Abstract
Ts65Dn mice have segmental trisomy for orthologs of about half of the genes on human chromosome 21, including Ets2. These mice develop anomalies of the cranial skeleton and thymus that parallel those in Down syndrome. Over-expression of the Ets2 transcription factor gene was posited to be sufficient to produce these craniofacial and thymus deficits in transgenic mice that constitutively over-express a processed Ets2 transcript under a promiscuous promoter [Sumarsono et al., 1996; Wolvetang et al., 2003]. Evaluation of trisomic mice with varying copy numbers of a properly regulated Ets2 gene indicated increased dosage of Ets2 was not sufficient to produce effects on thymus and most of the cranial anomalies seen in Ts65Dn mice. However, mesoderm-derived cranial skeletal elements are significantly more affected in Ts65Dn, Ets2+/− compared to Ts65Dn littermates suggesting a differential interaction of Ets2-related processes with mesoderm-derived and neural crest-derived formative tissues. Our results support the growing evidence for interactions among multiple genes contributing to developmental perturbations resulting in variation in complex DS phenotypes.
Keywords: Down syndrome, Ts65Dn, Ets2, thymus, craniofacial
INTRODUCTION
Trisomy 21 is a complex genetic insult and the only autosomal aneuploidy with a high frequency of postnatal survival in humans, affecting 1 in 750 live births. The diverse Down syndrome (DS) phenotypes include traits that are expressed in nearly all individuals with DS including mental retardation, distinct facial morphology and Alzheimer-like pathology, in addition to numerous other traits that may or may not be expressed in an individual with DS [Van Cleve et al., 2006; Van Cleve and Cohen 2006].
The specific genetic mechanisms responsible for producing the diverse complex phenotypes present in individuals with DS are unknown. Numerous genes have been proposed, with the “Down syndrome critical region” (DSCR) model serving as the locus of debate. In general, this hypothesis posits that a dosage-sensitive gene or small subset of human chromosome (Hsa) 21 genes is responsible for many specific phenotypes of DS [Delabar et al., 1993; Korenberg et al., 1994]. This hypothesis was formulated from the study of a sample of individuals with trisomy for only part of Hsa21, who shared triplication of specific genes in a region extending from D21S55 to BCEI and also showed specific craniofacial anomalies. Direct testing of the role of 33 genes from this region demonstrated that `DSCR' is a misnomer; these genes are not sufficient and are largely unnecessary to produce several prototypic phenotypes that were posited to map to this region [Olson et al., 2004; Olson et al., 2007].
Sumarsono and colleagues [Sumarsono et al. 1996] generated several lines of Ets2 transgenic mice that constitutively expressed this transcription factor at relatively high levels in all tissues throughout development. These mice had skeletal anomalies that the authors equated to those in trisomy 16 mice and in humans with DS. Qualitative comparisons between 19-day fetal Ets2 transgenic mice and normal littermates indicated that the skull anomalies result from changes in both endochondral and intramembranous ossification. Additional morphological differences were noted in the vertebral column. Subsequent analysis of Ets2 transgenic mice demonstrated anomalies of thymus size, cellularity, T cell maturation and increased apoptosis [Wolvetang et al., 2003]. Taken together, these observations suggested that overexpression of Ets2 is responsible (sufficient) for the skeletal and thymus anomalies in DS [Sumarsono et al., 1996].
Ets2 is found on Hsa21 band q22.3. It is a prototype of the ETS family of transcription factors that activate or repress the expression of genes that are involved in a number of biological processes including cellular proliferation, differentiation, development, transformation and apoptosis [Seth and Watson 2005]. Ets2 is essential for development of trophoblasts and is involved in establishment of the AP axis and of paraxial mesoderm [Georgiades and Rossant 2006; Raouf and Seth 2000; Ristevski et al., 2002]. Historically considered a proto-oncogene, misregulation of Ets2 is associated with cancer in the normal population [Papas et al., 1990; Seth and Watson 2005]. However, recent studies indicate that increased dosage of Ets2 contributes substantially to a significantly reduced risk of cancer in persons with DS [Sussan et al., 2008].
Ts65Dn mice are trisomic for a distal segment of mouse chromosome 16 that contains orthologs of 108 Hsa21 genes, including Ets2 [Davisson et al., 1993]. Mice with a null allele of Ets2 have a single functional copy of the gene. Crossing these lines will generate mice that are still trisomic for the distal segment of MMU16, but have only two copies of Ets2 (hereafter, Ts65Dn, Ets2+/− mice). Here we provide analyses of the thymus and three-dimensional skull morphology of the Ts65Dn, Ets2+/− transgenic mouse model, comparing it to euploid and to Ts65Dn littermates to determine the contributions of the Ets2 transcription factor to production of trisomic phenotypes. If dosage imbalance of Ets2 plays an important role in the patterning of DS craniofacial morphology and thymus anomalies, then Ts65Dn, Ets2+/− mice should be significantly different from Ts65Dn mice and more similar to euploid mice. Our results demonstrate that the role of Ets2 in the developing DS thymus and skull phenotype is more complex than previously proposed.
