Abstract
Ingestion or inhalation of botulinum neurotoxin (BoNT) results in botulism, a severe and frequently fatal disease. Current treatments rely on antitoxins, which while effective cannot reverse symptoms once BoNT has entered the neuron. For treatments that can reverse intoxication, interest has focused on developing inhibitors of the enzymatic BoNT light chain (BoNT Lc). Such inhibitors typically mimic substrate and bind in or around the substrate cleavage pocket. To explore the full range of binding sites for serotype A light chain (BoNT/A Lc) inhibitors, we created a library of non-immune llama single domain VHH antibodies displayed on the surface of the yeast Saccharomyces cerevisiae. Library selection on BoNT/A Lc yielded 15 yeast displayed VHH with equilibrium dissociation constants (KD) from 230 to 0.03 nM measured by flow cytometry. Eight of 15 VHH inhibited the cleavage of substrate SNAP25 by BoNT/A Lc. The most potent VHH (Aa1) had a solution KD for BoNT/A Lc of 1.47 × 10-10 M, an IC50 of 4.7 × 10-10 M, and was resistant to heat denaturation and reducing conditions. To understand the mechanism by which Aa1 inhibited catalysis, the X-ray crystal structure of the BoNT/A Lc - Aa1 VHH complex was solved at 2.6 Å resolution. The structure reveals that the Aa1 VHH binds in the alpha-exosite of the BoNT/A Lc, far from the active site for catalysis. The study validates the utility of non-immune llama VHH libraries as a source of enzyme inhibitors and identifies the BoNT/A Lc alpha-exosite as a target for inhibitor development.
Keywords: botulinum neurotoxin type A, llama single VHH, single domain antibody, alpha-exosite, Naïve yeast-displayed library
INTRODUCTION
Human botulism is caused by Clostridium botulinum neurotoxin (BoNT) serotypes A, B, E, and F, and is characterized by flaccid muscle paralysis. The illness, when not immediately fatal, requires prolonged hospitalization in an intensive care unit. Besides causing naturally occurring botulism, BoNTs are also classified by the Centers for Disease Control and Prevention as one of the six highest-risk threat agents for bioterrorism 1. The symptoms of botulism are caused by BoNT 2, the most poisonous substance known 3. The crystal structure of BoNT 4 shows three functional domains comprised of a light chain and two heavy chain segments 4; 5; 6. The C-terminal portion of the heavy chain (Hc) is the cell binding domain, which docks the toxin to ganglioside receptors and a protein receptor(s) on presynaptic neurons resulting in toxin endocytosis 7; 8; 9. The translocation domain (Hn), at the N-terminal portion of the heavy chain, mediates escape of the toxin light chain (Lc) from the endosome 10. Depending on serotype, the Lc cleaves one or more members of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex of proteins involved in synaptic vesicle docking thereby inhibiting neurotransmitter release 11; 12.
The multi-domain structure of BoNT and its mechanism of action provide a number of ways to prevent and treat botulism. The mainstay of treatment for botulism is antitoxin 13. Antibody products, such as equine antitoxin and human botulism immunoglobulin, are used to treat adult 14; 15 and infant botulism 16, respectively. Antitoxin appears to work primarily by clearing toxin from the circulation before it can accumulate inside the neuron 17, but can also prevent BoNT entry into neurons by binding to the Hc 18. In addition, antibody may be able to inhibit translocation and catalysis by binding to the Hn and/or Lc, riding into the cell on BoNT, and then interfering with the function of these domains 10; 19. The recent visualization of the protein and ganglioside receptor binding sites on the BoNT Hc may also permit the design of small molecule drugs that can block toxin uptake 20; 21; 22.
A limitation of the above therapeutics is that they do not work once the toxin has entered the neuron, and therefore cannot be used to reverse paralysis. Thus, there is considerable interest in developing inhibitors of the translocation and catalytic domains 23; 24. Since the window to prevent translocation is relatively short, most attention has been focused on molecules that prevent the catalytic domain from cleaving their SNARE substrate. Such inhibitors typically mimic substrate and bind in or around the substrate cleavage pocket 25; 26. The crystal structure of the substrate synaptosome-associated protein of 25,000 daltons (SNAP25) complexed to the BoNT/A Lc showed the extended nature of ligand recognition and identified potential exosites of substrate binding that are away from the catalytic active site 27. While such exosites have been targeted for inhibitor development 28; 29, no such inhibitors have been reported for BoNT.
To explore the range of binding sites for potential BoNT/A Lc inhibitors, we generated and selected a non-immune camelid (llama) library of single domain VHH antibodies for binding to the BoNT/A Lc. Such single domain antibodies have been postulated as more able to bind into enzymatic cavities, and a number of enzyme inhibitors have been generated after immunizing camelids with enzyme antigens 30; 31. In this work, a number of inhibitory VHH were obtained and a selected complex characterized by x-ray diffraction validated the alpha-exosite as a viable target for BoNT/A inhibitor development.
RESULTS
Generation and initial characterization of single domain antibodies to BoNT/A Lc
To generate a panel of single domain antibodies binding the BoNT/A Lc, a non-immune llama single domain library was constructed for display on the surface of Saccharomyces cerevisiae. Briefly, whole blood was isolated from llamas without prior immunization and RNA prepared. After first strand cDNA synthesis, llama specific primers annealing to the VH and VHH leader sequence genes and to the CH2 gene were used to PCR amplify the VH and VHH gene repertoires. VHH repertoires were separated from VH repertoires by running the PCR fragments on a gel and excising the smaller band. The VHH gene repertoire was reamplified and cloned into the vector pYD2 for display as a C-terminal fusion to the AgaII protein on the surface of Saccharomyces cerevisiae. After transformation, a library of size of 6.1 × 107 transformants with a VHH sized insert was obtained. DNA sequencing of 50 VHH genes picked at random revealed 44 unique sequences, indicating that the library was diverse.
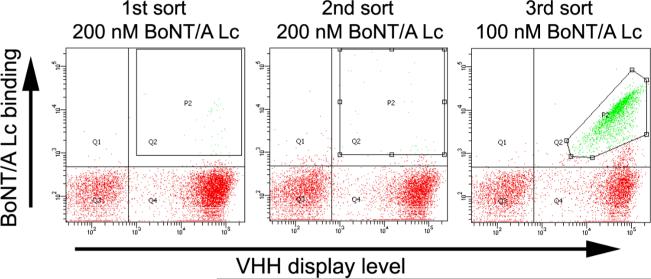
To generate BoNT/A Lc specific single domain antibodies, the library was induced to display VHH on yeast surface and incubated with recombinant BoNT/A Lc. After staining with anti-BoNT/A Lc mAbs and a mAb directed to the C-terminal SV5 epitope tag, yeasts displaying VHH and bound to BoNT/A Lc were flow sorted and collected (Figure 1). After amplification by growth in liquid culture, surface display was induced and the staining, sorting, and growth cycle repeated twice more (Figure 1). After three rounds of sorting, collected yeasts were plated and 48 individual colonies were analyzed for binding to BoNT/A Lc. The VHH gene of each binding clone was sequenced revealing the presence of 15 unique VHH, two pairs of which (Aa1 and A23; Aa12 and A10) were clonally related based on the VHH complementarity determining region 3 (CDR3) sequence (Table 1). The affinity of each of the yeast displayed VHH for BoNT/A Lc was determined by flow cytometry and found to range from a low of 230 nM (clone A8) to a high of 30 pM (clone Aa1) with an average KD of 56 nM (Table 1).
Figure 1. Selection of yeast displayed VHH by flow cytometry.
Dot-plots of flow cytometry sorting of VHH displaying yeast labeled with BoNT/A Lc are shown. For each of the three rounds of sorting, the concentration of BoNT/A Lc used to stain yeast is indicated. BoNT/A Lc binding is indicated on the Y-axis and the VHH display level on the X-axis. The sort gates used for yeast collection are indicated and the yeast in these gates are colored green.
Table 1.
