Abstract
Poly(ADP-ribose) polymerases (PARPs) catalyze the transfer of multiple adenine diphosphate ribose (ADP-ribose) units from nicotinamide adenine dinucleotide (NAD) to substrate proteins. There are seventeen PARPs in humans. Several PARPs, such as PARP-1 and Tankyrase-1, are known to play important roles in DNA repair, transcription, mitosis, and telomere length maintenance. To better understand the functions of PARPs at a molecular level, it is necessary to know what substrate proteins PARPs modify. Here we report clickable NAD analogs that can be used to label PARP substrate proteins. The clickable NAD analogs have a terminal alkyne group which allows the conjugation of fluorescent or affinity tags to the substrate proteins. Using this method, PARP-1 and tankyrase-1 substrate proteins were labeled by a fluorescent tag and visualized on SDS-PAGE gel. Using a biotin affinity tag, we were able to isolate and identify a total of 79 proteins were identified as potential PARP-1 substrates. These include known PARP-1 substrate proteins, including histones and heterogeneous nuclear ribonucleoproteins. About 40% of the proteins were also identified in recent proteomic studies as potential PARP-1 substrates. Among the identified potential substrates, we further demonstrated that tubulin and three mitochondrial proteins, TRAP1 (TNF receptor-associated protein 1), citrate synthase, and GDH (glutamate dehydrogenase 1), are substrates of PARP-1 in vitro. These results demonstrate that the clickable NAD analog is useful for labeling, in-gel detection, isolation, and identification of the substrate proteins of PARPs and will help to understand the biological functions of PARPs.
Introduction
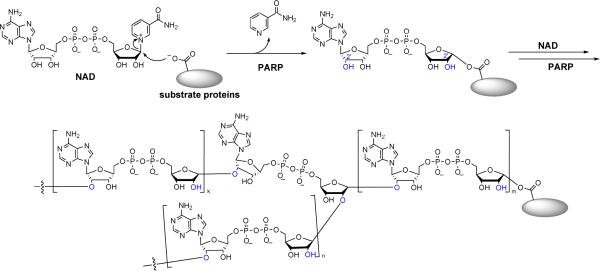
Poly(ADP-ribosyl)ation is the addition of multiple ADP-ribosyl groups from NAD to substrate proteins1–3. The first ADP-ribosyl group is added to the carboxylate side chain of Glu or Asp residues of the substrate proteins, followed by addition of more ADP-ribosyl groups to the 2'-OH groups of the two ribose rings (Figure 1), leading to long and branched poly(ADP-ribose) chains (PAR) that can contain hundreds ADP-ribose units. The large size and the enormous negative charges of PAR chains can affect protein structure and function and thus regulate the biological processes that the substrate proteins are involved in. The enzymes that catalyze poly(ADP-ribosylation) are termed poly(ADP-ribose) polymerases, or PARPs. Seventeen PARPs have been identified in humans.1, 4 Of the 17 PARPs identified, only two of them, PARP-1 and Tankyrase-1 are relatively well studied. PARP-1 is found to be required for DNA repair/genome maintenance5–7 and transcriptional regulation of certain genes8–11 by poly(ADP-ribosyl)ation of itself and many nucleosomal or nucleosome-associated proteins. It is also responsible for the induction of cell death under extreme stress (such as excessive DNA damage)12, 13 or pathological conditions (stroke, ischemia, diabetes).14–16 Tankyrase-1 is known to be required for mitosis and telomere length maintenance by modifying the nuclear mitotic apparatus protein (NuMA)17–20 and a telomere-binding protein TRF1.21, 22 The important biological functions of PARP-1 and Tankyrase-1 suggest that other PARPs could be likewise important. However, very little is known about other PARPs.
Figure 1.
Poly(ADP-ribosyl)ation reaction catalyzed by PARPs.
To understand the biological function of different PARPs, it is necessary to find out what substrate proteins are modified by different PARPs, similar to the efforts that have been invested in identifying kinase substrates.23, 24 At present, 32P-NAD and PAR antibodies are the major methods used to detect protein poly(ADP-ribosyl)ation. For isolation and identification of substrate proteins of PARPs, using PAR antibodies is the preferred option because the use of 32P-NAD does not provide an affinity tag for isolation and purification. Recently, macro domain that can bind to ADP-ribose was used to isolate ADP-ribosylated proteins.25 A major concern about using PAR antibodies and macro domains to identify substrate proteins is that the immunoprecipitation step is executed under native conditions and thus some non-substrate proteins that interacts with a poly(ADP-ribosyl)ated protein will also be pulled down. This will give false positive results. Thus new and improved methods to label and identify PARP substrate proteins are still needed.
Here, we report the use of clickable alkyne-tagged NAD analogs for labeling and identifying PARP substrate proteins (Figure 2). The terminal alkyne attached to the adenine ring of NAD can be conjugated via click chemistry to many other functional tags, such as fluorescent tags for in-gel visualization and affinity tags for purification, while its small size will make sure that the NAD analog will still be accepted by PARPs as the co-substrate. We demonstrate that one of the clickable NAD analogs, 6-alkyne-NAD, can be used to label the substrate proteins of PARPs for in-gel visualization and identification. In addition to known PARP-1 substrate proteins, over 70 unknown potential substrate proteins from cell lysate were identified. Among these unknown potential substrate proteins, we demonstrated that four of them are indeed PARP-1 substrates in vitro. These results demonstrate that the clickable NAD analog will be very useful for studying the biological function of PARPs by facilitating the in-gel visualization, isolation and identification of substrate proteins.
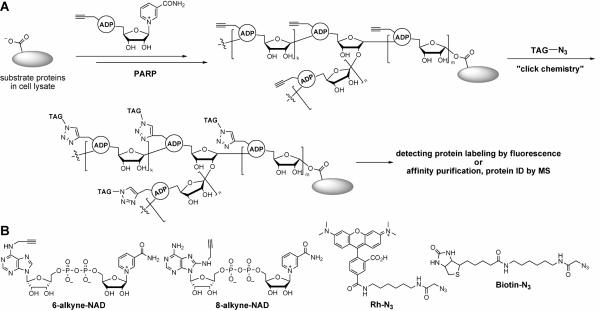
Figure 2.
(A) Labeling of poly(ADP-ribosyl)ated proteins with NAD analogs. NAD analogs bearing an alkyne group will be used in PARP-catalyzed reactions. An affinity tag can be added using click chemistry after the substrate protein is labeled. The labeled protein can then be affinity purified, separated on 1D/2D protein gel, and then the sequence identified by MS. (B) Structure of 6-/8-alkyne-NAD, Rh-N3, and Biotin-N3, which were used in labeling reactions.
Results and Discussion
Labeling of PARP-1 auto-modification
The synthesis of the alkyne-tagged NAD analogs has been reported.26 Here we tested whether these alkyne-tagged NAD analogs can be accepted by PARPs as a co-substrate. We first tested the best studied PARP family member, PARP-1. PARP-1 is a 110-kDa protein consisting of three domains: an N-terminal DNA binding domain, a C-terminal catalytic domain, and a central auto-modification domain. The DNA binding domain can bind to various forms of DNA strand breaks, which stimulate its catalytic activity leading to the poly(ADP-ribosyl)ation of PARP-1 itself and other target proteins, including histones27, 28, DEK,29 and p5330–33.
