuhr W, Heilmann C, Gotz F, et al. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65(3):890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozitskaya S, Cho SH, Dietrich K, Marre R, Naber K, Ziebuhr W. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect Immun. 2004;72(2):1210–1215. doi: 10.1128/IAI.72.2.1210-1215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers KL, Rupp ME, Fey PD. The presence of icaADBC is detrimental to the colonization of human skin by Staphylococcus epidermidis. Appl Environ Microbiol. 2008;74(19):6155–6157. doi: 10.1128/AEM.01017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowden MG, Chen W, Singvall J, et al. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology. 2005;151(Pt 5):1453–1464. doi: 10.1099/mic.0.27534-0. [DOI] [PubMed] [Google Scholar]
- 33.Tormo MA, Knecht E, Gotz F, Lasa I, Penades JR. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology. 2005;151(Pt 7):2465–2475. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 34.Vacheethasanee K, Temenoff JS, Higashi JM, et al. Bacterial surface properties of clinically isolated Staphylococcus epidermidis strains determine adhesion on polyethylene. J Biomed Mater Res. 1998;42(3):425–432. doi: 10.1002/(sici)1097-4636(19981205)42:3<425::aid-jbm12>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 35.Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24(5):1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 36.Heilmann C, Thumm G, Chhatwal GS, Hartleib J, Uekotter A, Peters G. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology. 2003;149(Pt 10):2769–2778. doi: 10.1099/mic.0.26527-0. [DOI] [PubMed] [Google Scholar]
- 37.Qin Z, Ou Y, Yang L, et al. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153(Pt 7):2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 38▪.Rice KC, Mann EE, Endres JL, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci USA. 2007;104(19):8113–8118. doi: 10.1073/pnas.0610226104. First manuscript demonstrating function of eDNA in staphylococcal biofilm development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mann EE, Rice KC, Boles BR, et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE. 2009;4(6):e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clarke SR, Foster SJ. Surface adhesins of Staphylococcus aureus. Adv Microb Physiol. 2006;51:187–224. doi: 10.1016/S0065-2911(06)51004-5. [DOI] [PubMed] [Google Scholar]
- 41.Pei L, Flock JI. Lack of fbe, the gene for a fibrinogen-binding protein from Staphylococcus epidermidis, reduces its adherence to fibrinogen coated surfaces. Microb Pathog. 2001;31(4):185–193. doi: 10.1006/mpat.2001.0462. [DOI] [PubMed] [Google Scholar]
- 42.Pei L, Flock JI. Functional study of antibodies against a fibrogenin-binding protein in Staphylococcus epidermidis adherence to polyethylene catheters. J Infect Dis. 2001;184(1):52–55. doi: 10.1086/321003. [DOI] [PubMed] [Google Scholar]
- 43.Sellman BR, Timofeyeva Y, Nanra J, et al. Expression of Staphylococcus epidermidis SdrG increases following exposure to an in vivo environment. Infect Immun. 2008;76(7):2950–2957. doi: 10.1128/IAI.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brennan MP, Loughman A, Devocelle M, et al. Elucidating the role of Staphylococcus epidermidis serine-aspartate repeat protein G in platelet activation. J Thromb Haemost. 2009;7(8):1364–1372. doi: 10.1111/j.1538-7836.2009.03495.x. [DOI] [PubMed] [Google Scholar]
- 45.Arrecubieta C, Lee MH, Macey A, Foster TJ, Lowy FD. SdrF, a Staphylococcus epidermidis surface protein, binds type I collagen. J Biol Chem. 2007;282(26):18767–18776. doi: 10.1074/jbc.M610940200. [DOI] [PubMed] [Google Scholar]
- 46.Arrecubieta C, Toba FA, von Bayern M, et al. SdrF, a Staphylococcus epidermidis surface protein, contributes to the initiation of ventricular assist device driveline-related infections. PLoS Pathog. 2009;5(5):e1000411. doi: 10.1371/journal.ppat.1000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clarke SR, Harris LG, Richards RG, Foster SJ. Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect Immun. 2002;70(12):6680–6687. doi: 10.1128/IAI.70.12.6680-6687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48▪.Christner M, Franke GC, Schommer NN, et al. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol Microbiol. 75(1):187–207. doi: 10.1111/j.1365-2958.2009.06981.x. First paper demonstrating that Embp can function as adhesin and accumulation factor in biofilm development. [DOI] [PubMed] [Google Scholar]