Properties of BoNT/A Lc binding VHH antibodies.
| VHH clone name | Sequences of CDR3 | Yeast-displayed VHH KD for BoNT/A Lc (nM) |
|---|---|---|
| A26 | EVSSGQPAVTTFWEDMYDY | 8.7 |
| A3 | YRRRHRCSAFGIANEYDY | 6.63 |
| A16 | DDPLVGRGWDGAEGYDY | 4.04 |
| Aa1a,b | DEDVTPRVMGVIPHADH | 0.03 |
| A23a,b | DEDVTPRGMGVIPYAEY | 16.94 |
| A10 c | DDGEYVIPSDQNEYEF | 76.34 |
| Aa12 c | DDGEYVIPSDQNEYEF | 30.27 |
| Aa6 a | SSDYRWSRQPFEFEN | 0.52 |
| Aa9 a | DFDTPWGASGRYDY | 4.08 |
| A8 a | DEDLLPSFVSDFDY | 229.9 |
| A21 | DLGSVGPGAEYDY | 60.74 |
| A19 a | DSYVDYEDDRLK | 4.65 |
| Aa8 a | HWDYGLGPE | 112.09 |
| Aa5 a | VSTDWTTDY | 207 |
| Aa11 | WSLEEQY | 76.08 |
VHH which inhibit the catalytic activity of BoNT/A Lc.
VHH Aa1 and A23 have the same CDR1 sequence and highly related CDR3 sequence.
VHH A10 and Aa12 have the same CDR3 sequence but different CDR1 and CDR2 sequences.
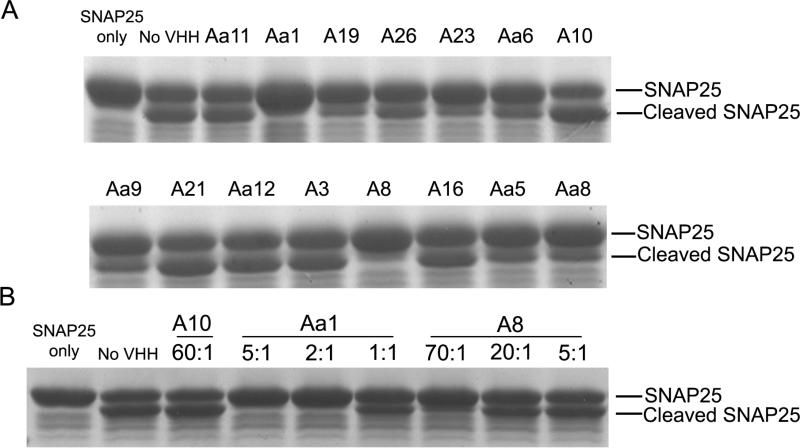
To determine whether any of the VHH inhibited the cleavage of SNAP25 by BoNT/A Lc, each VHH was subcloned for expression in E. coli and purified by IMAC to greater than 90% purity. Each purified VHH was then evaluated for its ability to prevent the cleavage of a GST-SNAP25141-206 fusion protein as determined by SDS-PAGE (Figure 2a). Eight of fifteen clones (Aa1, A19, A23, Aa6, Aa9, A8, Aa5, and Aa8) showed partial or complete inhibition of cleavage (Figure 2A). Not surprisingly, higher affinity VHH inhibited cleavage at lower molar ratios to BoNT/A Lc compared to lower affinity VHH (Figure 2B).
Figure 2. SDS-PAGE analysis of VHH inhibition of GST-SNAP cleavage by BoNT/A Lc.
(A) Inhibitory effect of 15 unique VHH fragments. Each VHH was incubated in a 50 fold molar excess over BoNT/A Lc for 3 min followed by the addition of GST-SNAP25141-206. After 10 min of incubation, SNAP25 cleavage was analyzed by SDS-PAGE. (B) Effect of molar ratio on GST-SNAP25 cleavage. Two inhibitory and one non-inhibitory VHH were incubated with varying fold molar excesses over BoNT/A Lc (1:1 to 70:1) for 30 min followed by the addition of GST-SNAP25141-206. After 10 min of incubation, SNAP25 cleavage was analyzed by SDS-PAGE.
Aa1 VHH binding affinity and potency of BoNT/A Lc inhibition
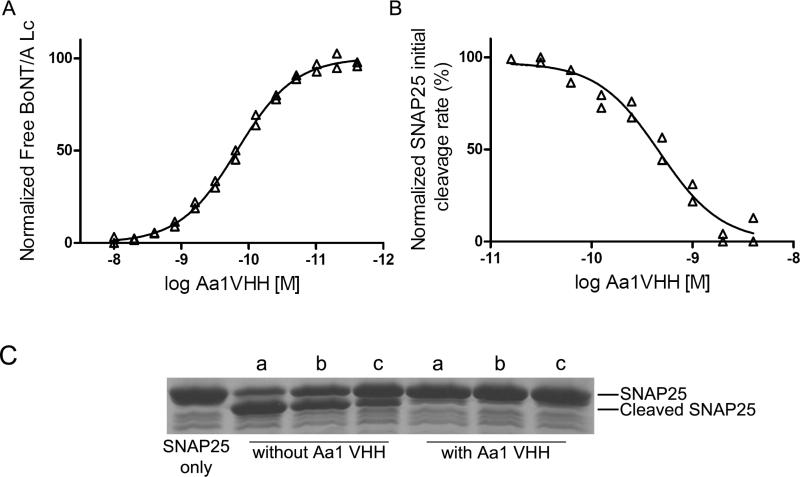
The solution KD and binding kinetics of the Aa1 VHH were determined by flow fluorimetry in a KinExA instrument (Figure 3A). The KD was measured to be 1.47 × 10-10 M (95% confidence interval 1.32 × 10-10 M 1.66 × 10-10 M), with a kon as 4.39 × 107 M-1s-1 and a koff calculated to be 6.66 × 10-3 s-1. The VHH solution KD is within 5 fold of the KD measured for the yeast displayed VHH by flow cytometry. To determine the potency of BoNT/A Lc inhibition, the IC50 of the Aa1 VHH was measured by FRET using as a substrate CFP and YFP connected through SNAP25 residues 141-206 (YsCsY) 32. The initial cleavage rate as a function of VHH concentration was used to calculate the IC50 which was determined to be 4.7 × 10-10 M with a 95% confidence interval of 3.7 × 10-10 M to 6.0 × 10-10 M (Figure 3B).
Figure 3. Characterization of the Aa1 VHH fragment.
(A) Solution KD. The solution KD of the purified Aa1 VHH fragment was measured by flow fluorimetry in a KinExA instrument. (B) Aa1 VHH fragment IC50 for GST-SNAP25141-206 cleavage by BoNT/A Lc. The indicated Aa1 VHH concentration was incubated with BoNT/A Lc and the FRET substrate YsCsY and the initial rate of cleavage determined from the change in the YFP fluorescence reading. IC50 was determined by fitting the initial rate and log Aa1 VHH concentration to a sigmoidal dose-response (variable slope) model. (C) SDS-PAGE analysis of the impact of reducing agents on Aa1 VHH inhibition of GST-SNAP cleavage by BoNT/A Lc. The Aa1 VHH was incubated with no reducing agent (a), 20 mM glutathione reduced (b), or 14 mM β-mercaptoethanol (c) for 15 min at 37°C followed by addition of BoNT/A Lc and GST-SNAP25141-206. After 15 min, cleavage was analyzed by SDS-PAGE.
Stability of the Aa1 VHH fragment
The ability of the Aa1 VHH to withstand the intracellular reducing environment was determined by incubating the Aa1 VHH with either 20 mM glutathione or 14 mM β-mercaptoethanol at 37°C, followed by incubation with the BoNT/A Lc and GST-SNAP25141-206. The Aa1 VHH was still able to prevent BoNT/A Lc cleavage of SNAP25 after a 15 min incubation with either reducing agent (Figure 3C).