MATERIALS AND METHODS
Animal Husbandry
B6EiC3Sn a/A-Ts(1716)65Dn (herein Ts65Dn) mice were purchased from The Jackson Laboratory and maintained as an advanced intercross by crossing to (C57Bl/6J × C3H/HeJ)F1 mice. Ts65Dn mice carrying a null allele of Ets2 (herein Ts65Dn, Ets2+/− mice, F. Li and M.C. Ostrowski, Ohio State University, ms. in preparation) were backcrossed for six or more generations onto C57BL/6J (B6) before being used in these experiments. Mice were typed as described elsewhere [Sussan et al., 2008]. All animal husbandry procedures were approved by the Institutional Animal Care and Use Committee. The study sample for cranial morphology included Ts65Dn, Ets2+/− mice (N=6; trisomic for a large segment of Mmu16, but disomic for Ets2), Ts65Dn mice (N=10; trisomic for a large segment of Mmu16 including Ets2) and their euploid littermates (N=16; having two copies of all genes including Ets2). Table I provides the sample sizes for the comparisons of thymi.
Table I.
Ets2 dosage does not affect body mass nor mass of the thymus in trisomic or gene-targeted mice at eight weeks of age. Standard deviations (s.d.) are provided in parentheses)
| Genotype (Ets2 copy number) | Euploid (2) | Ts65Dn (3) | Ets2+/− (1) | Ts65Dn, Ets2+/− (2) |
|---|---|---|---|---|
| Avg. thymus mass, mg (s.d.) | 47.3 (15.3) | 52.0 (17.3) | 52.8 (15.1) | 49.6 (19.6) |
| Avg. scaled thymus mass (s.d.)a | 2.15(1.83) | 2.62(0.78) | 2.71(0.92) | 2.35(0.70) |
| Avg. body mass, g (s.d.) | 22.33(3.38) | 19.8b (2.32) | 20.71(3.84) | 20.93(3.93) |
| Sample size | 15 | 8 | 16 | 5 |
Scaled thymus mass is (mg thymus) / (g body mass × 10−3).
Ts65Dn body mass is significantly less than euploid (p = 0.04, Students t-test). There is no difference with other groups or between other genotypes and euploid.
Analysis of Thymus
Thymus glands were removed, weighed and fixed in 4% paraformaldehye for histology. After determining homogeneity of variance, thymus weights scaled for body mass were compared using ANOVA. Histological sections were stained with hematoxylin and eosin for microscopic examination. FACS was used to evaluate thymocyte maturation and apoptosis index.
For FACS analysis, thymi were removed from 4–5 week old mice and dissociated with a plunger. Cells were suspended in complete Dulbecco's Modified Eagle's medium (Invitrogen Corp., Carlsbad, CA), then passed through a 100 □m filter using a 20 gauge needle. Cells were washed and resuspended in staining buffer (phosphate buffered saline pH7.4, 5% fetal bovine serum, 0.02% sodium azide) at a concentration of 2×107 cells/ml. Fifty μl of cell suspension was stained with 10 μl of 20 μg/ml CD4-FITC and/or 40 μg/ml CD8-PE (BD Biosciences Pharmingen, San Diego, CA) for 20 min on ice. Cells were then washed twice in staining buffer and resuspended in 400 μl staining buffer. Cells were analyzed on a Becton Dickinson FACScan. For the apoptosis assay, cells were collected as described above, stained with annexin V-FITC and/or propidium iodide (BD Biosciences Pharmingen) as described by the manufacturer, and analyzed on a FACScan.
Imaging
Adult mice used for the craniofacial study were sacrificed at 8 weeks of age and carcasses were skinned and eviscerated. Heads and limbs were frozen for preservation prior to imaging and data collection. Micro-computed tomography (micro-CT) images of all skulls were acquired at the Center for Quantitative Imaging at Pennsylvania State University (www.cqi.psu.edu) using an OMNI-X Universal HD600 industrial x-ray high resolution computed tomography system (Bio-Imaging Research Inc., Lincolnshire IL). We developed a scanning protocol to maximize the power of the imaging system while acquiring images of a maximum number of skulls in an efficient manner without compromising resolution. Six mouse skulls were encased in cotton batting and placed nose-to-foramen magnum in hard plastic tubing (18 mm in diameter). Four plastic tubes, each containing six mice, were secured jointly for imaging. Since cotton and plastic are CT-transparent, images of twenty-four mice were acquired in the coronal plane with slice thicknesses of 0.048 mm (z dimension) and with pixel sizes of 0.04 mm (x and y dimensions) during a single session. Individual mice were cropped from the larger scan volume for data collection using ImageJ (http://rsb.info.nih.gov/ij/). All primary data are available on request.