- 49.Soderquist B, Andersson M, Nilsson M, et al. Staphylococcus epidermidis surface protein I (SesI): a marker of the invasive capacity of S. epidermidis? J Med Microbiol. 2009;58(Pt 10):1395–1397. doi: 10.1099/jmm.0.008771-0. [DOI] [PubMed] [Google Scholar]
- 50.Shahrooei M, Hira V, Stijlemans B, Merckx R, Hermans PW, Van Eldere J. Inhibition of Staphylococcus epidermidis biofilm formation by rabbit polyclonal antibodies against the SesC protein. Infect Immun. 2009;77(9):3670–3678. doi: 10.1128/IAI.01464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51▪.Mack D, Fischer W, Krokotsch A, et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178(1):175–183. doi: 10.1128/jb.178.1.175-183.1996. Purification of polysaccharide intercellular adhesin (PIA) and its function in biofilm accumulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qin Z, Yang X, Yang L, et al. Formation and properties of in vitro biofilms of ica-negative Staphylococcus epidermidis clinical isolates. J Med Microbiol. 2007;56(Pt 1):83–93. doi: 10.1099/jmm.0.46799-0. [DOI] [PubMed] [Google Scholar]
- 53.Rohde H, Frankenberger S, Zahringer U, Mack D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur J Cell Biol. 2010;89(1):103–111. doi: 10.1016/j.ejcb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 54▪.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Gotz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20(5):1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. Discovery of the icaADBC operon and its relationship to biofilm formation. [DOI] [PubMed] [Google Scholar]
- 55▪.Conlon KM, Humphreys H, O’Gara JP. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol. 2002;184(16):4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002. First paper describing the function of IcaR in repression of icaADBC transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56▪.Gerke C, Kraft A, Submuth R, Schweitzer O, Gotz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273:18586–18593. doi: 10.1074/jbc.273.29.18586. Biochemical characterization of IcaAD and its function as an N-acetylglucosaminyltransferase. [DOI] [PubMed] [Google Scholar]
- 57.Gotz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43(6):1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 58.Kristian SA, Birkenstock TA, Sauder U, Mack D, Gotz F, Landmann R. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J Infect Dis. 2008;197(7):1028–1035. doi: 10.1086/528992. [DOI] [PubMed] [Google Scholar]
- 59▪.Vuong C, Kocianova S, Voyich JM, et al. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279(52):54881–54886. doi: 10.1074/jbc.M411374200. Describes the function and importance of IcaB as a deacetylase. [DOI] [PubMed] [Google Scholar]
- 60.Li H, Xu L, Wang J, et al. Conversion of Staphylococcus epidermidis strains from commensal to invasive by expression of the ica locus encoding production of biofilm exopolysaccharide. Infect Immun. 2005;73(5):3188–3191. doi: 10.1128/IAI.73.5.3188-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rupp ME, Ulphani JS, Fey PD, Bartscht K, Mack D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999;67(5):2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62▪.Rupp ME, Ulphani JS, Fey PD, Mack D. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect Immun. 1999;67(5):2656–2659. doi: 10.1128/iai.67.5.2656-2659.1999. First manuscript describing the importance of biofilm formation in the pathogenesis of a biomaterial-related infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olson ME, Garvin KL, Fey PD, Rupp ME. Adherence of Staphylococcus epidermidis to biomaterials is augmented by PIA. Clin Orthop Relat Res. 2006;451:21–24. doi: 10.1097/01.blo.0000229320.45416.0c. [DOI] [PubMed] [Google Scholar]
- 64.Ganeshnarayan K, Shah SM, Libera MR, Santostefano A, Kaplan JB. Poly-N-acetylglucosamine matrix polysaccharide impedes fluid convection and transport of the cationic surfactant cetylpyridinium chloride through bacterial biofilms. Appl Environ Microbiol. 2009;75:1308–1314. doi: 10.1128/AEM.01900-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65▪.Vuong C, Voyich JM, Fischer ER, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6(3):269–275. doi: 10.1046/j.1462-5822.2004.00367.x. Manuscript demonstrating that PIA is antiphagocytic. [DOI] [PubMed] [Google Scholar]