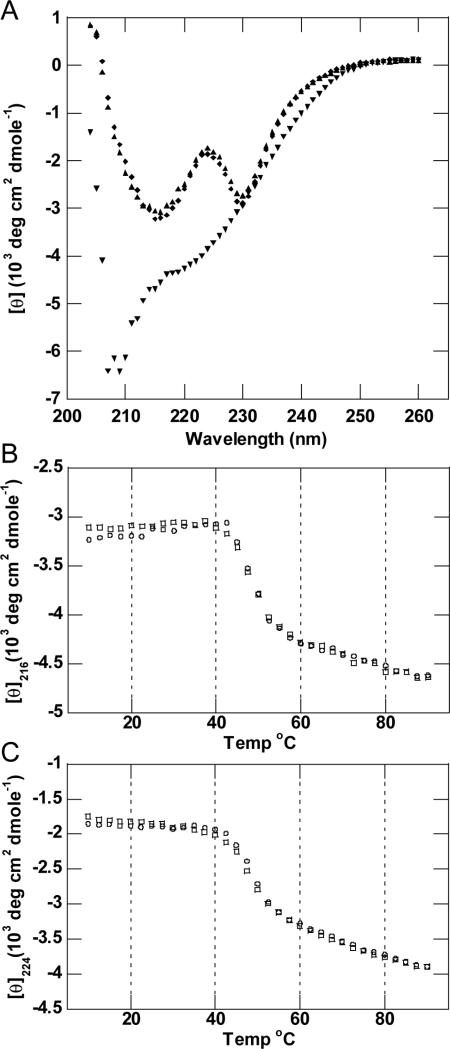
CD was utilized to test the thermostability of the Aa1 VHH fragment. The far UV CD spectrum of the Aa1 VHH protein displayed negative minima at 216 and 230 nm and a maximum at 224 nm (Figure 4A). These features in the CD spectrum are consistent with the mixture of β-sheet and α-helical secondary structure observed in immunoglobulin domains. To investigate the melting temperature and potential reversibility of thermal denaturation, changes in the protein conformation were monitored by CD at wavelengths of 216 and 224 nm (Figures 4B & 4C). These melting profiles revealed two-state unfolding kinetics with Tms of ~49°C as determined by the minima of the first derivative of θ versus T plots (data not shown). The protein sample incubated at 90°C visually appeared identical to the protein at 10°C and did not show any evidence of aggregation or precipitation. Wavelength spectra recorded at 90°C looked significantly different from the spectrum at 10°C and revealed a minimum at ~208 nm. The differences in spectra obtained at 10°C to 90°C are consistent with a transition to mostly random coil with a small component of residual secondary structure. The Aa1 VHH protein was then cooled slowly and allowed to refold (Figures 4B & 4C). The refolding profile coincided well with the temperature induced unfolding profile, and wavelength spectra recorded at 10°C before and after the melting experiment were superimposable (Figure 4A), indicating the protein refolded properly.
Figure 4. Thermal denaturation and refolding of Aa1 VHH.
(A) Far UV CD spectra of Aa1 VHH obtained at 10°C (◆) before melting, 90°C (▼) after melting, and 10°C (▲) following the melting and refolding of the protein. (B) (C) Thermal denaturation (○) and refolding (□) data of Aa1 VHH obtained by CD spectroscopy at a wavelength of 216 nm (panel B) and 224 nm (panel C).
The CD melting experiment was also performed in the presence of 1mM TCEP to explore the importance of the single disulfide bond connecting both halves of the immunoglobulin β-sandwich. The wavelength spectrum recorded at 10°C before any heating was essentially identical to the spectrum in the absence of TCEP (Supplemental). The melting profile recorded at 216 nm was similar to the non-reduced profiles with the primary melting transition occurring ~50°C, although the elipticity trend following this transition had a positive slope, unlike the non-reduced sample (Supplemental Figure S1 and Figures 4B & 4C). In this case, the refolding CD profile of the reduced Aa1 VHH was very different from the melting profile, and the wavelength spectrum recorded at 10°C after the melting experiment confirmed that the structure was significantly perturbed. These data suggest that the disulfide linkage within the single immunoglobulin domain is essential for the refolding process following thermal denaturation.
Structure of BoNT/A Lc - Aa1 VHH complex
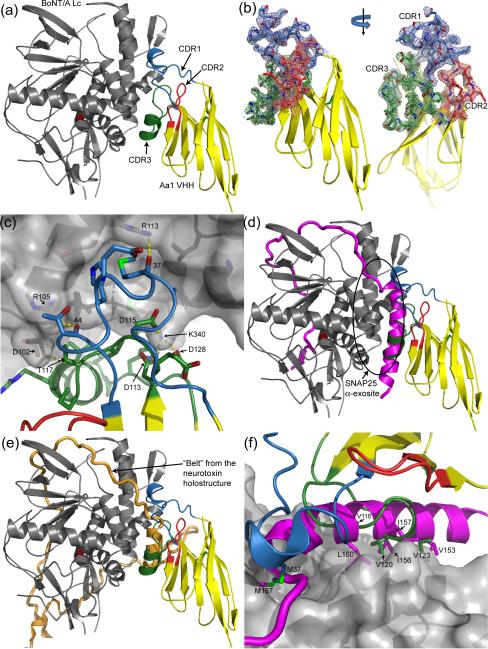
To understand the mechanism by which Aa1 inhibits BoNT/A Lc, the X-ray crystal structure of the BoNT/A Lc425 - Aa1 VHH complex was determined at 2.6 Å resolution (Figure 5A). The asymmetric unit contains a single BoNT/A Lc endopeptidase bound by the Aa1 VHH fragment in a 1:1 stoichiometry. The VHH fragment consists of a single immunoglobulin domain with three CDRs which were well defined by the experimental electron density (Figure 5B). Binding by the llama antibody fragment is driven by CDR1 and CDR3 interactions resulting in the burial of ~619 Å2 of solvent exposed surface area from the interface of each protein (Figure 5A; calculated by PISA) 33. The structure of CDR1 appears to be unique as it is 11 amino acids larger than most VHH structures (eight amino acids longer than the longest VHH structure in the PDB) giving it a topology not seen in other VHH structures (Figure 6, Supplemental Figure S2). A single sidechain interaction between Arg68 of CDR2 and a symmetry related endopeptidase molecule exists in the crystal lattice, but is likely to be a crystallization artifact and is unlikely to contribute to the observed binding affinity measured by FACS. The paratope formed by CDR1 and CDR3 is convex in shape facilitating interactions with a relatively concave epitope located on the opposite side of the endopeptidase active site. This interaction is stabilized by several hydrogen bonds (VHH residues 37, 44, 117, and 124 interactions with BoNT/A Lc residues 113,105,102, and 356, respectively) and salt bridges (Lc Lys340 interactions with VHH Asp residues 113, 115, and 128) between Aa1 and the endopeptidase, and numerous van der Waals and hydrophobic interactions (Figure 5C).
Figure 5. Structure of the BoNT/A Lc - Aa1 VHH complex.
(A) BoNT/A Lc endopeptidase in gray complexed with the Aa1 VHH fragment in yellow with the CDR1, CDR2, and CDR3 regions colored blue, red, and green, respectively. The catalytic zinc is depicted as a red sphere in all figures. (B) Surface representation of the BoNT/A Lc highlighting the Aa1 VHH binding site. Six hydrogen bonds between the Lc and the Aa1 VHH fragment are indicated with yellow dashes. (C) The SNAP25 natural substrate colored in magenta from PDB code 1XTG superimposed onto the BoNT/A Lc – Aa1 VHH complex. The α-helical portion of SNAP25 that binds to the BoNT/A Lc α-exosite coincides with the α-helical tips of CDR1 and CDR3. (D) The same superposition from panel (C) highlighting the amino acid conservation between the SNAP25 α-exosite binding region and the Aa1 VHH fragment. (E) The “belt” from the BoNT holostructure colored orange (from PDB code 3BTA), superimposed onto the BoNT/A Lc – Aa1 VHH complex. The α-helical tips of CDR1 and CDR3 coincide with an α-helical portion of the “belt” in a fashion similar to the SNAP25 / VHH superposition shown in panel (C).