Landmark Data Collection and Morphometric Analyses
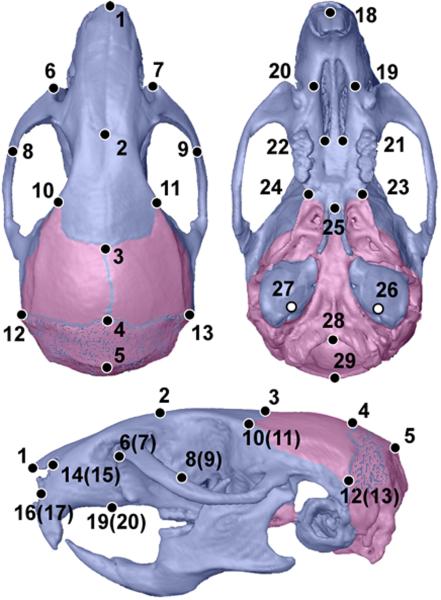
Three-dimensional coordinate locations of twenty-seven biologically relevant cranial landmarks were recorded for all of the mice (Figure 1). Detailed descriptions of these landmarks are provided on the landmark collection page at the Richtsmeier laboratory website (http://www.getahead.psu.edu/landmarks_new.html). Coordinate locations of all landmarks were recorded from the three-dimensional reconstructions of the micro-CT scans of the mice using eTDIPS, 3D reconstruction and visualization software for medical images (http://www.cc.nih.gov/cip/software/etdips/). With eTDIPS, landmarks are located according to the three orthogonal planes of the 3D reconstruction of the specimen. Previous analyses by our lab have demonstrated the accuracy and precision of this data collection method for CT scans [Corner et al., 1992; Richtsmeier et al., 1995]. To eliminate measurement error, two data collection trials were completed for the images of each specimen and the averages of those trials were used for analyses.
Figure 1.
Three-dimensional reconstruction of micro-CT images of an adult mouse skull indicating the contributions of neural crest (blue) and mesoderm (pink) to the skull, following [Jiang et al., 2002]. The interparietal bone which represents a combined derivative is shown as stippled. Twenty-nine landmarks were collected from the skull using etDIPS software. Landmark identifications: 1, nasale; 2, nasion; 3, bregma; 4, pari; 5, paro 6(7) infis; 8(9) jti; 10(11) fsq; 12(13) pto; 14(15) maxna; 16(17) maxi; 18, ids; 19(20) ipm; 21(22) apf; 23(24) mxph; 25, pns; 26(27) iam; 28, basion; 29, opisthion. Landmark definitions can be found at www.getahead.psu.edu.
Differences in skull shape were analyzed using the 3D landmark coordinate data and Euclidean Distance Matrix Analysis or EDMA [Lele and Richtsmeier 2001]. EDMA is a coordinate system-free method for statistically evaluating differences in size and shape of biological objects. EDMA converts 3D landmark data into linear distances, compiling a matrix of all linear distances between unique landmark pairs. For each sample, an average form is estimated using the linear distance data and differences in three-dimensional size and shape are statistically compared as a matrix of ratios of all like linear distances in the two samples. For this portion of the study, three separate inter-sample comparisons of shape and size were completed. The first two comparisons were: 1) Ts65Dn and euploid mice; 2) Ts65Dn, Ets2+/− and euploid littermates. The third comparison involved statistical evaluation of the contrasts between each trisomic model and euploid mice. This was accomplished by statistically comparing Ts65Dn-with-euploid contrasts to Ts65Dn, Ets2+/− -with-euploid contrasts using already established methods [Lele and Richtsmeier 2001]. The null hypothesis for each comparison, that there is no difference in shape contrasts consists of a matrix of 1's. A ratio greater or less than 1 for any linear distance indicates that that the two samples are not similar for that measure. Confidence intervals for the null hypothesis of similarity in shape are estimated using 10,000 pseudo-samples generated from the data using a non-parametric bootstrapping algorithm. For each linear distance the null hypothesis is rejected if the 90% confidence interval produced from the bootstrapping method does not include 1.0. Rejection of the null hypothesis enables localization of differences to specific landmarks and linear distances.
Additional statistical tests for differences in global shape for anatomical regions were evaluated using an alternate non-parametric bootstrapping procedure. For this aspect of the analysis, subsets of landmarks were identified that summarize regions with specific embryonic tissue origins (e.g., mesoderm-derived or neural crest-derived; Figure 1) as previously identified [Jiang et al., 2002; Noden and Trainor 2005]. Twenty landmarks located on neural crest-derived bones (or segments) were further divided into anatomically-relevant subsets, including palate, midface and nasal regions (see Figure 1 and Table III) Tests of shape difference using specific landmark subsets ensured that the sample size exceeded the number of landmarks being evaluated within a subset, a requirement for this statistical test [see Lele and Richtsmeier 2001]. The null hypothesis of similarity in shape was evaluated for each region using 10,000 bootstraps and rejected when p ≤ 0.05. EDMA software is available for download from http://www.getahead.psu.edu.