- 66.Begun J, Gaiani JM, Rohde H, et al. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 2007;3(4):e57. doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol. 2008;322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 68.Conlon KM, Humphreys H, O’Gara JP. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J Bacteriol. 2004;186(18):6208–6219. doi: 10.1128/JB.186.18.6208-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Handke LD, Slater SR, Conlon KM, et al. σB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can J Microbiol. 2007;53(1):82–91. doi: 10.1139/w06-108. [DOI] [PubMed] [Google Scholar]
- 70▪.Knobloch JK, Jager S, Horstkotte MA, Rohde H, Mack D. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative σ factor σB by repression of the negative regulator gene icaR. Infect Immun. 2004;72(7):3838–3848. doi: 10.1128/IAI.72.7.3838-3848.2004. Demonstrates that σB regulates icaADBC transcription through IcaR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sadykov MR, Olson ME, Halouska S, et al. Tricarboxylic acid cycle-dependent regulation of Staphylococcus epidermidis polysaccharide intercellular adhesin synthesis. J Bacteriol. 2008;190(23):7621–7632. doi: 10.1128/JB.00806-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tormo MA, Marti M, Valle J, et al. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J Bacteriol. 2005;187(7):2348–2356. doi: 10.1128/JB.187.7.2348-2356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang L, Li M, Dong D, et al. SarZ is a key regulator of biofilm formation and virulence in Staphylococcus epidermidis. J Infect Dis. 2008;197(9):1254–1262. doi: 10.1086/586714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li M, Villaruz AE, Vadyvaloo V, Sturdevant DE, Otto M. AI-2-dependent gene regulation in Staphylococcus epidermidis. BMC Microbiol. 2008;8(4) doi: 10.1186/1471-2180-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang C, Li M, Dong D, et al. Role of ClpP in biofilm formation and virulence of Staphylococcus epidermidis. Microbes Infect. 2007;9(11):1376–1383. doi: 10.1016/j.micinf.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 76.Xu L, Li H, Vuong C, et al. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun. 2006;74(1):488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77▪.Vuong C, Kidder JB, Jacobson ER, Otto M, Proctor RA, Somerville GA. Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J Bacteriol. 2005;187(9):2967–2973. doi: 10.1128/JB.187.9.2967-2973.2005. First manuscript demonstrating the link between central metabolism and PIA synthesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cerca N, Brooks JL, Jefferson KK. Regulation of the intercellular adhesin locus regulator (icaR) by SarA, σB, and IcaR in Staphylococcus aureus. J Bacteriol. 2008;190(19):6530–6533. doi: 10.1128/JB.00482-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Conlon KM, Humphreys H, O’Gara JP. Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol Lett. 2002;216(2):171–177. doi: 10.1111/j.1574-6968.2002.tb11432.x. [DOI] [PubMed] [Google Scholar]
- 80.Jefferson KK, Pier DB, Goldmann DA, Pier GB. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J Bacteriol. 2004;186(8):2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jeng WY, Ko TP, Liu CI, et al. Crystal structure of IcaR, a repressor of the TetR family implicated in biofilm formation in Staphylococcus epidermidis. Nucleic Acids Res. 2008;36(5):1567–1577. doi: 10.1093/nar/gkm1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82▪.Rohde H, Burdelski C, Bartscht K, et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55(6):1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. Mechanistic description of protein-mediated biofilm accumulation. [DOI] [PubMed] [Google Scholar]