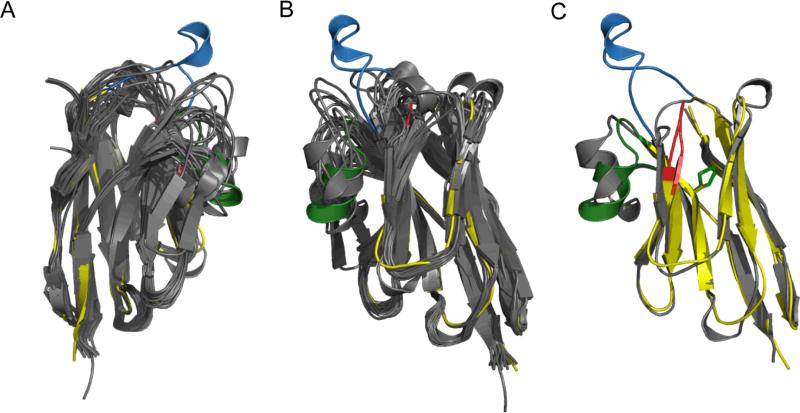
Figure 6. Structural Comparison between Aa1 VHH and the available VHH structures in the PDB databank.
(A) Structural alignment of the available VHH fragment structures in the PDB colored gray with Aa1 VHH colored yellow with the CDRs 1, 2, 3, colored blue, red and green, respectively. The unique CDR1 of Aa1 VHH forms an extended loop with a small α-helix at the tip. All structural alignments were performed using the combinatorial extension (CE) method 70 and the PDB codes are listed in the amino acid sequence alignment of Supplemental Figure S2. (B) 180° rotation (along the Y-axis) of the superposition shown in panel (A). (C) CE structural alignment of the VHH fragment from PDB code 1F2X colored gray and Aa1 VHH colored the same as in panels (A) and (B) with an RMSD of 2.0 Å.
Superposition of this BoNT/A Lc425- Aa1 VHH structure with the structure of SNAP25 bound to the Lc enzyme (PDB code 1XTG; 27) reveals that the Aa1 paratope coincides with an alpha helical portion of the SNAP25 substrate (Figure 5D). Both of these compete for a groove on the surface of the endopeptidase termed the α-exosite, which is occupied by the heavy chain belt of the neurotoxin holostructure (Figure 5E; PDB code 3BTA) 4. The CDR1 and CDR3 both form small alpha helices that appear to mimic the secondary structure of SNAP25 bound at the α-exosite. Although there is no primary sequence homology, the substrate mimicry is extended to tertiary structure because a number of hydrophobic residues found on the VHH fragment coincide with similar residues found on SNAP25 (Figure 5F). These residues include Met37 on CDR1 of the VHH fragment which occupies the position of Met167 from SNAP25; Val116 from CDR3 which coincides with Leu160 of SNAP25; Val120 of CDR3 which occupies the space of Ile156 and Ile157 from SNAP25; and Val123 of CDR3 which replaces Val153 of the natural substrate. Because mutagenesis studies have indicated that the α-exosite is important for proper binding and cleavage of SNAP25 27; 34; 35, binding by Aa1 likely inhibits substrate cleavage by precluding efficient binding of SNAP25 and positioning of its scissile bond.
DISCUSSION
The results demonstrate the ability to isolate a panel of potent inhibitors of BoNT/A Lc enzymatic activity from a non-immune library of VHH displayed on yeast. While other enzyme inhibitors have been isolated from immune VHH phage displayed libraries, we believe this to be the first example of the successful use of non-immune VHH repertoires to generate such inhibitors, and also the first example of the display and selection of VHH libraries on yeast. As with non-immune scFv and Fab repertoires displayed on filamentous phage 36 or yeast 37, non-immune VHH libraries offer the ability to generate ligands without the need to make a new library for every antigen. In addition, immunization can result in a limited repertoire of antibodies directed at immunodominant epitopes, whereas non-immune libraries may provide more diverse epitopic coverage 38.
While we cannot find prior reports of yeast displayed VHH libraries, at least twelve previous publications describe generation of antigen binding VHH from non-immune camelid libraries displayed on filamentous phage (see Tables 1 and 2 in Wesolowski et al., 39 and Monegal et al., 40). VHH phage library size in these publications ranged from 5.4 × 108 to 5 × 109 with an average of 5.2 VHH generated to each of the 23 antigens used for selection. Affinities of 31 of these VHH were reported for ten different antigens and ranged from 0.15 nM to 7750 nM with a mean of 875 nM and a median of 138 nM. In contrast, from our library of 6.1 × 107 VHH displayed on yeast, 15 BoNT/A Lc binding VHH were isolated with an average KD of 56 nM and a median KD of 16.9 nM. While the affinities for the yeast displayed VHH were measured with the VHH on the yeast surface, these are likely to be comparable to the KD measured for native VHH as we previously reported for yeast displayed scFv and native scFv 41. Theory 42, and results in the case of phage displayed non-immune antibody fragment repertoires 36; 43; 44, indicate that larger libraries yield more antibodies of higher affinity than smaller libraries. In contrast, both the numbers of VHH isolated and their average affinities from the relatively small yeast displayed library compare favorably to those from much larger phage displayed VHH libraries (see above). One potential explanation is that yeast displayed libraries are functionally larger than phage displayed libraries, perhaps as a result of using a eukaryotic versus prokaryotic expression system for a eukaryotic protein. This conclusion was the one drawn by the authors of the only example comparing the quantity and quality of antibody fragments from phage versus yeast libraries 45. In this study, three times more antibody fragments were isolated from the yeast displayed repertoire compared to the same repertoire displayed on phage.
Table 2.
Summary of Crystallographic Data
| BoNT/A Lc425 – Aa1 VHH | |
|---|---|
| PBD Code | 3K3Q |
| Space group | C2 |
| Unit Cell (Å) | a=168.2, b=48.8, c=103.6 |
| X-ray Source | APS GM/CA, 1.03 Å λ |
| Resolution Limits (Å) | 40.0-2.6 (2.64-2.60) |
| Reflections | |
| Total Collected | 64,075 |
| Unique | 22,641 |
| Redundancy | 2.8 (1.9) |
| I/σ | 7.3 (2.2) |
| Completeness (%) | 96.5 (83.9) |
| Rsym (%) | 11.7 (31.4) |
| Refinement | |
| Resolution Range (Å) | 40.0 – 2.6 |
| R-work (%) | 21.8 |
| R-free (%) | 25.6 |
| Ramachandran | |
| Favored | 388 |
| Allowed | 86 |
| Generous | 0 |
| Disfavored | 0 |
Data in parentheses are for the highest resolution shell.
R-free was computed using 5% of the data.
Ramachandran statistics do not include Gly and Pro residues.
An emerging feature of single domain camelid (camels, dromedaries, and llamas) VHH is their propensity to recognize concave epitopes by relatively convex shaped paratopes. The Aa1 VHH binds via alpha helices in the CDR1 and CDR3 to the BoNT/A Lc α-exosite groove in a manner similar to an α-helix in the BoNT/A belt and to the α-helix in the SNAP25 substrate. In fact, a number of the amino acid side chains in the VHH which contact the BoNT/A Lc are the same contact side chains in SNAP25. Such conservation of ligand binding sites are a feature of a number of protein-protein interactions including hormones and their receptors 46 and the common binding site in human immunoglobulin for Protein A, Protein G, rheumatoid factor, and neonatal Fc receptor 47. In some instances, many of the same contact residues are used in the contacts between receptor and ligand 47; 48. These same sites are also sites bound by ligands isolated from peptide and polypeptide libraries 48; 49. Such sites also have obvious functional significance; in the case of SNAP25 and the Aa1 VHH, this work experimentally validates the α-exosite as a target for inhibiting the enzymatic activity of the BoNT/A Lc, and perhaps BoNT/C and BoNT/E, which also cleave SNAP25.