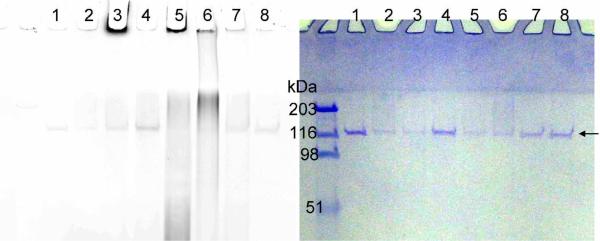
We expressed and purified full length PARP-1 using baculoviral expression system in Sf-9 cells.8 The purified protein was incubated with 6-alkyne-NAD and 8-alkyne-NAD in the presence of salmon sperm DNA (ssDNA), an activator of PARP-1 enzymatic activity. Next, click chemistry was initiated to conjugate the fluorescent tag, Rh-N3, to the modified protein. The reaction mixture was then resolved by SDS-PAGE and the fluorescent labeling was detected. The fluorescent image and the corresponding Coomassie stained gel are shown in Figure 3. With 100 μM 6-alkyne-NAD, a strong fluorescent band was observed at the top of the gel (Figure 3, lane 3), suggesting that PARP-1 was extensively poly(ADP-ribosyl)ated. Correspondingly, the intensity of the unmodified PARP-1 band was decreased compared with the control in lane 1. When 100 μM NAD was included in the labeling reaction with 6-alkyne-NAD, in addition to the top fluorescent band, a smear of fluorescence was also observed above the unmodified PARP-1 protein band (Figure 3, lane 5). This smear is likely PARP-1 modified to a lesser degree. A smear of fluorescence signal below 116 kDa was also observed. This is likely due to the hydrolysis of poly(ADP-ribose) chain resulting in poly(ADP-ribose) polymers that are not covalently bound to proteins. The linkage between different ADP-ribose units is a relatively labile ester bond. This result suggests that the presence of normal NAD does not decrease the labeling efficiency with 6-alkyne-NAD and may in fact increase it. This is possibly because 6-alkyne NAD is not a good substrate for initiation of poly(ADP-ribosyl)ation, but after a poly(ADP-ribose) chain is formed with the better substrate NAD, then 6-alkyne-NAD becomes a better substrate for subsequent elongation of the poly(ADP-ribose) chain, leading to higher labeling efficiency. Without ssDNA (lane 7), the fluorescence signal is comparable to the negative controls (lanes 1 and 2), consistent with the low activity of PARP-1 in the absence of DNA strand breaks. The other analog, 8-alkyne-NAD, can also label PARP-1 auto-modification. However, it is less efficient than 6-alkyne-NAD. Even in the presence of normal NAD, the extensive poly(ADP-ribosyl)ation band at the top of the gel is weak with 8-alkyne-NAD (Figure 3, lane 6). It is possible that attaching the alkyne group at position 8 of the adenine ring produces steric clash when it binds to PARP-1.
Figure 3.
Labeling of PARP-1 auto-(ADP-ribosyl)ation with 6- or 8-alkyne-NAD. The panel on the left shows the image of Rhodamine fluorescence and the panel on the right is the same gel stained with Coomassie blue. The arrow points at the position of unmodified PARP-1. All lanes contain PARP-1 (0.15 μM) and all lanes except 7 and 8 contain ssDNA (0.25 μg/μL). Lane 1, control without NAD; lane 2, control with normal NAD (100 μM); lane 3, 6-alkyne-NAD (100 μM); lane 4, 8-alkyne-NAD (100 μM); lane 5, 6-alkyne-NAD (100 μM) and NAD (100 μM); lane 6, 8-alkyne-NAD (100 μM) and NAD (100 μM); lane 7, control with 6-alkyne-NAD (100 μM), NAD (100 μM), and no ssDNA; lane 8, control with 8-alkyne-NAD (100 μM), NAD (100 μM), and no ssDNA. The fluorescence signal below 116 kDa in lane 5 and 6 is likely due to the hydrolysis of poly(ADP-ribose) chain resulting in poly(ADP-ribose) polymers that are not covalently bound to proteins. The linkage between different ADP-ribose units is a relatively labile ester bond.
To compare the labeling results with clickable NAD analogs to that obtained with 32P-NAD, we carried out similar labeling experiments with 32P-NAD (Supporting Information, Figure S1). Comparing the results shown in Figure 3 and Figure S1, the labeling results obtained with 6-alkyne-NAD is very similar to that obtained with 32P-NAD, particularly when 100 μM normal NAD was present (Figure 3 lane 5 and Figure S1 lane 3). This result suggests that 6-alkyne-NAD can be used to label and detect PARP-1 auto-modification.
We further demonstrated that two PARP-1 truncations, PARP-1(374–524, the automodification domain) and PARP-1(374–1014, the automodification domain plus the catalytic domain), can be labeled with PARP-1 full length and 6-alkyne-NAD (Supporting Information, Figure S2). In the presence of 100 μM NAD and 6-alkyne NAD, the labeling of PARP-1(374–1014) reached maximum levels in about 6 min (Supporting Information, Figure S3). The labeling of as little as 2.5 pmol of PARP-1(374–1014) can be detected (Supporting Information, Figure S4).
Labeling of known substrate proteins of PARP-1 and Tankyrase-1
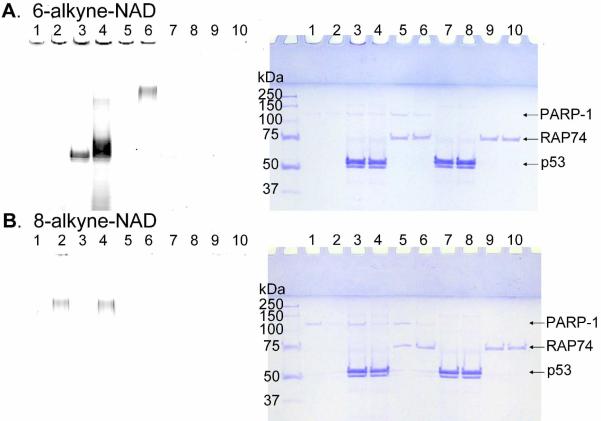
It has been reported that PARP-1 can modify many other proteins,34 even though the major substrate is PARP-1 itself.2 To make sure that 6-alkyne-NAD is a robust substrate for PARP-1, we tested whether modification of other substrate proteins can be detected. For this purpose, we chose to use p53 and TFIIF, both of which have previously been reported to be PARP-1 targets30–33, 35, 36, 37. The RAP74 subunit of TFIIF or p53 was incubated with full length PARP-1 and 6- or 8-alkyne-NAD, followed by click chemistry to conjugate Rh-N3. The results shown in Figure 4 demonstrate that p53 can be poly(ADP-ribosyl)ated by PARP-1 when 6-alkyne-NAD was used, both in the absence and presence of normal NAD (Figure 4A, lanes 3 and 4). The smear over p53 protein band is poly(ADP-ribosyl)ated p53. The RAP74 subunit of TFIIF can be labeled with 6-alkyne-NAD when normal NAD was present (Figure 4A, lane 6), but not when normal NAD was not present (Figure 4A, lane 5). Comparing lane 6 to lane 2 in Figure 4A, the extra smear on the top is poly(ADP-ribosyl)ated RAP74. With 8-alkyne-NAD, neither p53 nor RAP74 was labeled (Figure 4B).
Figure 4.
Labeling of p53 and RAP74 subunit of TFIIF by PARP-1 using 6-alkyne-NAD (A) and 8-alkyne-NAD (B). The panel on the left shows the image of Rhodamine fluorescence recorded by Typhoon 9400 Variable Mode Imager, and the panel on the right is the same gels stained with Coomassie blue. In addition to 6- or 8-alkyne NAD (100 μM), the following were present in different lanes: 1 and 2, PARP-1 (0.15 μM); 3 and 4, PARP-1 (0.15 μM) and p53 (2.8 μM); 5 and 6, PARP-1 (0.15 μM) and RAP74 (0.59 μM); 7 and 8, p53 (2.8 μM); 9 and 10, RAP74 (0.59 μM). ssDNA were present in all lanes, and normal NAD (100 μM) were present in lanes 2, 4, 6, 8, and 10. The fluorescence signal in the loading wells likely came from poly(ADP-ribosyl)ated PARP-1.
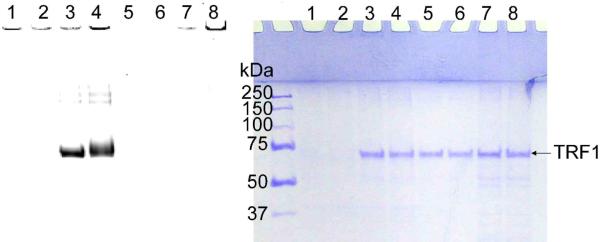
To further confirm that 6-alkyne-NAD is a robust substrate for labeling poly(ADP-ribosyl)ated proteins, we wanted to find out whether substrate proteins of other PARPs can also be labeled. For this purpose, we used tankyrase-1, which can regulate telomere length and mitosis by modifying TRF1 and NuMA, respectively. Using the insect cell expression system developed by de Lange and coworkers, we expressed and purified tankyrase-1 and TRF1. The labeling reactions were then carried out, similarly to those above. Consistent with the reported results obtained with 32P-NAD,21 we observed efficient TRF1 modification in the presence of tankyrase-1 and 6-alkyne-NAD (Figure 5A, lanes 3 and 4), but not in the absence of tankyrase-1 (Figure 5A, lanes 5–8). These results suggest that 6-alkyne-NAD is a robust substrate, and may be applicable to other PARPs.