Table III.
Results of the nonparametric bootstrap testing of the null hypothesis of no difference in shape for various subsets of cranial landmarks analyzed in this study. For these analyses, euploid mice with Ets2+/+ genotype were used as our “Euploid” group. The null hypothesis of similarity in shape (for contrast 1 and 2; or similarity in shape difference for contrast 3) of regions defined by subsets of cranial landmarks was rejected if p ≤ 0.05. Significant contrasts are marked with an “*”.
|
P-values for inter-sample comparisons |
||||
|---|---|---|---|---|
| Anatomical region depicted by subset | Landmarks in Subset | Euploid compared with Ts65Dn | Euploid compared with Ts65Dn, Ets2+/− | Ts65Dn/Euploid contrast compared with Ts65Dn Ets2+/−/Euploid contrast |
| Mesoderm derivatives | 4,5,28,29 | 0.331 | 0.026* | 0.005* |
|
| ||||
| Maximum width of neurocranium (Brachycephaly) | 3,10–13 | 0.144 | 0.213 | 0.821 |
|
| ||||
| Midface | 2,8–11, 23,24 | 0.008* | 0.012* | 0.637 |
|
| ||||
| Neural crest-Mesoderm Boundary | 3,4,12,13 | 0.244 | 0.216 | 0.801 |
|
| ||||
| Neural Crest derivatives | 1–3, 6–11,1421,23–27 | 0.030* | 0.022* | 0.119 |
|
| ||||
| Palate | 18,21–25 | 0.055 | 0.244 | 0.101 |
|
| ||||
| Nasal | 1,2,6,7,19,20 | 0.000* | 0.001* | 0.722 |
RESULTS
Thymus
Ts65Dn mice have 3 copies of the Ets2 gene and expression is upregulated by about 50% at the RNA and protein levels [Kahlem et al., 2004]. As expected from previous studies, body mass was significantly reduced in Ts65Dn mice compared to their euploid littermates (Table I). However, Ets2 dosage did not create a significant difference in body mass on either a trisomic or euploid background.
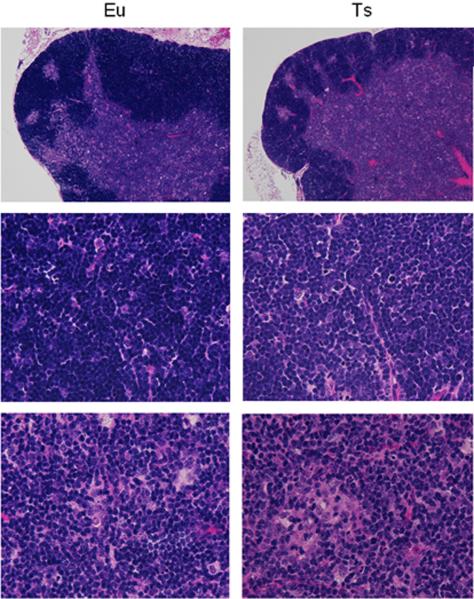
Thymus morphology was grossly normal in Ts65Dn at eight weeks of age with a clear corticomedullary interface (Figure 2). We compared the mass of the thymus as a function of Ets2 copy number (and expression level) in euploid and trisomic mice. Scaled thymus weights normalized to body mass were compared to test for differences between and among mice of genotypes: euploid; Ets2+/−; Ts65Dn; and Ts65Dn,Ets2+/− (2, 1, 3 and 2 copies of Ets2, respectively) using ANOVA with multiple comparisons and pairwise comparisons using t-tests. ANOVA showed no difference in thymus weights among groups. Examination of p-values for pairwise comparisons indicates that Ts65Dn mice have thymus weight similar to those of their euploid littermates (Table I). This result indicates that increased expression of properly regulated Ets2 due to gene dosage is not sufficient to cause the thymus phenotype seen in Ets2 transgenic mice, even in conjunction with trisomy for approximately 100 additional genes orthologous to those on Hsa21. Further, no significant difference in thymus size was seen between Ts65Dn, Ets2+/− and euploid mice, nor did down-regulation of Ets2 in Ets2+/− mice have a significant effect on thymus size.
Figure 2.
Thymic morphology of three month old Ts65Dn mice is grossly the same as euploid. Hematoxylin and eosin stains of thymic sections, showing the outer cortex and the inner medulla. Top panels, low power view. Middle panels, cortex. Bottom panels, medulla.