The BoNT Lc is unique among endopeptidases in that it requires relatively long substrates for efficient catalytic efficiency due to its recognition strategy. Exosites far from the scissile bond determine substrate specificity, and mutation of these sites, or truncation of the substrate regions that normally interact with these exosites substantially decrease the catalytic efficiency 27. Inhibiting catalysis by binding to exosites can potentially overcome specificity issues of active site inhibitors, which may bind to and inhibit proteases with similar structures. Exosite inhibitors have been developed against a number of proteases including matrix metalloproteinase 2 50, β-amyloid precursor cleaving enzyme, coagulation factor VIIa 51, activated protein C 28, and pregnancy associated plasma protein A 52. Structurally, these inhibitors include scFv antibodies, DNA aptamers, and peptides isolated from combinatorial libraries. In the case of intracellular enzymes, like the BoNT/A Lc, getting polypeptides like VHH and recombinant antibodies into cells is a therapeutic challenge requiring a gene therapy or drug delivery approach. However, the high thermal stability and resistance to reducing conditions may make the VHH optimal for such approaches. Alternatively, exosites defined structurally by VHH ligands can be used as targets for small molecule drug development 53. Development of small molecules interrupting the large binding surface typical of protein-protein interactions has proven a challenge, however the energetics of protein-protein binding are frequently dominated by interactions between a few amino acids on receptor and ligand whose size is comparable to the size of small molecules 47; 54. Such “hot spots” 54; 55 can be identified by a number of alanine-scanning techniques and used as targets for small molecule drug development 56; 57.
In summary, we have shown that non-immune llama VHH libraries displayed on yeast can provide a panel of ligands binding and inhibiting the enzymatic activity of BoNT/A Lc. One of these experimentally defines the α-exosite as a potential target for the development of drugs that can be used to treat botulism after the toxin has entered the neuron, which may prove to be more specific for the neurotoxin
MATERIALS AND METHODS
Oligonucleotides for library construction
Strains, media, antibodies, and toxins
Saccharomyces cerevisiae strain EBY100 was maintained in YPD medium. EBY100 transformed with expression vector pYD2 41 was selected on SD-CAA medium. VHH yeast surface display was induced as described. E. coli DH5α, was used for cloning and preparation of plasmid DNA. Pure BoNT/A1 (Hall hyper) was purchased from Metabiologics (Madison, WI). Mouse anti-SV5 antibody was purified from hybridoma supernatant using Protein G and directly labeled with Alexa-647 using a kit (Molecular Probes). Human BoNT/A antibodies 3D12 and AR2 were purified from Chinese hamster ovary cell (CHO) supernatants 41; 58.
Expression and purification of proteins
BoNT/A Lc (residues 1-448) was amplified from a synthetic BoNT/A1 gene 59, subcloned into an IPTG inducible pET15b vector, and expressed in BL21 (DE3) cells at 18°C overnight. The cells were broken with Bugbuster Master (Novagen), and hexahistidine tagged BoNT/A Lc was purified by immobilized metal affinity chromatography (IMAC) using Ni-NTA agarose (Qiagen) followed by cation exchange chromatography. BoNT/A Lc425, (residues 1-425) was constructed and expressed similarly. BoNT/A Lc425 was purified by IMAC. The SNAP25141-206-pGEX-2T construct was a gift from Dr. Joseph Barbieri (Medical College of Wisconsin, Milwaukee, WI) 60 and was expressed in BL21 (DE3) cells with 0.5 mM IPTG at 18°C overnight. GST-SNAP25 was purified on a glutathione-sepharose 4B column (Clontech).
Naïve llama VHH library construction
Total RNA was isolated from 40 ml fresh blood from three llamas using PAX gene Blood RNA kit (PreAnalytiX, QIAGEN/BD). The cDNA was synthesized by RT-PCR using a ThermoScript RT-PCR Kit (Invitrogen), with oligo dT and random hexamer as primers. The gene fragment encoding the heavy chain variable domains was PCR amplified with primers annealing at the leader sequence and at the CH2 exon of the llama heavy chains, (leader-L01 and CH2-L01; see Oligonucleotides for library construction in the Supplemental Materials for the sequences). The PCR product showed two bands, ~550bp and 800bp, on an agarose gel and the 550bp fragment (VHH-CH2 without CH1) was gel extracted and reamplified with primers GAP5-VHHBack and GAP3-VHHForward. Approximately, 22 μg of VHH gene and 65 μg of NcoI-NotI-digested pYD2 vector was used to transform lithium acetate treated EBY100 cells 61. Library size was determined by plating the transformation mixture on SD-CAA plates. VHH display was induced by culturing in SG-CAA media.
Selection of BoNT/A Lc binding VHH
For flow sorting, 200 nM of BoNT/A Lc was used for the first two rounds, and 100 nM was used for the third round. The volume of the incubation was chosen to ensure that BoNT/A Lc was in at least a fivefold excess over the number of VHH (assuming 5 × 105 VHH/yeast), and the incubation times were 30 min. All the following washing and staining steps were performed at 4°C using ice-cold FACS buffer (phosphate-buffered saline (pH 7.4), 0.5% bovine serum albumin). After incubation with BoNT/A Lc and washing in FACS buffer, the yeast sample was incubated with the human BoNT/A Lc -specific IgG1 monoclonal antibodies (mAb) ING2 and 5A20.4 (59 and R. Levy, unpublished) which bind non-overlapping epitopes (1 μg/ml), followed by incubation with 1 μg/ml PE-labeled goat anti-human Fc antibody (Jackson Immunogenetic) and 1 μg/ml Alexa-647-labelled anti-SV5 mAb. Cells were sorted on a FACSAria II. The VHH expressing (SV5 positive) and BoNT/A Lc binding population were gated for collection. Collected cells were grown and induced for the next round of sorting. Yeast from the third round of sorting were plated on SD-CAA plates, and colonies on the plates were picked and grown in 96 deep-well plates. The individual clones were then induced and screened for BoNT/A Lc binding and binding clones were identified by DNA sequencing.
Expression and purification of VHH
The genes of the selected clones that bound to BoNT/A Lc were subcloned into the expression vector pSYN1 62, transformed into E. coli TG1 cells and expression of VHH induced as described using 0.2 mM IPTG 62. Periplasmic proteins were extracted by osmotic shock and hexahistidine tagged VHH proteins were purified by IMAC on Ni-NTA agarose 62. To increase the expression level for crystal growing, the Aa1 clone was subcloned into pET20b (Novagen), and transformed into Rosetta (DE3) (Novagen). The expression and purification were performed as above.
Measurement of yeast-displayed VHH affinity for BoNT/A Lc
The equilibrium dissociation constant (KD) of yeast displayed VHH was measured by flow cytometry as previously described for yeast displayed single chain Fv (scFv) 41; 63. Briefly, yeast displaying VHH were incubated with one of six different BoNT/A Lc concentrations spanning the expected KD. BoNT/A Lc binding was detected by incubating with mAbs ING2 and 5A20.4 followed by PE-anti-human Fc and Alexa-647 anti-SV5 mAb. To determine KD, the fluorescence and BoNT/A Lc concentration data was then fit into equation:
Where Fback is the fluorescence intensity when there is no BoNT/A Lc, Fmax is the fluorescence intensity when binding is saturated, and [L] is the concentration of BoNT/A Lc.