Figure 5.
Labeling of TRF1 by tankyrase-1 using 6-alkyne-NAD. The panel on the left shows the image of Rhodamine fluorescence, and the panel on the right is the same gels stained with Coomassie blue. Lanes 1 and 2: tankyrase-1 suspension (0.09 μg/μL) with 6-alkyne-NAD (100 μM); 3 and 4: tankyrase-1 suspension (0.09 μg/μL) and TRF1 (1.1 μM) with 6-alkyne-NAD (100 μM); 5 and 6: TRF1 (1.1 μM) with 6-alkyne-NAD (100 μM); 7 and 8: suspension of the insoluble fraction of non-infected SF9 cell lysate (0.15 μg/μL, negative control) and TRF1 (1.1 μM) with 6-alkyne-NAD (100 μM). Lanes 2, 4, 6, and 8 contained normal NAD (100 μM).
Kinetics of PARP-1 labeling with 6-alkyne-NAD
To quantify how efficient 6-alkyne-NAD is as a PARP substrate, we measured the steady state kinetics of NAD, 6-alkyne-NAD, and 8-alkyne-NAD in the PARP-1 and Tankyrase-1 auto-modification reactions. Previously, the kinetics assays were typically done by measuring the PARP-1 auto-modification using radio-labeled NAD38, 39. Recently, a chromogenic NAD analog was also reported.40 However, both methods cannot be directly applied to the kinetic assay with 6-alkyne-NAD. Thus we developed a HPLC-based assay to measure the release of nicotinamide. To correct for the NADase acitivity (hydrolysis of NAD to ADP-ribose and nicotinamide), we also meansured the release of ADP-ribose, 6-alkyne-ADP-ribose, or 8-alkyne-ADP-ribose. The kcat and Km values for the PARP activity and NADase activity are shown in Table 1. Compared with NAD, the kcat//Km value of 6-alkyne-NAD is 12-fold and 4-fold lower for PARP-1 and Tankyrase-1, respectively. In contrast, 8-alkyne-NAD is a much worse substrate for both PARP-1 and Tankyrase 1, consistent with the labeling results (Figure 3 and Figure 4). Although 6-alkyne-NAD has a lower catalytic efficiency compared with NAD, the labeling efficiency will not be affected significantly because in practice a much higher 6-alkyne-NAD concentration can be used, particularly when comparing to 32P-NAD, the concentration of which in labeling reactions is limited to a few μM by the commercially available 32P-NAD.
Table 1.
Kinetics of PARP-1 and Tankyrase-1 with NAD, 6- and 8-alkyne-NAD.
| PARPs | Substrates | PARP activity | NADase activity | ||||
|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (s−1M–1) | kcat (s−1) | Km (μM) | kcat/Km (s−1M−1) | ||
| PARP-1 | NAD | 5.2 ± 0.1 | 97 ± 7 | 5.4 × 104 | 0.99 ±0.06 | 203 ±33 | 4.88 × 103 |
| 6-alkyne-NAD | 0.73 ± 0.04 | 147 ± 23 | 5.0 × 103 | 0.94 ±0.1 | 1891 ± 273 | 497 | |
| 8-alkyne-NAD* | N/A | N/A | 0.51 | N/A | N/A | 11.90 | |
| Tankyrase-1 | NAD | 0.51 ± 0.04 | 1125 ± 133 | 453 | 0.54 ± 0.02 | 607 ± 29 | 890 |
| 6-alkyne-NAD* | N/A | N/A | 122 | N/A | N/A | 252 | |
| 8-alkyne-NAD* | N/A | N/A | 0.29 | N/A | N/A | 3.41 | |
(The reaction rate of PARP-1 or Tankyrase-1 with 6- or 8-alkyne-NAD was linear with the 6- or 8-alkyne-NAD concentration used (10 μM − 1 mM). Thus kcat and Km could not be measured and only kcat/Km was obtained.)
Identification of substrate proteins of PARP-1 using 6-alkyne-NAD
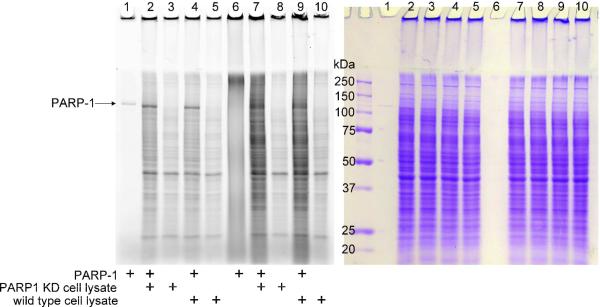
Since 6-alkyne-NAD is a robust substrate for PARP-1, we then tried to use 6-alkyne-NAD to identify substrate proteins of PARP-1 from cell lysate. We used both MCF-7 wild type and PARP-1 knockdown (KD) cell lysate as the source of the PARP-1 substrate proteins. After the lysate was incubated with 6-alkyne-NAD, click chemistry was used to conjugate the Rh-N3 fluoroscent dye. The fluorescence image shown in Figure 6 suggests that incubation of the cell lysates with PARP-1 and 6-alkyne-NAD indeed led to the poly(ADP-ribosyl)ation of proteins in the cell lysates.
Figure 6.
Labeling of MCF-7 wild type and PARP-1 KD cell lysate with PARP1 and 6-alkyne-NAD. The panel on the left shows the image of Rhodamine fluorescence, and the panel on the right is the same gels stained with Coomassie blue. PARP-1 (0.075 μM), MCF-7 wild type cell lysate (2 μg/μL) and PARP-1 KD cell lysate (2 μg/μL) were used in the labeling reactions. All lanes contain 6-alkyne-NAD (100 μM) and ssDNA (0.25 μg/μL). Lanes 6–10 contained normal NAD (100 μM).
To identify the proteins that are modified by PARP-1, an affinity tag, Biotin-N3, was used for purification and isolation. MCF-7 PARP-1 KD cell lysate was used as the source of the PARP-1 substrate proteins. In the PARP-1 KD cells, because PARP-1 level is decreased (Supporting Information, Figure S5),41 fewer PARP-1 substrate proteins should be poly(ADP-ribosyl)ated. Thus, more substrate proteins may be available for labeling in the in vitro reactions containing recombinant PARP-1 and 6-alkyne-NAD. For negative controls, we used 100 μM PJ34 (a PARP-1 inhibitor) or without 6-alkyne-NAD in the reactions.
After incubation of PARP-1 and 6-alkyne-NAD with MCF-7 PARP1 KD cell lysate, click chemistry was carried out to conjugate Biotin-N3. Controls were treated similarly. Following the published protocol,42 labeled proteins were affinity purified and isolated using streptavidin beads, and the samples were prepared and analyzed by nanoLC-MS/MS. It should be noted that proteins from the labeling reactions were denatured and precipitated out with acetone, then solubilized with SDS buffer for affinity purification with streptavidin beads. Under the denaturing conditions, non-covalent interactions between protein-protein or protein-DNA/RNA are minimized, unlike the use of PAR antibodies or macrodomains that use native conditions. Therefore, this affinity purification and identification by nanoLC-MS/MS will give fewer false positive results, making the results more reliable.