Ets2 transgenic mice demonstrated a delay in thymocyte maturation and increased apoptosis [Wolvetang et al., 2003]. We assessed thymocytes from Ts65Dn and euploid mice with CD4-FITC and CD8-PE antibodies, and measured fluorescence using flow cytometry. No significant difference was observed in the ratios of CD4−/CD8−, CD4+/CD8−, CD4−/CD8+ and CD4/CD8 cells in Ts65Dn mice (Table II). The number of apoptotic thymus cells determined by annexin V staining was also not significant between the two genotypes (data not shown). Taken together, the results of these analyses in trisomic mice show that increased expression of Ets2 in a spatially- and temporally-appropriate manner in Ts65Dn mice does not produce thymus phenotypes like those observed when a processed Ets2 cDNA is expressed promiscuously throughout development. It is unlikely, therefore, that Ets2 transgenic mice represent the contributions of Ets2 to anomalies of the thymus in DS, but rather produce an artifact peculiar to this transgene.
Table II.
Comparative assessment of thymocyte development in Ts65Dn and euploid mice. No significant differences are observed between groups in the ratios of cell types observed.
| CD4−/CD8−(%) | CD4+/CD8−(%) | CD4−/CD8+(%) | CD4+/CD8+(%) | |
|---|---|---|---|---|
| Euploid | 1.3 +/− 0.4 | 3.6 +/− 2.4 | 1.7 +/− 0.6 | 93.4 +/− 2.5 |
| Ts65Dn | 1.6 +/− 0.6 | 4.2 +/− 2.9 | 2.4 +/− 0.7 | 91.7 +/− 2.9 |
Craniofacial Skeleton
Ts65Dn, Ets2+/− mice differed significantly from euploid littermates in ways that parallel contrasts established previously between Ts65Dn mice and their euploid littermates [Richtsmeier et al., 2000]. Using the landmark subsets (see Figure 1, Table III) designed to analyze particular anatomical and developmental regions of the skull (Table III), nonparametric statistical tests of the null hypothesis of similarity in shape found that both Ts65Dn and Ts65Dn, Ets2+/− mice differed significantly from euploid littermates for a number of the same subsets at our chosen level of significance (p≤0.05; Table III). Although the magnitude of these contrasts (measured as the ratio of like linear distances between euploid and the two groups of aneuploid mice, data not shown) is greater in Ts65Dn, Ets2+/− mice, statistical testing showed that the majority of these differences in magnitude were not statistically significant. Comparison of the results of significance tests of euploid and Ts65Dn contrasts with euploid and Ts65Dn, Ets2+/− contrasts (columns 1 and 2 of the reported p-values in Table III, respectively) indicates similar changes in craniofacial phenotypes of Ts65Dn and Ts65Dn,Ets2+/− mice for all regions of the skull excepting those elements derived from mesoderm.
A direct test of similarity of craniofacial shape change in the Ts65Dn and Ts65Dn,Ets2+/− mice is evaluated by statistical comparison of the two sets of contrasts (the Ts65Dn-with-euploid comparison and Ts65Dn, Ets2+/− -with-euploid comparison, column 3 of the reported p-values in Table III). With the exception of the mesoderm-derived skeletal subset (see below) these results indicate that the 3D morphology of skulls of Ts65Dn trisomic mice differ from euploid littermates following similar patterns of shape change whether they have two or three copies of Ets2.
The p-values reported in Table III represent the results of tests of similarity in form for craniofacial regions defined by a subset of landmarks. In that sense, it is a test of global similarity in shape for a defined cranial region. Our analytical methods also allow testing for significance of localized shape differences defined by specific linear measures. The following sections detail the results of confidence interval tests for differences of specific linear distances in Ts65Dn, Ets2+/− mice as compared to euploid littermates, and the differences in the phenotypic effects of trisomy in Ts65Dn and Ts65Dn, Ets2+/− mice.
Neural Crest-Derived Skeletal Elements
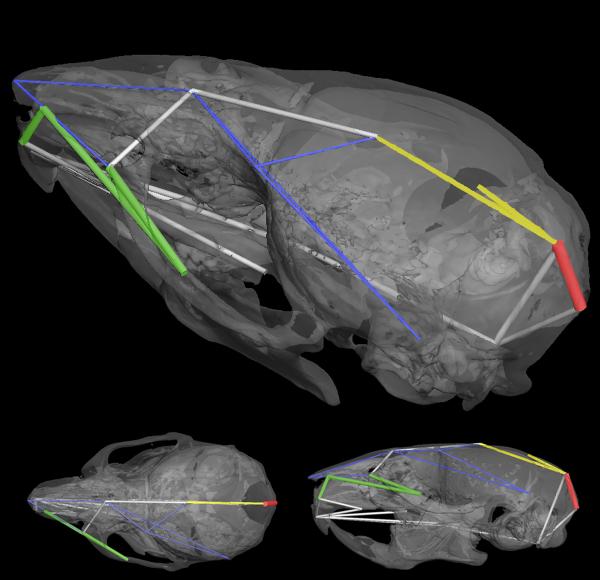
Differences in the neural-crest derived regions of the Ts65Dn, Ets2+/− skull compared to euploid littermates parallel the differences between Ts65Dn mice and euploid littermates as described by [Richtsmeier et al., 2000]. Our analyses indicate an overall reduction in size of the Ts65Dn, Ets2+/− skull along the cranio-caudal axis with the nasal region and midface showing a greater reduction than the neurocranium (Figure 3), a pattern similar to what is seen in Ts65Dn mice.