Measurement of the Aa1 VHH solution phase affinity at equilibrium and binding kinetics
The Aa1 VHH solution KD for BoNT/A Lc was measured by flow fluorimetry in a KinExA instrument as previously described for IgG 41; 63. Briefly, Aa1 VHH was serially diluted into a constant concentration of BoNT/A Lc. After reaching equilibrium, samples were passed over a flow cell with a 4 mm column of Azlactone beads (Sapidyne Instruments) covalently coated with Aa1 VHH to capture the free BoNT/A Lc which was quantitated by flowing Alexa-647 labeled BoNT/A Lc mAb 5A20.4 over the beads. The equilibrium titration data were fit to a reversible binding model using GraphPad Prism Software to determine the KD. To measure the association rate constant (kon), an Aa1 VHH and BoNT/A Lc reaction mixture was passed over a flow cell with Aa1 VHH coated beads. The amount of BoNT/A Lc bound to the beads was quantitated as described above. The exponential decrease in the concentration of free BoNT/A Lc as a function of time was fit to a standard bimolecular rate equation using the KinExA Pro software to determine the kon64. The dissociation rate constant (koff) was calculated as KD × kon.
Cleavage of SNAP25 by BoNT/A Lc
BoNT/A Lc and VHH proteins were combined in 50 mM Tris buffer, pH 8.0 and GST-SNAP25141-206 was added to initiate the reaction. The final concentration was approximately 20 nM for BoNT/A Lc and 5 μM for GST-SNAP25141-206. The concentration of each VHH was at least 50 fold higher than the BoNT/A Lc concentration. The reaction was run at room temperature for 10 min or for the indicated time, stopped by adding SDS-PAGE loading buffer, heated for 10 min at 99°C, then analyzed by SDS-PAGE. The gel was stained in 0.1% Coomassie Blue R-250.
IC50 measurement
The Aa1 50% inhibitory concentration (IC50) for BoNT/A Lc was determined by using a fluorescent resonance energy transfer (FRET) based cleavage assay with YsCsY as substrate as previously described 32. Reactions were performed in black 96-well plates (Corning) in assay buffer (10 mM HEPES, 150 mM K glutamate, 0.01% Tween20, pH 7.2) (modified from 65). YsCsY was mixed with two-fold serially diluted Aa1 VHH, with the estimated IC50 equal to the median VHH concentration. After pre-incubation at 30°C for 15 min, BoNT/A Lc was added to the wells to initiate the reaction. The final concentration was 0.5 μM for YsCsY and 400 pM for BoNT/A Lc. Fluorescence was measured in the monochromatic mode with excitation at 425 nm and emission at 525 nm in a fluorescence reader (Spectra Max Gemini, Molecular Devices). Initial rates were determined from the change in YFP fluorescence (525 nm); the fluorescent data from the first 40 sec was fit to a simple linear regression model Y = RX + C (where Y = YFP fluorescence, R = slope, X = time, and C = y-intercept) and the “-R” value was taken as the initial rate (the R value was negative since the signal at 525 nm gets weaker with substrate cleavage). The Aa1 IC50 was determined by fitting the initial rate and log Aa1 concentration to a sigmoidal dose-response (variable slope) model (GraphPad Prism).
CD spectroscopy
All circular dichroism (CD) experiments were performed on an Aviv model 202SF circular dichroism spectrometer with a 0.2 cm path length cell containing 18-20 μM Aa1 VHH protein in PBS buffer. Wavelength data were collected in 1 nm steps with an averaging time of 15 sec. Thermal denaturation/refolding experiments were run from 10 to 90°C in 2.5 degree steps, at a two-degree/minute rate of change with 1.5 min equilibration and 30 sec data averaging at each temperature. Tm values were obtained from the minima of the first derivative of θ versus T plots.
Crystallization and structure determination
The BoNT/A Lc425 - Aa1 VHH complex was formed by mixing at a 1:1.5 molar ratio followed by purification using size exclusion chromatography on a Superdex 200 column (GE Healthcare), with running buffer of 100 mM NaCl and 10 mM HEPES (pH 7.0). The complex eluate was then concentrated to 13 mg/mL by centrifugal ultrafiltration (Amicon). Crystals were grown by sitting nanodrop vapor diffusion at 20°C using 13 mg/ml total protein sample. Crystals grew in 4 days with a precipitant/well solution containing 25% ethylene glycol and were flash frozen in liquid nitrogen directly from the sitting drop. Diffraction data were collected at the Advanced Photon Source GM/CA CAT beamline 23ID-B (Argonne, Chicago IL). Reflections were processed using HKL2000 66 and the initial structure solution was obtained by molecular replacement with Phaser 67 using the light chain from PDB code 3BTA (residues 2:425) and a polyalanine model of the llama VHH domain from PDB code 1I3V as search models. Cycles of refinement and manual rebuilding were performed with CNS 68 and Coot, respectively 69. Crystallographic details are listed in Table 2 and structure figures were generated with Pymol Molecular Graphics System (www.pymol.org).
Supplementary Material
Acknowledgments
Funding: This work was partially supported by National Institutes of Health cooperative agreements U01 AI056493 (J.D.M.) and U01 AI075502 (B.A.W.), Defense Threat Reduction Agency contract 1-07-C-0030 (J.D.M.), the Centers for Disease Control and Prevention contract 200-2006-16697 (J.D.M.), the Pacific Southwest Regional Center of Excellence U54 AI065359 (R.C.S. and J.D.M.), and the Great Lakes Regional Center of Excellence U54 AI057153 (B.A.W.).
The abbreviations used are
- BoNT
botulinum neurotoxin
- BoNT/A
botulinum neurotoxin serotype A
- BoNT/A Lc
botulinum neurotoxin serotype A light chain residues 1-448
- BoNT/A Lc425
truncated BoNT/A Lc containing residues 1-425
- CD
circular dichroism
- CDR
complementarity determining region
- Fab
antigen binding fragment of immunoglobulin with variable domain and first constant domain
- FACS
fluorescent activated cell sorting
- FRET
fluorescence resonance energy transfer
- Hc
the C-terminal portion of the botulinum neurotoxin heavy chain
- Hn
the N-terminal portion of the botulinum neurotoxin heavy chain
- IC50
50% inhibitory concentration
- IgG
immunoglobulin G
- IPTG
isopropyl-β-D-thiogalactopyranoside
- IMAC
immobilized metal affinity chromatography
- KD
dissociation equilibrium constant
- kon
association rate constant
- koff
dissociation rate constant
- mAb
monoclonal antibody
- MFI
mean fluorescent intensity
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- scFv
single chain format of antibody variable regions
- SD-CAA
selective growth dextrose casamino acids media
- SG-CAA media
selective growth galactose casamino acids media
- SNARE
Soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- SNAP25
synaptosome-associated protein of 25,000 daltons
- VH
heavy chain variable region
- VHH
camelid heavy chain variable region derived from heavy chain only antibody
- YsCsY
cyan fluorescent protein (CFP) and yellow fluorescent 206
Footnotes
Accession codes
The coordinates and structure factors have been deposited in the RCSB Protein Data Bank under the accession code 3K3Q.
We thank Angela Walker and Olve Peersen for critical review of the manuscript. We also thank the staff of the GM/CA-CAT beamline at the Advanced Photon Source for assistance with the minibeam, and also Joshua Price and the laboratory of Jeffery Kelly at TSRI for the use and assistance of the CD spectrometer.
REFERENCES
- 1.Arnon SA, Schecter R, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Hauer J, Layton M, Lillibridge S, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Swerdlow DL, Tonat K. Botulinum toxin as a biological weapon. JAMA. 2001;285:1059–1070. doi: 10.1001/jama.285.8.1059. [DOI] [PubMed] [Google Scholar]
- 2.Control C. f. D. Botulism in the United States, 1899-1998. handbook for epidemiologists, clinicians,and laboratory workers. Atlanta, Georgia U.S. Department of Health and Human Services, Public Health Service. 1998 downloadable at http://www.bt.cdc.gov/agent/botulism/index,asp.