About three hundred proteins were identified from each sample, including negative controls. A protein is considered a potential substrate proteins if more than 2 peptides are identified in ≥ 2 out of 3 samples and is more abundant in the 3 experimental samples than in the 3 negative controls. The quantification is based on exponentially modified protein abundance index (emPAI) values.43 Among the ~300 proteins identified, 79 proteins with emPAI ratio (emPAI value in experimental sample / emPAI value in negative control) greater than 1.20 were identified as potential PARP-1 substrates (Table S1, Supporting Information). For proteins with emPAI ratios smaller than 2.0, up to three common peptide ions for each protein found in experimental samples and control samples were compared using extracted ion chromatograms.43 A protein was only considered a positive hit if the extracted ion chromatograms confirmed that the protein was more abundant in experimental samples than in control samples, The top 45 identified proteins are shown in Table 2. Among the 45 proteins, 6 of them are known substrate proteins of PARP-1, such as PARP-1 (auto-modification), histones, and heterogeneous nuclear ribonucleoproteins. In addition, 18 of the 45 proteins have been identified as potential poly(ADP-ribosyl)ated proteins in a proteomic study using PAR antibody.44 These results demonstrate that the strategy of using 6-alkyne-NAD to label and identify PARP-1 substrate proteins works. Not all known substrate proteins of PARP-1 were identified. One possibility is that these proteins have low concentrations in cells and thus are difficult to identify. Alternatively, these proteins may be conditional substrates that can only be poly(ADP-ribosyl)ated under certain cellular conditions.
Table 2.
Potential substrate proteins of PARP-1 identified from MS using the labeling strategy with 6-alkyne-NAD. Top 45 potential substrate proteins of PARP-1 (emPAI ratio >1.50) were listed. References are given for known substrate proteins of PARP-1 and potential PARP-1 substrate proteins identified in other proteomic studies. Full list of identified potential substrate proteins of PARP-1 is given in Supporting Information.
| Protein Accession Name | Description | MW (Da) | Known substrates of PARP-1 | Potential PARP-1 substrates identified in other proteomic studies |
|---|---|---|---|---|
| ATP5L_HUMAN | ATP synthase subunit g | 11421 | ||
| BAP31_HUMAN | B-cell receptor-associated protein 31 | 28031 | ||
| CH10_HUMAN | 10 kDa heat shock protein | 10925 | ||
| COX41_HUMAN | Cytochrome c oxidase subunit 4 isoform 1 | 19621 | ||
| DDX17_HUMAN | Probable ATP-dependent RNA helicase DDX17 | 72953 | reported in44 | |
| DDX5_HUMAN | Probable ATP-dependent RNA helicase DDX5 | 69618 | reported in44 | |
| DHE3_HUMAN | Glutamate dehydrogenase 1 | 61701 | reported in44 | |
| EF1G_HUMAN | Elongation factor 1-gamma | 50429 | ||
| ETFA_HUMAN | Electron transfer flavoprotein subunit alpha | 35400 | ||
| FUBP2_HUMAN | Far upstream element-binding protein 2 | 73443 | ||
| FUS_HUMAN | RNA-binding protein FUS | 53622 | ||
| GLYM_HUMAN | Serine hydroxymethyltransferase | 56414 | ||
| GSTK1_HUMAN | Glutathione S-transferase kappa 1 | 25594 | ||
| H2B1C_HUMAN | Histone H2B type 1-C/E/F/G/I | 13811 | reviewed in49 | |
| H31T_HUMAN | Histone H3.1t | 15613 | reviewed in49 | |
| HNRPL_HUMAN | Heterogeneous nuclear ribonucleoprotein L | 64720 | reported in44 | |
| HNRPM_HUMAN | Heterogeneous nuclear ribonucleoprotein M | 77749 | reviewed in49 | |
| HP1B3_HUMAN | Heterochromatin protein 1-binding protein 3 | 61454 | ||
| HXK1_HUMAN | Hexokinase-1 | 103561 | reported in44 | |
| IDH3A_HUMAN | Isocitrate dehydrogenase [NAD] subunit alpha | 40022 | ||
| IDHP_HUMAN | Isocitrate dehydrogenase [NADP] | 51333 | ||
| IMB1_HUMAN | Importin subunit beta-1 | 98420 | reported in44 | |
| KU70_HUMAN | ATP-dependent DNA helicase 2 subunit 1 | 70084 | reviewed in49 | |
| LETM1_HUMAN | LETM1 and EF-hand domain-containing protein 1 | 83986 | ||
| MATR3_HUMAN | Matrin-3 | 95078 | reported in44 | |
| MPCP_HUMAN | Phosphate carrier protein | 40525 | reported in44 | |
| NPM_HUMAN | Nucleophosmin | 32726 | reviewed in49 | |
| PARP1_HUMAN | Poly [ADP-ribose] polymerase 1 | 113811 | reviewed in49 | |
| PHB_HUMAN | Prohibitin | 29843 | reported in44 | |
| QCR2_HUMAN | Cytochrome b-c1 complex subunit 2 | 48584 | reported in44 | |
| RBM39_HUMAN | RNA-binding protein 39 | 59628 | ||
| RL19_HUMAN | 60S ribosomal protein L19 | 23565 | reported in44 | |
| RL26L_HUMAN | 60S ribosomal protein L26-like 1 | 17246 | reported in44 | |
| RL7_HUMAN | 60S ribosomal protein L7 | 29264 | reported in44 | |
| RS14_HUMAN | 40S ribosomal protein S14 | 16434 | reported in44 | |
| RS2_HUMAN | 40S ribosomal protein S2 | 31590 | reported in44 | |
| RS24_HUMAN | 40S ribosomal protein S24 | 15413 | reported in44 | |
| RS3_HUMAN | 40S ribosomal protein S3 | 26842 | ||
| SFRS1_HUMAN | Splicing factor, arginine/serine-rich 1 | 27842 | reported in44 | |
| SFRS6_HUMAN | Splicing factor, arginine/serine-rich 6 | 39677 | ||
| SFRS7_HUMAN | Splicing factor, arginine/serine-rich 7 | 27578 | ||
| TBB2C_HUMAN | Tubulin beta-2C chain | 50255 | reported in44 | |
| TMED9_HUMAN | Transmembrane emp24 domain-containing protein 9 | 27374 | ||
| TR150_HUMAN | Thyroid hormone receptor-associated protein 3 | 108658 | ||
| UBIQ_HUMAN | Ubiquitin | 8560 |
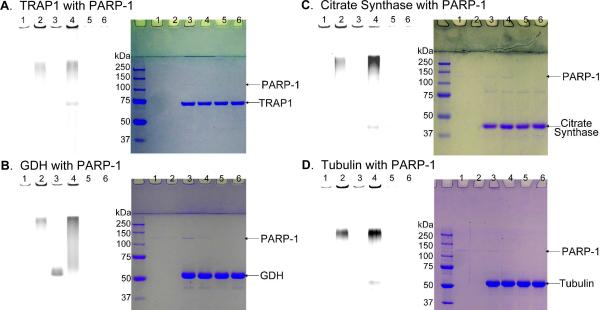
Among the proteins identified, our attention is drawn to several mitochondrial proteins. Although most known PARP-1 substrates are nuclear proteins, PARP-1's mitochondrial localization has been confirmed recently.45 This suggests that PARP-1 potentially has more substrate proteins in mitochondria. Consistent with this, several mitochondria proteins have been identified as potential PARP-substrates in a recent proteomic study.46 To further confirm that identified potential substrate proteins are indeed substrates of PARP-1, we chose tubulin and three mitochondrial proteins TRAP1 (TNF receptor-associated protein 1), citrate synthase, and GDH (glutamate dehydrogenase) to further validate the results. Tubulin (emPAI ratio 3.86) is a structural protein and was previously identified as a potential PARP substrate.44 TRAP1 (emPAI ratio 1.32) is a mitochondrial heat shock protein 75 with antioxidant and antiapoptotic functions.47 It has been shown to protect cells from apoptosis induced by DNA damaging reagents. GDH (emPAI ratio 2.47) catalyzed the conversion of glutamate to α-ketoglutarate, which feeds into the tricarboxylic acid (TCA) cycle for energy production. Citrate synthase (emPAI ration 1.25) is an enzyme in the TCA cycle. Tubulin and GDH were both identified in a previous proteomic studies as potential PARP substrates but they have not been confirmed in in vitro enzyme assays.44 TRAP1 and citrate synthase have not been known to be PARP-1 substrates. TRAP1 can be expressed recombinantly48 while GDH, citrate synthase, and tubulin are commercially available. Thus we tested whether these proteins can be poly(ADP-riboyl)ated by PARP-1 in vitro using 6-alkyne-NAD. Comparing lanes 3 and 4 with lanes 1 and 2 (Figure 6, A, B, C, and D), it is clear that all four proteins were poly(ADP-ribosyl)ated by PARP-1 when normal NAD was present. GDH was modified even when no normal NAD was present. These results further confirm that the proteins we identified as PARP-1 substrates should be highly reliable. Thus, the strategy of labeling, isolating, and identifying PARP-1 substrate proteins with 6-alkyne-NAD is successful. The substrate proteins that we identified for PARP-1 should be very useful for understanding the biological function of PARP-1 and the strategy should be similarly applicable to other PARPs.