- 83.Banner MA, Cunniffe JG, Macintosh RL, et al. Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are comprised of the accumulation-associated protein. J Bacteriol. 2007;189(7):2793–2804. doi: 10.1128/JB.00952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, Herr AB. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci USA. 2008;105(49):19456–19461. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Macintosh RL, Brittan JL, Bhattacharya R, et al. The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J Bacteriol. 2009;191(22):7007–7016. doi: 10.1128/JB.00764-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun D, Accavitti MA, Bryers JD. Inhibition of biofilm formation by monoclonal antibodies against Staphylococcus epidermidis RP62A accumulation-associated protein. Clin Diagn Lab Immunol. 2005;12(1):93–100. doi: 10.1128/CDLI.12.1.93-100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vautor E, Abadie G, Pont A, Thiery R. Evaluation of the presence of the bap gene in Staphylococcus aureus isolates recovered from human and animals species. Vet Microbiol. 2008;127(3–4):407–411. doi: 10.1016/j.vetmic.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 88.Ubeda C, Tormo MA, Cucarella C, et al. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol Microbiol. 2003;49(1):193–210. doi: 10.1046/j.1365-2958.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- 89.Marti M, Trotonda MP, Tormo-Mas MA, et al. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 2010;12(1):55–64. doi: 10.1016/j.micinf.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 90.Trotonda MP, Manna AC, Cheung AL, Lasa I, Penades JR. SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J Bacteriol. 2005;187(16):5790–5798. doi: 10.1128/JB.187.16.5790-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beenken KE, Dunman PM, McAleese F, et al. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186(14):4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yao Y, Sturdevant DE, Otto M. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J Infect Dis. 2005;191(2):289–298. doi: 10.1086/426945. [DOI] [PubMed] [Google Scholar]
- 93.Resch A, Rosenstein R, Nerz C, Gotz F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol. 2005;71(5):2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94▪.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6(3):199–210. doi: 10.1038/nrmicro1838. Excellent review describing metabolic states found within biofilms. [DOI] [PubMed] [Google Scholar]
- 95.Rani SA, Pitts B, Beyenal H, et al. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J Bacteriol. 2007;189(11):4223–4233. doi: 10.1128/JB.00107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nagarajan V, Smeltzer MS, Elasri MO. Genome-scale transcriptional profiling in Staphylococcus aureus: bringing order out of chaos. FEMS Microbiol Lett. 2009;295(2):204–210. doi: 10.1111/j.1574-6968.2009.01595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abdelal AT. Arginine catabolism by microorganisms. Annu Rev Microbiol. 1979;33:139–168. doi: 10.1146/annurev.mi.33.100179.001035. [DOI] [PubMed] [Google Scholar]
- 98.Makhlin J, Kofman T, Borovok I, et al. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J Bacteriol. 2007;189(16):5976–5986. doi: 10.1128/JB.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fuchs S, Pane-Farre J, Kohler C, Hecker M, Engelmann S. Anaerobic gene expression in Staphylococcus aureus. J Bacteriol. 2007;189(11):4275–4289. doi: 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kohler C, von Eiff C, Liebeke M, et al. A defect in menadione biosynthesis induces global changes in gene expression in Staphylococcus aureus. J Bacteriol. 2008;190(19):6351–6364. doi: 10.1128/JB.00505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu Y, Weiss EC, Otto M, Fey PD, Smeltzer MS, Somerville GA. Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect Immun. 2007;75(9):4219–4226. doi: 10.1128/IAI.00509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barcelona-Andres B, Marina A, Rubio V. Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J Bacteriol. 2002;184(22):6289–6300. doi: 10.1128/JB.184.22.6289-6300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Griswold A, Chen YY, Snyder JA, Burne RA. Characterization of the arginine deiminase operon of Streptococcus rattus FA-1. Appl Environ Microbiol. 2004;70(3):1321–1327. doi: 10.1128/AEM.70.3.1321-1327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gruening P, Fulde M, Valentin-Weigand P, Goethe R. Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J Bacteriol. 2006;188(2):361–369. doi: 10.1128/JB.188.2.361-369.