- 3.Gill MD. Bacterial toxins: a table of lethal amounts. Microbiol. Rev. 1982;46:86–94. doi: 10.1128/mr.46.1.86-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacy DB, Tepp W, Cohen AC, DasGupta BR, Stevens RC. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nature Struct. Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- 5.Montecucco C, Schiavo G. Structure and function of tetanus and botulinum neurotoxins. Quart. Rev. Biophys. 1995;28:423–472. doi: 10.1017/s0033583500003292. [DOI] [PubMed] [Google Scholar]
- 6.Simpson LL. Kinetic studies on the interaction between botulinum toxin type A and the chloinergic neuromuscular junction. J. Pharmacol. Expt. Ther. 1980;212:16–21. [PubMed] [Google Scholar]
- 7.Dolly JO, Black J, Williams RS, Melling J. Acceptors for botulinum neurotoxin reside on motor nerve terminals and mediate its internalization. Nature. 1984;307:457–460. doi: 10.1038/307457a0. [DOI] [PubMed] [Google Scholar]
- 8.Dong M, Yeh F, Tepp WH, Dean C, Johnson EA, Janz R, Chapman ER. SV2 is the protein receptor for botulinum neurotoxin A. Science. 2006;312:592–6. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- 9.Mahrhold S, Rummel A, Bigalke H, Davletov B, Binz T. The synaptic vesicle protein 2C mediates the uptake of botulinum neurotoxin A into phrenic nerves. FEBS Lett. 2006;580:2011–4. doi: 10.1016/j.febslet.2006.02.074. [DOI] [PubMed] [Google Scholar]
- 10.Fischer A, Montal M. Single molecule detection of intermediates during botulinum neurotoxin translocation across membranes. Proc Natl Acad Sci U S A. 2007;104:10447–52. doi: 10.1073/pnas.0700046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schiavo G, Benfenati F, B. P, Rossetto O, Polverino d. L., P., DasGupta BR, Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 12.Schiavo G, Rossetto O, Catsicas S, Polverino d. L., P., DasGupta BR, Benfenati F, Montecucco C. Identification of the nerve terminal targets of botulinum neurotoxin serotypes A, D, and E. J. Biol. Chem. 1993;268:23784–23787. [PubMed] [Google Scholar]
- 13.Franz DR, Pitt LM, Clayton MA, Hanes MA, Rose KJ. Efficacy of prophylactic and therapeutic administration of antitoxin for inhalation botulism. In: DasGupta BR, editor. Botulinum and Tetanus Neurotoxins: Neurotransmission and Biomedical Aspects. Plenum Press; New York: 1993. pp. 473–476. [Google Scholar]
- 14.Black RE, Gunn RA. Hypersensitivity reactions associated with botulinal antitoxin. Am. J. Med. 1980;69:567–570. doi: 10.1016/0002-9343(80)90469-6. [DOI] [PubMed] [Google Scholar]
- 15.Hibbs RG, Weber JT, Corwin A, Allos BM, Abd el Rehim MS, Sharkawy SE, Sarn JE, McKee KTJ. Experience with the use of an investigational F(ab')2 heptavalent botulism immune globulin of equine origin during an outbreak of type E origin in Egypt. Clin. Infect. Dis. 1996;23:337–340. doi: 10.1093/clinids/23.2.337. [DOI] [PubMed] [Google Scholar]
- 16.Arnon SS, Schechter R, Maslanka SE, Jewell NP, Hatheway CL. Human botulism immune globulin for the treatment of infant botulism. N Engl J Med. 2006;354:462–71. doi: 10.1056/NEJMoa051926. [DOI] [PubMed] [Google Scholar]
- 17.Ravichandran E, Gong Y, Al Saleem FH, Ancharski DM, Joshi SG, Simpson LL. An initial assessment of the systemic pharmacokinetics of botulinum toxin. J Pharmacol Exp Ther. 2006;318:1343–51. doi: 10.1124/jpet.106.104661. [DOI] [PubMed] [Google Scholar]
- 18.Hall YH, Chaddock JA, Moulsdale HJ, Kirby ER, Alexander FC, Marks JD, Foster KA. Novel application of an in vitro technique to the detection and quantification of botulinum neurotoxin antibodies. J Immunol Methods. 2004;288:55–60. doi: 10.1016/j.jim.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi T, Joshi SG, Al-Saleem F, Ancharski D, Singh A, Nasser Z, Simpson LL. Localization of the sites and characterization of the mechanisms by which anti-light chain antibodies neutralize the actions of the botulinum holotoxin. Vaccine. 2009;27:2616–24. doi: 10.1016/j.vaccine.2009.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, Stevens RC. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444:1096–100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]
- 21.Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–5. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- 22.Stenmark P, Dupuy J, Imamura A, Kiso M, Stevens RC. Crystal structure of botulinum neurotoxin type A in complex with the cell surface co-receptor GT1b-insight into the toxin-neuron interaction. PLoS Pathog. 2008;4:e1000129. doi: 10.1371/journal.ppat.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickerson TJ, Janda KD. The use of small molecules to investigate molecular mechanisms and therapeutic targets for treatment of botulinum neurotoxin A intoxication. ACS Chem Biol. 2006;1:359–69. doi: 10.1021/cb600179d. [DOI] [PubMed] [Google Scholar]
- 24.Fischer A, Nakai Y, Eubanks LM, Clancy CM, Tepp WH, Pellett S, Dickerson TJ, Johnson EA, Janda KD, Montal M. Bimodal modulation of the botulinum neurotoxin protein-conducting channel. Proc Natl Acad Sci U S A. 2009;106:1330–5. doi: 10.1073/pnas.0812839106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burnett JC, Ruthel G, Stegmann CM, Panchal RG, Nguyen TL, Hermone AR, Stafford RG, Lane DJ, Kenny TA, McGrath CF, Wipf P, Stahl AM, Schmidt JJ, Gussio R, Brunger AT, Bavari S. Inhibition of metalloprotease botulinum serotype A from a pseudo-peptide binding mode to a small molecule that is active in primary neurons. J Biol Chem. 2007;282:5004–14. doi: 10.1074/jbc.M608166200. [DOI] [PubMed] [Google Scholar]
- 26.Park JG, Sill PC, Makiyi EF, Garcia-Sosa AT, Millard CB, Schmidt JJ, Pang YP. Serotype-selective, small-molecule inhibitors of the zinc endopeptidase of botulinum neurotoxin serotype A. Bioorg Med Chem. 2006;14:395–408. doi: 10.1016/j.bmc.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 27.Breidenbach MA, Brunger AT. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature. 2004;432:925–9. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- 28.Muller J, Isermann B, Ducker C, Salehi M, Meyer M, Friedrich M, Madhusudhan T, Oldenburg J, Mayer G, Potzsch B. An exosite-specific ssDNA aptamer inhibits the anticoagulant functions of activated protein C and enhances inhibition by protein C inhibitor. Chem Biol. 2009;16:442–51. doi: 10.1016/j.chembiol.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y, Eigenbrot C, Liang WC, Stawicki S, Shia S, Fan B, Ganesan R, Lipari MT, Kirchhofer D. Structural insight into distinct mechanisms of protease inhibition by antibodies. Proc Natl Acad Sci U S A. 2007;104:19784–9. doi: 10.1073/pnas.0708251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci U S A. 2006;103:4586–91. doi: 10.1073/pnas.0505379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratier L, Urrutia M, Paris G, Zarebski L, Frasch AC, Goldbaum FA. Relevance of the diversity among members of the Trypanosoma cruzi trans-sialidase family analyzed with camelids single-domain antibodies. PLoS One. 2008;3:e3524. doi: 10.1371/journal.pone.0003524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pires-Alves M, Ho M, Aberle KK, Janda KD, Wilson BA. Tandem fluorescent proteins as enhanced FRET-based substrates for botulinum neurotoxin activity. Toxicon. 2009;53:392–9. doi: 10.1016/j.toxicon.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Rossetto O, Schiavo G, Montecucco C, Poulain B, Deloye F, Lozzi L, Shone CC. SNARE motif and neurotoxins. Nature. 1994;372:415–6. doi: 10.1038/372415a0. [DOI] [PubMed] [Google Scholar]
- 35.Washbourne P, Pellizzari R, Baldini G, Wilson MC, Montecucco C. Botulinum neurotoxin types A and E require the SNARE motif in SNAP-25 for proteolysis. FEBS Lett. 1997;418:1–5. doi: 10.1016/s0014-5793(97)01328-8. [DOI] [PubMed] [Google Scholar]