Conclusions
We have demonstrated that the clickable 6-alkyne-NAD is an efficient substrate that can be used to label poly(ADP-ribosyl)ated proteins. In contrast, 8-alkyne NAD is not an efficient substrate for PARPs. Using the alkyne functional group, fluorescent or affinity tags can be conveniently installed on the substrate proteins to facilitate in-gel visualization and identification of the substrate proteins. Over 70 potential novel substrate proteins of PARP-1 have been identified using the labeling strategy with 6-alkyne-NAD, including many mitochondrial proteins. Tubulin, TRAP1, citrate synthase, and GDH were further proved to be PARP-1 substrate proteins in vitro. We believe 6-alkyne-NAD will greatly facilitate the identification of the substrate proteins of the 17 PARPs in humans and help to understand their biological functions.
Experimental
General methods
Reagents were obtained from Aldrich in the highest purity available and used as supplied. Kinetics experiments were carried out on a SHIMADZU LCMS-QP8000α with a Sprite TARGA C18 column (40 × 2.1 mm, 5 μm, Higgins Analytical, Inc.) monitoring at 260 nm. Solvents were 50 mM ammonium acetate pH 5.4 (buffer A) and 50% methanol in water (buffer B). Rhodamine fluorescence signal from protein gel was recorded by Typhoon 9400 Variable Mode Imager (GE Healthcare Life Sciences) with setting of Green(532 nm)/555BP20 PMT450V (normal sensitivity), and analyzed by ImageQuant TL v2005. The syntheses of 6-alkyne-NAD, 8-alkyne-NAD, Rh-N3 and N-(6-aminohexyl)-azidoacetamide (for synthesis of Biotin-N3) have been reported.26, 50 The synthesis of Biotin-N3 is given in the Supporting Information. ssDNA was purchased from Sigma and used after sonication. RAP74 (one subunit of TFIIF) was purchased from ProteinOne (Bethesda, MD). GDH (from bovine liver, type II) and citrate synthase (from porcine heart) were purchased from Sigma. Tubulin (from bovine brain) was purchased from Cytoskeleton (Denver, CO).
Expression of PARP-1 full length and truncations
FLAG-tagged human PARP-1 (PARP-1-FLAG) was purified from SF9 insect cells by FLAG M2 affinity chromatography following published procedures.8 The PARP-1 truncation (374–524) was PCR-amplified using primers 5'-CACCGCCTCGGCTCCTGCTGCT-3' and 5'-TTATTTCATTCTCTTTTCAGATTTGTT-3'. The PARP-1 truncation (374–1014) was PCR-amplified using primers 5'-CACCGCCTCGGCTCCTGCTGCT-3' and 5'-TTACCACAGGGAGGTCTTAAAAT-3'. Amplified products were cloned using TOPO and Gateway cloning technology (Invitrogen Corporation, Carlsbad, CA) into pDEST-F1 (PARP-1(374–1014)) and pDEST-566 (PARP-1(374–524)) for expression. PARP-1(374–524) and PARP-1(374–1014) were expressed in E. coli BL21 pRARE2 strain. Cells were first cultured in LB media at 37 °C and 200 rpm to OD600 ~0.5 in MaxQ 5000 shaker (Barnstead International, Dubuque, Iowa). The temperature was then changed to 15 °C and the expression was induced by adding isopropyl β-D-thiogalactoside (IPTG) to 0.1 mM. After incubation at 15 °C for 24 hr, cells were harvested by centrifugation at 6370 g for 10 min. After resuspended in lysis buffer (20 mM Tris-HCl, 500 mM NaCl, 30 mM imidazole, pH 8.0) at 4 °C, cells were lysed by using an EmulsiFlex-C3 cell disruptor (AVESTIN, Inc., Canada), and cell lysate was clarified by centrifugation at 48400 g for 30 min at 4 °C. Protein was purified by Bio-RAD BioLogic DuoFlow FPLC using a HisTrap HP 5 mL column (GE Healthcare) with a linear gradient of imidazole from 30 mM to 500 mM in elution buffer (20 mM Tris-HCl, 500 mM NaCl, pH 8.0). Fractions containing PARP-1 proteins were collected and dialyzed to 25 mM Tris, pH 6.0, 50 mM NaCl (for PARP-1(374–1014)) or to 25 mM Tris, pH 8.0, 50 mM NaCl, 1 mM DTT (for PARP1(374–524)) at 4 °C. Protein concentrations were determined by Bradford assay or by comparing Coomassie brilliant blue staining with BSA standards after SDS-PAGE.
Expression of p53
GST-tagged human p53 in pGEX-2T vector was transformed into E. coli BL21 pRARE2 strain. Cells were first cultured in LB media at 37 °C and 200 rpm to OD600 ~0.5. The temperature was then changed to 15 °C and the expression was induced by adding isopropyl β-D-thiogalactoside (IPTG) to 0.1 mM. After incubation at 15 °C for 24 hr, cells were harvested by centrifugation at 6370 g for 10 min. Cells were resuspended in GST binding buffer (4.3 mM Na3PO4, 1.47 mM K3PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.3) at 4 °C, and lysed by using an EmulsiFlex-C3 cell disruptor. Cell lysate was clarified by centrifugation at 48400 g for 30 min at 4 °C, and incubated with 1 mL GST-Bind Resin (Novagen) for 1 hr at 4 °C. The resin was washed with GST binding buffer thoroughly, and then incubated with 150 units Thrombin (GE Healthcare) for 12 hours at 4 °C. The supernatant was collected and protein was dialyzed into 25 mM Tris-HCl, pH 8.0, 50 mM NaCl, and 1 mM DTT at 4 °C.
Expression of tankyrase-1 and TRF1
His-tagged human tankyrase-1 and TRF1 in baculovirus vector were provided from Dr. de Lange, and proteins were purified from SF9 insect cells following the published procedure.21 Recombinant tankyrase-1 was found in the insoluble fraction during the purification. However, the suspension of insoluble fractions contains active tankyrase-1 and was used in the labeling reactions. The suspension of insoluble fraction from non-infected SF9 cells was used as the negative control.
Auto-modification of PARP-1 with 6- or 8-alkyne-NAD
PARP-1 (0.15 μM), ssDNA (0.25 μg/μL) and 6- or 8-alkyne-NAD (100 μM) with or without NAD (100 μM) in 10 μL reaction buffer (50 mM Tris-HCl, 4 mM MgCl2, 0.2 mM DTT, pH 8.0) were incubated at 37 °C for 30 min. Control experiments were done without ssDNA. Then the click chemistry step was carried out (described later).
Labeling of p53 and RAP74 by PARP-1 with 6- or 8-alkyne-NAD
PARP-1 (0.15 μM), p53 (2.8 μM) or RAP74 (0.59 μM), ssDNA (0.25 μg/μL) and 6- or 8-alkyne-NAD (100 μM) with or without NAD (100 μM) in 10 μL reaction buffer were incubated at 37 °C for 30 min (without NAD) or 6 min (with NAD). Shorter incubation time was used because if longer incubation time is used, the protein will be extensively poly(ADP-ribosyl)ated and cannot be distinguished from the automodified PARP-1. Control experiments were done without PARP-1 or p53 or RAP74. Then the click chemistry step was carried out.