Figure 3.
Ts65Dn, Ets2+/− mice are significantly different from their euploid littermates following patterns previously described for comparisons between Ts65Dn and euploid mice. 3D reconstruction of micro computed tomography scans of an adult mouse skull showing 3transparent views to aid visualization: upper panel is oblique view showing superior and left surface of skull, lower left shows a superior view of the skull, and lower right is a lateral view. Yellow lines are those linear distances that are increased by 5–8% in Ts65Dn, Ets2+/− mice relative to euploid mice. Blue lines indicate linear distances that are reduced by 3–8% in Ts65Dn, Ets2+/− mice compared to euploid littermates, white lines indicate those measures reduced by 8–10% in Ts65Dn, Ets2+/− mice, and green lines represent linear distances that are reduced by 10–16% in Ts65Dn, Ets2+/− relative to euploid mice. When Ts65Dn, Ets2+/− mice-with-euploid contrasts are compared to Ts65Dn-with-euploid contrasts, one linear distance (shown in red) is significantly more reduced in Ts65Dn, Ets2+/− by confidence interval. Since Ts65Dn and Ts65Dn, Ets2+/− crania differ from their respective euploid littermates following similar patterns, only a subset of the linear distances whose measures are statistically significantly different between Ts65Dn, Ets2+/− and euploid littermates are shown. The reader is directed to Richtsmeier et al. [2000] for additional information on the Ts65Dn cranial phenotype.
Mesoderm-Derived Skeletal Elements
Confidence intervals for linear distances within the mesoderm-derived portion of the skull indicate that Ts65Dn, Ets2+/− mice demonstrate reduced dorso-ventral dimensions in the caudal portion of the neurocranium as compared to euploid littermates (between landmarks 5 and 28; Figure 3). Cranio-caudal dimensions of the interparietal bone (distance from landmark 4-to-5) were increased in Ts65Dn, Ets2+/− mice. These changes mimic what occurs in the skulls of Ts65Dn mice.
Although the Ts65Dn and Ts65Dn, Ets2+/− mice exhibit similar patterns of reduction of various anatomical regions, statistical comparison of the Ts65Dn, Ets2+/− -with-euploid contrasts and the Ts65Dn-with-euploid contrasts indicate that the Ts65Dn, Ets2+/− mice are significantly more affected compared to Ts65Dn mice for the subset of landmarks representing the mesoderm-derived portion of the skull (p=0.005). Confidence interval testing of specific linear distances indicated that one linear distance is driving this overall difference in the effects of trisomy on mesoderm-derived elements. The supero-infero height of the occipital bone (5-to-29) is reduced by 16% in Ts65Dn, Ets2+/− mice relative to euploid, while this distance is reduced by 4% in Ts65Dn mice.
DISCUSSION
It has been known for at least a century that multiple factors play a role in the development of complex traits, and yet modern biology still holds fast to the implicit idea that complexity can be explained by reducing it to enumerable genes [Buchanan et al., 2009]. Recent tests of the Down syndrome “critical region” hypothesis failed to support the idea that triplication of a specific gene or set of genes is sufficient to produce a number of DS phenotypes, including characteristic alterations of the cranial skeleton [Olson et al., 2004; Olson et al., 2007]. Since the “critical region”, which includes Ets2, was not sufficient to cause characteristic cranial dysmorphology of DS, we used the Ts65Dn, Ets2+/− mouse to determine whether Ets2 is necessary for those effects. Our analysis of the craniofacial phenotype and thymus weight of Ts65Dn, Ets2+/− mice provides additional evidence against the “critical region” hypothesis and specifically, of a role for Ets2 as a single cause of these DS phenotypes. In Ts65Dn mice, returning the dosage imbalance of Ets2 to the normal two copies has little effect on the skeletal and thymus anomalies that reflect those noted in individuals with DS. Direct testing of the function of Ets2 using chromosomally engineered mice highlights the complexity of genetic interactions in the production of the final phenotypes in Down syndrome.