- 36.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization: Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 37.Feldhaus MJ, Siegel RW, Opresko LK, Coleman JR, Feldhaus JM, Yeung YA, Cochran JR, Heinzelman P, Colby D, Swers J, Graff C, Wiley HS, Wittrup KD. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003;21:163–70. doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 38.Amersdorfer P, Wong C, Smith T, Chen S, Deshpande S, Sheridan R, Marks JD. Genetic and immunological comparison of anti-botulinum type A antibodies from immune and non-immune human phage libraries. Vaccine. 2002;20:1640–8. doi: 10.1016/s0264-410x(01)00482-0. [DOI] [PubMed] [Google Scholar]
- 39.Wesolowski J, Alzogaray V, Reyelt J, Unger M, Juarez K, Urrutia M, Cauerhff A, Danquah W, Rissiek B, Scheuplein F, Schwarz N, Adriouch S, Boyer O, Seman M, Licea A, Serreze DV, Goldbaum FA, Haag F, Koch-Nolte F. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immunol. 2009;198:157–74. doi: 10.1007/s00430-009-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monegal A, Ami D, Martinelli C, Huang H, Aliprandi M, Capasso P, Francavilla C, Ossolengo G, de Marco A. Immunological applications of single-domain llama recombinant antibodies isolated from a naive library. Protein Eng Des Sel. 2009;22:273–80. doi: 10.1093/protein/gzp002. [DOI] [PubMed] [Google Scholar]
- 41.Razai A, Garcia-Rodriguez C, Lou J, Geren IN, Forsyth CM, Robles Y, Tsai R, Smith TJ, Smith LA, Siegel RW, Feldhaus M, Marks JD. Molecular evolution of antibody affinity for sensitive detection of botulinum neurotoxin type A. J Mol Biol. 2005;351:158–69. doi: 10.1016/j.jmb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Perelson AS, Oster GF. Theoretical studies of clonal selection: Minimal antibody repertoire size and reliability of self non-self discrimination. J. Theor. Biol. 1979;81:645–670. doi: 10.1016/0022-5193(79)90275-3. [DOI] [PubMed] [Google Scholar]
- 43.Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindquist E, Schier R, Hemingsen G, Wong C, Gerhart JC, Marks JD, Lindqvist E. Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc Natl Acad Sci U S A. 1998;95:6157–62. doi: 10.1073/pnas.95.11.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, McCafferty J, Hodits RA, Wilton J, Johnson KS. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nature Biotech. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 45.Bowley DR, Labrijn AF, Zwick MB, Burton DR. Antigen selection from an HIV-1 immune antibody library displayed on yeast yields many novel antibodies compared to selection from the same library displayed on phage. Protein Eng Des Sel. 2007;20:81–90. doi: 10.1093/protein/gzl057. [DOI] [PubMed] [Google Scholar]
- 46.Wells JA, de Vos AM. Hematopoietic receptor complexes. Annu Rev Biochem. 1996;65:609–34. doi: 10.1146/annurev.bi.65.070196.003141. [DOI] [PubMed] [Google Scholar]
- 47.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–83. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 48.Cunningham BC, Wells JA. Rational design of receptor-specific variants of human growth hormone. Proc Natl Acad Sci U S A. 1991;88:3407–11. doi: 10.1073/pnas.88.8.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kay BK, Kurakin A, Hyde-DeRuyscher R. From peptides to drugs via phage display. Drug Discovery Today. 1998;3:370–378. [Google Scholar]
- 50.Xu X, Chen Z, Wang Y, Bonewald L, Steffensen B. Inhibition of MMP-2 gelatinolysis by targeting exodomain-substrate interactions. Biochem J. 2007;406:147–55. doi: 10.1042/BJ20070591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberge M, Santell L, Dennis MS, Eigenbrot C, Dwyer MA, Lazarus RA. A novel exosite on coagulation factor VIIa and its molecular interactions with a new class of peptide inhibitors. Biochemistry. 2001;40:9522–31. doi: 10.1021/bi010592d. [DOI] [PubMed] [Google Scholar]
- 52.Mikkelsen JH, Gyrup C, Kristensen P, Overgaard MT, Poulsen CB, Laursen LS, Oxvig C. Inhibition of the proteolytic activity of pregnancy-associated plasma protein-A by targeting substrate exosite binding. J Biol Chem. 2008;283:16772–80. doi: 10.1074/jbc.M802429200. [DOI] [PubMed] [Google Scholar]
- 53.Sperandio O, Miteva MA, Segers K, Nicolaes GA, Villoutreix BO. Screening Outside the Catalytic Site: Inhibition of Macromolecular Inter-actions Through Structure-Based Virtual Ligand Screening Experiments. Open Biochem J. 2008;2:29–37. doi: 10.2174/1874091X00802010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–6. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 55.Moreira IS, Fernandes PA, Ramos MJ. Hot spots--a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68:803–12. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 56.Braisted AC, Oslob JD, Delano WL, Hyde J, McDowell RS, Waal N, Yu C, Arkin MR, Raimundo BC. Discovery of a potent small molecule IL-2 inhibitor through fragment assembly. J Am Chem Soc. 2003;125:3714–5. doi: 10.1021/ja034247i. [DOI] [PubMed] [Google Scholar]
- 57.Thanos CD, DeLano WL, Wells JA. Hot-spot mimicry of a cytokine receptor by a small molecule. Proc Natl Acad Sci U S A. 2006;103:15422–7. doi: 10.1073/pnas.0607058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nowakowski A, Wang C, Powers DB, Amersdorfer P, Smith TJ, Montgomery VA, Sheridan R, Blake R, Smith LA, Marks JD. Potent neutralization of botulinum neurotoxin by recombinant oligoclonal antibody. Proc Natl Acad Sci U S A. 2002;99:11346–50. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levy R, Forsyth CM, LaPorte SL, Geren IN, Smith LA, Marks JD. Fine and domain-level epitope mapping of botulinum neurotoxin type A neutralizing antibodies by yeast surface display. J Mol Biol. 2007;365:196–210. doi: 10.1016/j.jmb.2006.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen S, Barbieri JT. Unique substrate recognition by botulinum neurotoxins serotypes A and E. J Biol Chem. 2006;281:10906–11. doi: 10.1074/jbc.M513032200. [DOI] [PubMed] [Google Scholar]
- 61.Gietz RD, Schiestl RH. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:38–41. doi: 10.1038/nprot.2007.15. [DOI] [PubMed] [Google Scholar]
- 62.Schier R, Marks JD, Wolf EJ, Apell G, Wong C, McCartney JE, Bookman MA, Huston JS, Houston LL, Weiner LM, et al. In vitro and in vivo characterization of a human anti-c-erbB-2 single-chain Fv isolated from a filamentous phage antibody library. Immunotechnology. 1995;1:73–81. doi: 10.1016/1380-2933(95)00007-0. [DOI] [PubMed] [Google Scholar]
- 63.Garcia-Rodriguez C, Levy R, Arndt JW, Forsyth CM, Razai A, Lou J, Geren I, Stevens RC, Marks JD. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol. 2007;25:107–16. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- 64.Drake AW, Myszka DG, Klakamp SL. Characterizing high-affinity antigen/antibody complexes by kinetic- and equilibrium-based methods. Anal Biochem. 2004;328:35–43. doi: 10.1016/j.ab.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 65.Vaidyanathan VV, Yoshino K, Jahnz M, Dorries C, Bade S, Nauenburg S, Niemann H, Binz T. Proteolysis of SNAP-25 isoforms by botulinum neurotoxin types A, C, and E: domains and amino acid residues controlling the formation of enzyme-substrate complexes and cleavage. J Neurochem. 1999;72:327–37. doi: 10.1046/j.1471-4159.1999.0720327.x. [DOI] [PubMed] [Google Scholar]
- 66.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 67.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 69.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 70.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–47. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.