Labeling of TRAP1, GDH, citrate synthase, and tubulin by PARP-1 with 6-NAD
PARP-1 (0.05 μM), TRAP1 (1.0 μM, overexpressed and purified according to published procedures48) or GDH (3.1 μM) or citrate synthase (3.37 μM) or tubulin (3.64 μM), ssDNA (0.25 μg/μL) and 6-alkyne-NAD (100 μM) with or without NAD (100 μM) in 10 μL reaction buffer were incubated at 37 °C for 30 min (without NAD) or 6 min (with NAD). Control experiments were done without PARP-1 or without TRAP1 or GDH or citrate synthase or tubulin. Then the click chemistry step was carried out.
Labeling of TRF1 by tankyrase-1 with 6- or 8-alkyne-NAD
Tankyrase-1 pellet suspension (0.09 μg/μL) or non-infected SF9 cell pellet suspension (0.15 μg/μL), TRF1 (1.1 μM), and 6- or 8-alkyne-NAD (100 μM) with or without NAD (100 μM) in 10 μL reaction buffer were incubated at 37 °C for 30 min. Control experiments were done without tankyrase-1 or TRF1. Then the click chemistry step was carried out.
Click chemistry condition to conjugate Rh-N3 and detection of poly(ADP-ribosyl)ation by fluorescence
Rh-N3 (in DMF) was added to the above labeling reactions to a final concentration of 200 μM, followed by the addition of Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine51 (in DMF, final concentration 600 μM), CuSO4 (in water, final concentration 1 mM) and TCEP (in water, final concentration 1 mM).52 After the click chemistry was allowed to proceed at room temperature for 15 min, the reaction mixture was mixed with 10 μL 2× protein loading buffer and heated at 100 °C for 6 min. The samples were then resolved by SDS-PAGE using 12% acrylamide gel. Before staining with Coomassie blue, the fluorescence image of the gel was recorded by Typhoon 9400 imager. The image of protein gel after Coomassie blue staining was recorded with a digital camera (Canon PowerShot S3).
Kinetics of PARP-1 with NAD, 6-alkyne-NAD and 8-alkyne-NAD as substrates
PARP-1 (0.065 μM), ssDNA (0.05 μg/μL) with different concentrations of NAD, 6-alkyne-NAD or 8-alkyne-NAD (from 10 μM to 1 mM) in 30 μL reactions (50 mM Tris-HCl, 4 mM MgCl2, pH 8.0) were incubated at 25 °C for 1 min (with NAD) or 15 min (with 6-alkyne-NAD) or 60 min (with 8-alkyne-NAD). The reactions were quenched with 1 M HClO4, and then neutralized with 3 M K2CO3 5 min later. After centrifugation, the supernatant was analyzed by a SHIMADZU LCMS-QP8000α with a Sprite TARGA C18 column (40 × 2.1 mm, 5 μm, Higgins Analytical, Inc.) monitoring at 260 nm. Solvents were 50 mM ammonium acetate pH 5.4 (buffer A) and 50% methanol in water (buffer B). For PARP-1 with NAD reactions, compounds were eluted at a flow rate of 0.3 mL/min with 0% solvent B for 1 min, followed by a linear gradient of 0% to 3% solvent B over 14 min, and back to 0% solvent B over 2 min. Retention time of ADP-ribose, nicotinamide and NAD are 1.89 min, 3.52 min and 8.01 min, respectively. For PARP-1 with 6-alkyne-NAD or 8-alkyne-NAD reactions, compounds were eluted at a flow rate of 0.3 mL/min with 0% solvent B for 1 min, followed by a linear gradient of 0% to 1% solvent B over 5 min, then 1% to 50% solvent B over 5 min, and finally 50% solvent B for 1 min before equilibrating the column back to 0% solvent B over 2 min. Retention time of nicotinamide, 6-alkyne-ADP-ribose, 8-alkyne-ADP-ribose, 6-alkyne-NAD and 8-alkyne-NAD are 3.52 min, 8.42 min, 6.98 min, 10.59 min and 10.65 min, respectively. Reaction progress was monitored by the formation of nicotinamide, ADP-ribose, 6-alkyne-ADP-ribose and 8-alkyne-ADP-ribose. The quantification of nicotinamide, ADP-ribose, 6-alkyne-ADP-ribose and 8-alkyne-ADP-ribose produced in the reaction was analyzed by the integration of absorption peak monitored at 260 nm comparing with the plot of nicotinamide, ADP-ribose, 6-alkyne-ADP-ribose and 8-alkyne-ADP-ribose standards. The kcat and Km values were obtained by curve-fitting the V/[E] ~ [S] plot by KaleidaGraph. NADase activity was obtained from the formation of ADP-ribose, 6-alkyne-ADP-ribose and 8-alkyne-ADP-ribose. PARP activity was obtained from the formation of nicotinamide after deduction of ADP-ribose, 6-alkyne-ADP-ribose and 8-alkyne-ADP-ribose.
Kinetics of tankyrase-1 with NAD, 6-alkyne-NAD and 8-alkyne-NAD as substrates
Tankyrase-1 (0.03 μM) with different concentrations of NAD, 6-alkyne-NAD or 8-alkyne-NAD (from 10 μM to 1 mM) in 30 μL reactions (50 mM Tris-HCl, 4 mM MgCl2, pH 8.0) were incubated at 25 °C for 30 min (with NAD) or 2 hours (with 6-alkyne-NAD) or 13 hours (with 8-alkyne-NAD). Then the reactions were quenched and handled same as PARP-1 reactions for kinetics. NADase and PARP activity of tankyrase-1 were measured same as PARP-1 kinetics.
Cell lysate of MCF-7 wild type and PARP-1 KD cells
MCF-7 wild type and PARP-1 KD cells41 from ten 10 cm plates (90% confluency) were lysed by Dounce Homogenizer in 5 mL lysis buffer (25 mM Tris, 50 mM NaCl, 10% glycerol, pH 7.4) with 100 μL protease inhibitor cocktail (Sigma, Saint Louis, MO). The cell lysate was then centrifuged at 2000 g for 15 min at 4 °C. Under this condition, the pellet contained nuclei, mitochondria and other organelles. The pellet was solublized in 4 mL lysis buffer with 1% NP-40 and 100 μL protease inhibitor cocktail. After centrifugation at 14,000 g for 5 min at 4 °C, the supernatant was collected as cell lysate for later labeling reactions with PARP-1 and 6-alkyne-NAD.
Labeling of MCF-7 wild type and PARP-1 KD cell lysate by PARP-1 with 6-alkyne-NAD
PARP-1 (0.075 μM), MCF-7 wild type cell lysate (2 μg/μL) or PARP-1 KD cell lysate (2 μg/μL), ssDNA (0.25 μg/μL) and 6-alkyne-NAD (100 μM) with or without NAD (100 μM) in 10 μL reaction buffer with 0.5% NP-40 were incubated at 37 °C for 30 min. Control experiments were done without PARP-1 or without cell lysates. Then the click chemistry step was carried out.
Pull-down of substrate proteins of PARP-1 using 6-alkyne-NAD
Cell lysate of MCF-7 PARP-1 KD cells (2 mg) was incubated with PARP-1 (1.8 μg), 6-alkyne-NAD (10 nmol, final concentration 10 μM), and ssDNA (20 μg) in 1 mL reaction buffer at 37 °C for 30 min with gentle rotating. Control experiments were done with 100 μM PJ34 (a PARP-1 inhibitor) or without 6-alkyne-NAD. Then the click chemistry step was carried out with Biotin-N3 (in DMF, final concentration 16 μM), Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (in DMF, final concentration 200 μM), CuSO4 (in water, final concentration 800 μM) and TCEP (in water, final concentration 800 μM) at room temperature for 30 min. The reaction mixture was denatured and precipitated with 10 mL cold acetone. After centrifugation at 14,000 g for 5 min at 4 °C, the precipitate was further washed with 3 × 1 mL cold methanol to remove extra small molecule reagents. After solublizing the precipitate in 1 mL 2% SDS in PBS with heating at 90 °C for 10 min and centrifugation at 14,000 g for 5 min at room temperature, the supernatant was diluted to 10 mL by adding 9 mL PBS (final 0.2% SDS in PBS), and incubated with 100 μL streptavidin beads (Pierce Biotechnology, Rockford, IL) for 90 min at room temperature with gentle rotating. After centrifugation at 1000 g for 2 min at room temperature, the beads were further washed with 3 × 1 mL 0.2% SDS in PBS, 3 × 1 mL PBS, 3 × 1 mL (20 mM Tris, 500 mM KCl, pH 7.4), 3 × 1 mL (20 mM Tris, pH 7.4), and then incubated with 400 μL 6 M urea in PBS with 9.5 mM TCEP for 20 min at 35 °C with gentle rotating. Iodoacetamide (20 μL 400 mM in water) was added to the suspension of beads with further incubation for 20 min at 35 °C with gentle rotating. After dilution with 760 μL PBS and removal of supernatant, the beads were incubated with 2 μg Trypsin (Promega, Madison, WI) in 200 μL 2 M urea in PBS and 1 mM CaCl2 at 37 °C for 8 hr with gently rotating. The supernatant was collected and combined with washings of 2 × 300 μL water from beads. Then the digested peptides in above solution were purified with Sep-Pak Vac C18 cartridge (Waters Corporation, Milford, MA) and analyzed by nanoLC-MS/MS. Each experiment was done individually three times, and proteins identified in the samples but not in the control experiments were listed in Table 2.