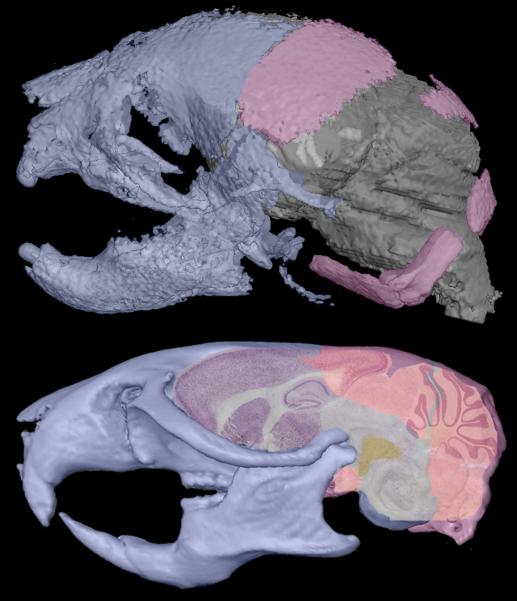
If Ets2 was necessary to produce the DS-like craniofacial and thymus anomalies in Ts65Dn mice, then Ts65Dn, Ets2+/− mice would be more similar to euploid littermates, showing a reversal of the effects of triplicated Ets2 in Ts65Dn mice. Our comparative analyses of Ts65Dn and Ts65Dn, Ets2+/−mice demonstrate minimal differences in the effects of trisomy on the thymus and the cranial skeleton with or without the additional copy of Ets2. Further, the number and magnitude of significant euploid-to-Ts65Dn, Ets2+/− contrasts are approximately equivalent to differences noted in the euploid-to-Ts65Dn contrasts. In our analysis, the exception is localized to specific dimensions of bones derived from mesoderm where trisomic-euploid shape differences of the Ts65Dn, Ets2+/− skull were of significantly greater magnitude than those estimated for the same skull portion of the Ts65D-euploid comparison. There is a possibility that over-expression of Ets2 plays a critical role during development of mesodermal-derived components of the skull, but we should also consider that skull size and shape is influenced, though not determined, by genes and that bones of the skull respond directly to changes in soft tissue structures that surround them. Localized changes in skull morphology reflect changes in brain morphology [Richtsmeier et al., 2006]. That part of the skull that forms from mesoderm surrounds derivatives of the mid- and hindbrain, but the locus of the measures that are statistically different between euploid and both trisomic models lie close to the cerebellum throughout murine development (Figure 4), a central nervous system structure that is disproportionately affected in Down syndrome and in Ts65Dn mice [Baxter et al., 2000]. Whether our findings reflect a differential role of Ets2 in mesoderm derived elements of the skull, or in cerebellar development cannot be determined by our analysis. The role of Ets2 should be explored in additional mesoderm-derived structures affected in DS using larger samples, as well as different aspects of the developing brain.
Figure 4.
Morphological relationship of brain and skull in mice at day of birth (P0; top) and in adult mice (bottom). The top figure shows a 3D reconstruction of micro-computed tomography images of a P0 mouse skull overlying a 3D reconstruction of micro-magnetic resonance images of the P0 mouse brain. Bottom figure shows a lateral view of an adult mouse brain superimposed over a 3D reconstruction of microCT images of an adult mouse skull. The skull in both figures is colored for mesoderm and neural crest derivatives following Figure 1 (neural crest in blue, mesoderm in pink). In the P0 and adult mouse, the cerebellum is in direct contact with parts of the skull that show significant shape differences in the Ts65Dn, Ets2+/− mouse. The cerebellum has previously been demonstrated to be disproportionately affected in Ts65Dn mice [Baxter et al., 2000]. Not to scale.
The phenotypes associated with Down syndrome include structures associated with nearly every bodily system. Production of DS phenotypes is complex, requiring consideration not only of the genes at dosage imbalance, but of all genetic and nongenetic factors influencing formative cell populations and emerging phenotypic form. This study highlighted the potential importance of embryonic tissue-specific effects of Ets2 on the craniofacial skeleton and thymus, though the analyses were limited to adult mice. Thus the mechanisms by which Ets2 dosage imbalance affects specific cell populations and the structures that they form requires further investigation. To be relevant to DS, these and related investigations should include expression of the gene(s), and their networks in a well-defined temporal context.
ACKNOWLEDGMENTS
We thank Dr. Timothy Ryan and the Center for Quantitative Imaging at Pennsylvania State University for his excellent technical skills allowing acquisition of micro-computed tomography scans of the mice. This work was supported in part by NICHD/NCI grant HD 38384(Reeves) and NIDCR grant DE018500 (Richtsmeier).