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maghnouj A, de Sousa Cabral TF, Stalon V, Vander Wauven C. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor argR. J Bacteriol. 1998;180(24):6468–6475. doi: 10.1128/jb.180.24.6468-6475.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zuniga M, Champomier-Verges M, Zagorec M, Perez-Martinez G. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J Bacteriol. 1998;180(16):4154–4159. doi: 10.1128/jb.180.16.4154-4159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zuniga M, Perez G, Gonzalez-Candelas F. Evolution of arginine deiminase (ADI) pathway genes. Mol Phylogenet Evol. 2002;25(3):429–444. doi: 10.1016/s1055-7903(02)00277-4. [DOI] [PubMed] [Google Scholar]
- 108.Diep BA, Gill SR, Chang RF, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367(9512):731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 109.Goering RV, McDougal LK, Fosheim GE, Bonnstetter KK, Wolter DJ, Tenover FC. Epidemiologic distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J Clin Microbiol. 2007;45(6):1981–1984. doi: 10.1128/JCM.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Degnan BA, Fontaine MC, Doebereiner AH, et al. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect Immun. 2000;68(5):2441–2448. doi: 10.1128/iai.68.5.2441-2448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Degnan BA, Palmer JM, Robson T, et al. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. Infect Immun. 1998;66(7):3050–3058. doi: 10.1128/iai.66.7.3050-3058.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Diep BA, Stone GG, Basuino L, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197(11):1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 113.Freeman DJ, Falkiner FR, Keane CT. New method for detection of slime production by coagulase-negative staphylococci. J Clin Pathol. 1989;42:872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Handke LD, Conlon KM, Slater SR, et al. Genetic and phenotypic analysis of biofilm phenotypic variation in multiple Staphylococcus epidermidis isolates. J Med Microbiol. 2004;53(Pt 5):367–374. doi: 10.1099/jmm.0.05372-0. [DOI] [PubMed] [Google Scholar]
- 115.Ziebuhr W, Krimmer V, Rachid S, Lobner I, Gotz F, Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]
- 116▪.Boles BR, Thoendel M, Singh PK. Self-generated diversity produces “insurance effects” in biofilm communities. Proc Natl Acad Sci USA. 2004;101(47):16630–16635. doi: 10.1073/pnas.0407460101. Description of insurance hypothesis and generation of phenotypic variants during biofilm maturation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4(4):e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188(5):706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- 119.Vuong C, Kocianova S, Yao Y, Carmody AB, Otto M. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J Infect Dis. 2004;190(8):1498–1505. doi: 10.1086/424487. [DOI] [PubMed] [Google Scholar]
- 120.Kapatral V, Anderson I, Ivanova N, et al. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol. 2002;184(7):2005–2018. doi: 10.1128/JB.184.7.2005-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Koehler TM. Bacillus anthracis physiology and genetics. Mol Aspects Med. 2009;30(6):386–396. doi: 10.1016/j.mam.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ren SX, Fu G, Jiang XG, et al. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422(6934):888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 123.Lenz AP, Williamson KS, Pitts B, Stewart PS, Franklin MJ. Localized gene expression in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2008;74(14):4463–4471. doi: 10.1128/AEM.00710-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rigoulay C, Entenza JM, Halpern D, et al. Comparative analysis of the roles of HtrA-like surface proteases in two virulent Staphylococcus aureus strains. Infect Immun. 2005;73(1):563–572. doi: 10.1128/IAI.73.1.563-572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dubin G, Chmiel D, Mak P, Rakwalska M, Rzychon M, Dubin A. Molecular cloning and biochemical characterisation of proteases from Staphylococcus epidermidis. Biol Chem. 2001;382(11):1575–1582. doi: 10.1515/BC.2001.192. [DOI] [PubMed] [Google Scholar]