Protein identification by nanoLC/MS/MS analyses
The tryptic digest was reconstituted in 10 μL of 2% acetonitrile (ACN) with 0.5% formic acid (FA) for nanoLC-ESI-MS/MS analysis, which is carried out using a LTQ-Orbitrap Velos (Thermo-Fisher Scientific, San Jose, CA) mass spectrometer equipped with nano ion source. The Orbitrap is interfaced with an UltiMate3000 MDLC system (Dionex, Sunnyvale, CA). The nanoLC was carried out by Dionex UltiMate3000 MDLC system (Dionex, Sunnyvale, CA). An aliquot of tryptic peptide (3.0 μL) was injected onto a PepMap C18 trap column (5 μm, 300 μm × 5 mm, Dionex) at 20 μL/min flow rate for on-line desalting and then separated on a PepMap C-18 RP nano column (3 μm, 75μm × 15cm), and eluted in a 90 min gradient of 5% to 38% ACN in 0.1% FA at 300 nL/min., followed by a 3-min ramping to 95% ACN-0.1%FA and a 5-min holding at 95% ACN-0.1%FA. The column was re-equilibrated with 2% ACN-0.1%FA for 20 min prior to the next run. The eluted peptides are detected by Orbitrap through nano ion source containing a 10-μm analyte emitter (NewObjective, Woburn, MA). The Orbitrap Velos is operated in positive ion mode with nano spray voltage set at 1.5 kV and source temperature at 175 °C. Either internal calibration using the background ion signal at m/z 445.120025 as a lock mass or external calibration for FT mass analyzer is performed. The instrument is performed at parallel data-dependent acquisition (DDA) mode using FT mass analyzer for one survey MS scan for precursor ions followed by MS/MS scans on top 7 most intensive peaks with multiple charged ions above a threshold ion count of 5000 in LTQ mass analyzer. MS survey scans at a resolution of 60,000 (fwhm at m/z 400), for the mass range of m/z 375–1400. Dynamic exclusion parameters were set at repeat count 1 with a 20 s repeat duration, exclusion list size of 500, 30 s exclusion duration, and ±10 ppm exclusion mass width. Collision induced dissociation (CID) parameters were set at the following values: isolation width 2.0 m/z, normalized collision energy 35 %, activation Q at 0.25, and activation time 30 ms. All data are acquired under Xcalibur 2.1 operation software (Thermo-Fisher Scientific).
Data analysis
All MS and MS/MS raw spectra were processed using Proteome Discoverer 1.1 (PD1.1, Thermo) and the spectra from each DDA file are output as an MGF file for subsequent database search using in-house license Mascot Deamon (version 2.2.04, Matrix Science, Boston, MA). The human protein sequence database containing 20,349 sequence entries in the SwissProt database was downloaded on January 21, 2010 was used for database search and the search was performed to query to SwissProt database (taxonomy: human) with one missed cleavage site by trypsin allowed. The peptide tolerance was set to 10 ppm and MS/MS tolerance was set to 0.8 Da. A fixed carbamidomethyl modification of cysteine, variable modifications on methionine oxidation and deamindation of asparagine and glutamine were set. Data filtering parameters were as follows: (a) the peptide identity probability is 99.9%CI with 35 peptide score cutoff; (b) only top matching proteins with p<0.001 (expectation value) were considered in the analysis, and (c) at least two distinct peptides met above criteria hit for each proteins as a finally identified protein list and used for obtaining exponentially modified protein abundance index (emPAI) number for each identified proteins. After filtering, the emPAIs outputted directly from Mascot results for each identified proteins were used for estimation of relative protein abundance in three experimental samples versus the three negative control samples. A protein is considered a potential PARP-s substrate only if the emPAI ratio (experimental/control) is more than 1.20. If the emPAI ration is between 2.0 to 1.20, the emPAI-based relative protein quantification results were also manually validated by averaging precursor ion peak areas of up to three top score peptides (found in both experimental samples and control samples) for each protein using extracted ion chromatograms in PD1.1 software.
Supplementary Material
Figure 7.
Labeling of TRAP1 (A), GDH (B), citrate Synthase (C) and tubulin (D) by PARP-1 using 6-alkyne-NAD. The panel on the left shows the image of Rhodamine fluorescence, and the panel on the right is the same gels stained with Coomassie blue. All lanes contain 6-alkyne-NAD (100 μM) and ssDNA (0.25 μg/μL). Lanes 1 and 2: PARP-1 (0.05 μM); 3 and 4: PARP-1 (0.05 μM) with TRAP1 (1.0 μM, A) or GDH (3.1 μM, B) or citrate synthase (3.37 μM, C) or tubulin (3.64 μM, D); 5 and 6: TRAP1 (1.0 μM, A), GDH (3.1 μM, B), citrate synthase (3.37 μM, C), or tubulin (3.64 μM, D). Lanes 2, 4 and 6 contained normal NAD (100 μM).
ACKNOWLEDGMENT
H.J. and H.L. thank Donald Ruhl and Jhoanna Berrocal for help with protein expression in Sf9 cells. We thank Dr. Jun-Lin Guan (University of Michigan Medical School) for providing the p53 expression vector, Dr. Titia de Lange (Rockefeller University) for providing the Sf9 expression cells for Tankyrase-1 and TRF-1. This work is supported in part by Dreyfus Foundation (New Faculty Award to H.L.), NIH R01 GM086703 (H.L.), and NIH R01 DK069710 (W.L.K).