REFERENCES
- Baxter LL, Moran TH, Richtsmeier JT, Troncoso J, Reeves RH. Discovery and genetic localization of Down syndrome cerebellar phenotypes using the Ts65Dn mouse. Hum Mol Genet. 2000;9:195–202. doi: 10.1093/hmg/9.2.195. [DOI] [PubMed] [Google Scholar]
- Buchanan AV, Sholtis S, Richtsmeier JT, Weiss KM. What are genes “for” or where are traits “from”? What is the question? Bioessays. 2009;31:198–208. doi: 10.1002/bies.200800133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corner BD, Lele S, Richtsmeier JT. Measuring precision of three-dimensional landmark data. Journal of Quantitative Anthropology. 1992;3:347–359. [Google Scholar]
- Davisson MT, Schmidt C, Reeves RH, Irving NG, Akeson EC, Harris BS, Bronson RT. Segmental trisomy as a mouse model for Down syndrome. Prog Clin Biol Res. 1993;384:117–33. [PubMed] [Google Scholar]
- Delabar J, Theophile D, Rahmani Z, Chettouh Z, Blouin J, Prieur M, Noel B, Sinet P. Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur J Hum Genet. 1993;1:114–124. doi: 10.1159/000472398. [DOI] [PubMed] [Google Scholar]
- Georgiades P, Rossant J. Ets2 is necessary in trophoblast for normal embryonic anteroposterior axis development. Development. 2006;133:1059–68. doi: 10.1242/dev.02277. [DOI] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–16. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- Kahlem P, Sultan M, Herwig R, Steinfath M, Balzereit D, Eppens B, Saran NG, Pletcher MT, South ST, Stetten G, Lehrach H, Reeves RH, Yaspo ML. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res. 2004;14:1258–67. doi: 10.1101/gr.1951304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg J, Chen X, Schipper R, Sun Z, Gonsky R, Gerwehr S, Carpenter N, Daumer C, Dignan P, Disteche C, Graham J, Hugdins L, McGillivray B, Miyazaki K, Ogasawara N, park J, Pagon R, Pueschel S, Sack G, Say B, Schuffenhauer S, Soukup S, Yamanaka T. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci. 1994;91:4997–5001. doi: 10.1073/pnas.91.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele S, Richtsmeier JT. An invariant approach to the statistical analysis of shapes. Chapman and Hall-CRC Press; London: 2001. [Google Scholar]
- Noden D, Trainor P. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LE, Richtsmeier JT, Leszl J, Reeves RH. A chromosome 21 critical region does not cause specific Down syndrome phenotypes. Science. 2004;306:687–90. doi: 10.1126/science.1098992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LE, Roper RJ, Sengstaken CL, Peterson EA, Aquino V, Galdzicki Z, Siarey R, Pletnikov M, Moran TH, Reeves RH. Trisomy for the Down syndrome `critical region' is necessary but not sufficient for brain phenotypes of trisomic mice. Hum Mol Genet. 2007;16:774–82. doi: 10.1093/hmg/ddm022. [DOI] [PubMed] [Google Scholar]
- Papas TS, Watson DK, Sacchi N, Fujiwara S, Seth AK, Fisher RJ, Bhat NK, Mavrothalassitis G, Koizumi S, Jorcyk CL, et al. ETS family of genes in leukemia and Down syndrome. Am J Med Genet Suppl. 1990;7:251–61. doi: 10.1002/ajmg.1320370751. [DOI] [PubMed] [Google Scholar]
- Raouf A, Seth A. Ets transcription factors and targets in osteogenesis. Oncogene. 2000;19:6455–63. doi: 10.1038/sj.onc.1204037. [DOI] [PubMed] [Google Scholar]
- Richtsmeier J, Paik C, Elfert P, Cole T, Dahlman H. Precision, repeatability and validation of the localization of cranial landmarks using computed tomography scans. Cleft Palate and Craniofacial Journal. 1995;32:217–27. doi: 10.1597/1545-1569_1995_032_0217_pravot_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- Richtsmeier JT, Aldridge K, Deleon VB, Panchal J, Kane AA, Marsh JL, Yan P, Cole TM., 3rd Phenotypic integration of neurocranium and brain. J Exp Zoolog B Mol Dev Evol. 2006;306B:360–378. doi: 10.1002/jez.b.21092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richtsmeier JT, Baxter LL, Reeves RH. Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Dev Dyn. 2000;217:137–45. doi: 10.1002/(SICI)1097-0177(200002)217:2<137::AID-DVDY1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Ristevski S, Tam PP, Hertzog PJ, Kola I. Ets2 is expressed during morphogenesis of the somite and limb in the mouse embryo. Mech Dev. 2002;116:165–8. doi: 10.1016/s0925-4773(02)00126-0. [DOI] [PubMed] [Google Scholar]
- Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–78. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Sumarsono S, Wison T, Tymms M, Venter D, Corrick C, Kola R, Lahoud M, Papas T, Seth A, Kola I. Down's syndrome-like skeletal abnormalities in Ets2 transgenic mice. Nature. 1996;379:534–537. doi: 10.1038/379534a0. [DOI] [PubMed] [Google Scholar]
- Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down's syndrome. Nature. 2008;451:73–5. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- Van Cleve SN, Cannon S, Cohen WI. Part II: Clinical Practice Guidelines for adolescents and young adults with Down Syndrome: 12 to 21 Years. J Pediatr Health Care. 2006;20:198–205. doi: 10.1016/j.pedhc.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Van Cleve SN, Cohen WI. Part I: clinical practice guidelines for children with Down syndrome from birth to 12 years. J Pediatr Health Care. 2006;20:47–54. doi: 10.1016/j.pedhc.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Wolvetang EJ, Wilson TJ, Sanij E, Busciglio J, Hatzistavrou T, Seth A, Hertzog PJ, Kola I. ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum Mol Genet. 2003;12:247–55. doi: 10.1093/hmg/ddg015. [DOI] [PubMed] [Google Scholar]