- 126.Ohara-Nemoto Y, Ikeda Y, Kobayashi M, Sasaki M, Tajika S, Kimura S. Characterization and molecular cloning of a glutamyl endopeptidase from Staphylococcus epidermidis. Microb Pathog. 2002;33(1):33–41. doi: 10.1006/mpat.2002.0515. [DOI] [PubMed] [Google Scholar]
- 127.Farrell AM, Foster TJ, Holland KT. Molecular analysis and expression of the lipase of Staphylococcus epidermidis. J Gen Microbiol. 1993;139(2):267–277. doi: 10.1099/00221287-139-2-267. [DOI] [PubMed] [Google Scholar]
- 128.Longshaw CM, Farrell AM, Wright JD, Holland KT. Identification of a second lipase gene, gehD, in Staphylococcus epidermidis: comparison of sequence with those of other staphylococcal lipases. Microbiology. 2000;146(Pt 6):1419–1427. doi: 10.1099/00221287-146-6-1419. [DOI] [PubMed] [Google Scholar]
- 129.Simons JW, van Kampen MD, Riel S, Gotz F, Egmond MR, Verheij HM. Cloning, purification and characterisation of the lipase from Staphylococcus epidermidis – comparison of the substrate selectivity with those of other microbial lipases. Eur J Biochem. 1998;253(3):675–683. doi: 10.1046/j.1432-1327.1998.2530675.x. [DOI] [PubMed] [Google Scholar]
- 130.Teufel P, Gotz F. Characterization of an extracellular metalloprotease with elastase activity from Staphylococcus epidermidis. J Bacteriol. 1993;175(13):4218–4224. doi: 10.1128/jb.175.13.4218-4224.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chamberlain NR. Identification and partial characterisation of an extracellular activator of fatty acid modifying enzyme (FAME) expression in Staphylococcus epidermidis. J Med Microbiol. 1999;48(3):245–252. doi: 10.1099/00222615-48-3-245. [DOI] [PubMed] [Google Scholar]
- 132.Kaletta C, Entian KD, Kellner R, Jung G, Reis M, Sahl HG. Pep5, a new lantibiotic: structural gene isolation and prepeptide sequence. Arch Microbiol. 1989;152(1):16–19. doi: 10.1007/BF00447005. [DOI] [PubMed] [Google Scholar]
- 133.Schnell N, Entian KD, Schneider U, et al. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature. 1988;333(6170):276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 134.Mehlin C, Headley CM, Klebanoff SJ. An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J Exp Med. 1999;189(6):907–918. doi: 10.1084/jem.189.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vuong C, Durr M, Carmody AB, Peschel A, Klebanoff SJ, Otto M. Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: quorum-sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell Microbiol. 2004;6(8):753–759. doi: 10.1111/j.1462-5822.2004.00401.x. [DOI] [PubMed] [Google Scholar]
- 136.Korem M, Gov Y, Shirron N, Shuster A, Rosenberg M. Alcohol increases hemolysis by staphylococci. FEMS Microbiol Lett. 2007;269(1):153–159. doi: 10.1111/j.1574-6968.2006.00625.x. [DOI] [PubMed] [Google Scholar]
- 137.Cotton JL, Tao J, Balibar CJ. Identification and characterization of the Staphylococcus aureus gene cluster coding for staphyloferrin A. Biochemistry. 2009;48(5):1025–1035. doi: 10.1021/bi801844c. [DOI] [PubMed] [Google Scholar]
- 138.Cockayne A, Hill PJ, Powell NB, Bishop K, Sims C, Williams P. Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect Immun. 1998;66(8):3767–3774. doi: 10.1128/iai.66.8.3767-3774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nilsson M, Frykberg L, Flock JI, Pei L, Lindberg M, Guss B. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect Immun. 1998;66(6):2666–2673. doi: 10.1128/iai.66.6.2666-2673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.McCrea KW, Hartford O, Davis S, et al. The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology. 2000;146(Pt 7):1535–1546. doi: 10.1099/00221287-146-7-1535. [DOI] [PubMed] [Google Scholar]
- 141.Sadovskaya I, Vinogradov E, Flahaut S, Kogan G, Jabbouri S. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect Immun. 2005;73(5):3007–3017. doi: 10.1128/IAI.73.5.3007-3017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Peschel A, Jack RW, Otto M, et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with L-lysine. J Exp Med. 2001;193(9):1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. Incorporation of D-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem. 1995;270(26):15598–15606. doi: 10.1074/jbc.270.26.15598. [DOI] [PubMed] [Google Scholar]
- 144.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274(13):8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]