Footnotes
Supporting Information Available: Labeling of PARP-1 with 32P-NAD, labeling of PARP-1 truncations by PARP-1, the time course and the detection limit of the labeling reaction, confirmation of PARP KD in MCF-7 cells, synthesis of Biotin-N3, the full list of identified potential PARP-1 substrate proteins. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Schreiber V, Dantzer F, Ame J-C, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 2.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 3.Lin H. Nicotinamide adenine dinucleotide: beyond a redox coenzyme. Org. Biomol. Chem. 2007;5:2541–2554. doi: 10.1039/b706887e. [DOI] [PubMed] [Google Scholar]
- 4.Amé J-C, Spenlehauer C, Murcia G. d. The PARP superfamily. BioEssays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 5.Poirier GG, Murcia GD, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADPRibosyl)ation of Polynucleosomes Causes Relaxation of Chromatin Structure. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Murcia G, Huletsky A, Lamarre D, Gaudreau A, Pouyet J, Daune M, et al. Modulation of chromatin superstructure induced by poly(ADP-ribose) synthesis and degradation. J. Biol. Chem. 1986;261:7011–7017. [PubMed] [Google Scholar]
- 7.Althaus FR. Poly ADP-ribosylation: a histone shuttle mechanism in DNA excision repair. J. Cell Sci. 1992;102:663–670. doi: 10.1242/jcs.102.4.663. [DOI] [PubMed] [Google Scholar]
- 8.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-Dependent Modulation of Chromatin Structure and Transcription by Nucleosome Binding Properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Tulin A, Spradling A. Chromatin Loosening by Poly(ADP-Ribose) Polymerase (PARP) at Drosophila Puff Loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 10.Ju B-G, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A Topoisomerase II{beta}-Mediated dsDNA Break Required for Regulated Transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 11.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu S-W, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, et al. Mediation of Poly(ADP-Ribose) Polymerase-1-Dependent Cell Death by Apoptosis-Inducing Factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 13.Yu S-W, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, et al. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burkle A. Physiology and pathophysiologyof poly(ADP-ribosyl)ation. BioEssays. 2001;23:795–806. doi: 10.1002/bies.1115. [DOI] [PubMed] [Google Scholar]
- 15.Haince J-F, Rouleau M, Hendzel MJ, Masson J-Y, Poirier GG. Targeting poly(ADP-ribosyl)ation: a promising approach in cancer therapy. Trends Mol. Med. 2005;11:456–463. doi: 10.1016/j.molmed.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Virag L, Szabo C. The Therapeutic Potential of Poly(ADP-Ribose) Polymerase Inhibitors. Pharmacol. Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 17.Dynek JN, Smith S. Resolution of Sister Telomere Association Is Required for Progression Through Mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 18.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 19.Chang W, Dynek JN, Smith S. NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem. J. 2005;391:177–184. doi: 10.1042/BJ20050885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 21.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a Poly(ADP-Ribose) Polymerase at Human Telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 22.Smith S, de Lange T. Tankyrase promotes telomere elongation in human cells. Curr. Biol. 2000;10:1299–1302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- 23.Berwick DC, Tavare JM. Identifying protein kinase substrates: hunting for the organ-grinder's monkeys. Trends Biochem. Sci. 2004;29:227–232. doi: 10.1016/j.tibs.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 25.Dani N, Stilla A, Marchegiani A, Tamburro A, Till S, Ladurner AG, et al. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4243–4248. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogs and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- 27.Adamietz P, Rudolph A. ADP-ribosylation of nuclear proteins in vivo. Identification of histone H2B as a major acceptor for mono- and poly(ADP-ribose) in dimethyl sulfate-treated hepatoma AH 7974 cells. J. Biol. Chem. 1984;259:6841–6846. [PubMed] [Google Scholar]
- 28.Krupitza G, Cerutti P. Poly(ADP-ribosylation) of histones in intact human keratinocytes. Biochemistry. 1989;28:4054–4060. doi: 10.1021/bi00435a063. [DOI] [PubMed] [Google Scholar]
- 29.Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat. Struct. Mol. Biol. 2007;14:548–555. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- 30.Kanai M, Hanashiro K, Kim S-H, Hanai S, Boulares AH, Miwa M, et al. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell Biol. 2007;9:1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- 31.Kanai M, Tong W-M, Sugihara E, Wang Z-Q, Fukasawa K, Miwa M. Involvement of Poly(ADP-Ribose) Polymerase 1 and Poly(ADP-Ribosyl)ation in Regulation of Centrosome Function. Mol. Cell. Biol. 2003;23:2451–2462. doi: 10.1128/MCB.23.7.2451-2462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simbulan-Rosenthal CM, Rosenthal DS, Luo R, Smulson ME. Poly(ADP-ribosyl)ation of p53 during Apoptosis in Human Osteosarcoma Cells. Cancer Res. 1999;59:2190–2194. [PubMed] [Google Scholar]
- 33.Mendoza-Alvarez H, Alvarez-Gonzalez R. Regulation of p53 Sequence-specific DNA-binding by Covalent Poly(ADP-ribosyl)ation. J. Biol. Chem. 2001;276:36425–36430. doi: 10.1074/jbc.M105215200. [DOI] [PubMed] [Google Scholar]
- 34.Ogata N, Ueda K, Kawaichi M, Hayaishi O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J. Biol. Chem. 1981;256:4135–4137. [PubMed] [Google Scholar]
- 35.Kumari SR, Mendoza-Alvarez H, Alvarez-Gonzalez R. Functional Interactions of p53 with Poly(ADP-ribose) Polymerase (PARP) during Apoptosis following DNA Damage: Covalent Poly(ADP-ribosyl)ation of p53 by Exogenous PARP and Noncovalent Binding of p53 to the Mr 85,000 Proteolytic Fragment. Cancer Res. 1998;58:5075–5078. [PubMed] [Google Scholar]
- 36.Simbulan-Rosenthal CM, Rosenthal DS, Luo R, Samara R, Jung M, Dritschilo A, et al. Poly(ADP-ribosyl)ation of p53 In Vitro and In Vivo Modulates Binding to its DNA Consensus Sequence. Neoplasia. 2001;3:179–188. doi: 10.1038/sj.neo.7900155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rawling JM, Alvarez-Gonzalez R. TFIIF, a basal eukaryotic transcription factor, is a substrate for poly(ADP-ribosyl)ation. Biochem. J. 1997;324:249–253. doi: 10.1042/bj3240249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawaichi M, Ueda K, Hayaishi O. Multiple autopoly(ADP-ribosyl)ation of rat liver poly(ADP-ribose) synthetase. Mode of modification and properties of automodified synthetase. J. Biol. Chem. 1981;256:9483–9489. [PubMed] [Google Scholar]
- 39.Beneke S, Alvarez-Gonzalez R, Bürkle A. Comparative characterisation of poly(ADP-ribose) polymerase-1 from two mammalian species with different life span. Exp. Gerontol. 2000;35:989–1002. doi: 10.1016/s0531-5565(00)00134-0. [DOI] [PubMed] [Google Scholar]
- 40.Nottbohm Amanda C., Dothager Robin S., Putt Karson S., Hoyt Mirth T, Hergenrother Paul J. A Colorimetric Substrate for Poly(ADP-Ribose) Polymerase-1, VPARP, and Tankyrase-1. Angew. Chem. Int. Ed. 2007;46:2066–2069. doi: 10.1002/anie.200603988. [DOI] [PubMed] [Google Scholar]
- 41.Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, et al. Global Analysis of Transcriptional Regulation by Poly(ADP-ribose) Polymerase-1 and Poly(ADP-ribose) Glycohydrolase in MCF-7 Human Breast Cancer Cells. J. Biol. Chem. 2009;284:33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)-]a general method for mapping sites of probe modification in proteomes. Nat. Protocols. 2007;2:1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- 43.Schulze WX, Usadel B. Quantitation in mass-spectrometry-based proteomics. Annu. Rev. Plant Biol. 2010;61:491–516. doi: 10.1146/annurev-arplant-042809-112132. [DOI] [PubMed] [Google Scholar]
- 44.Gagne J-P, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucl. Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossi MN, Carbone M, Mostocotto C, Mancone C, Tripodi M, Maione R, et al. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J. Biol. Chem. 2009;284:31616–31624. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai Y, Chen Y, Watkins SC, Nathaniel PD, Guo F, Kochanek PM, et al. Identification of poly-ADP-ribosylated mitochondrial proteins after traumatic brain injury. J. Neurochem. 2008;104:1700–1711. doi: 10.1111/j.1471-4159.2007.05114.x. [DOI] [PubMed] [Google Scholar]
- 47.Landriscina M, Amoroso MR, Piscazzi A, Esposito F. Heat shock proteins, cell survival and drug resistance: The mitochondrial chaperone TRAP1, a potential novel target for ovarian cancer therapy. Gynecol. Oncol. 2010;117:177–182. doi: 10.1016/j.ygyno.2009.10.078. [DOI] [PubMed] [Google Scholar]
- 48.Felts SJ, Owen BAL, Nguyen P, Trepel J, Donner DB, Toft DO. The hsp90-related Protein TRAP1 Is a Mitochondrial Protein with Distinct Functional Properties. Journal Of Biological Chemistry. 2000;275:3305–3312. doi: 10.1074/jbc.275.5.3305. [DOI] [PubMed] [Google Scholar]
- 49.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPs, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 50.Jiang H, Congleton J, Liu Q, Merchant P, Malavasi F, Lee HC, et al. Mechanism-based small molecule probes for labeling CD38 on live cells. J. Am. Chem. Soc. 2009;131:1658–1659. doi: 10.1021/ja808387g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. Bioconjugation by Copper(I)-Catalyzed Azide-Alkyne [3 + 2] Cycloaddition. J. Am. Chem. Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 52.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective "Ligation" of Azides and Terminal Alkynes. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.